中国科学院微生物研究所、中国微生物学会主办
文章信息
- 陈英, 王培娟, 张文君, 杨丘旭, 杨瑶
- Chen Ying, Wang Peijuan, Zhang Wenjun, Yang Qiuxu, Yang Yao
- mCherry红色荧光标记乳酸菌的融合表达系统构建及应用
- Construction and application of mCherry red fluorescent protein fusion expression system in lactic acid bacteria
- 生物工程学报, 2019, 35(3): 492-504
- Chinese Journal of Biotechnology, 2019, 35(3): 492-504
- 10.13345/j.cjb.180306
-
文章历史
- Received: July 24, 2018
- Accepted: October 8, 2018
- Published: February 22, 2019
乳酸菌(Lactic acid bacteria,LAB)是一类能够利用碳水化合物发酵产生大量乳酸,降低发酵产品的pH值,不形成芽孢的革兰氏阳性、兼性厌氧细菌的总称[1-2]。大多数乳酸菌无毒、无害,对人体及动物体具有益生作用,因此被称为益生菌[3-5]。随着近20几年来分子生物学的发展,以乳酸菌作为表达载体的高效基因表达系统的构建和应用成为目前的研究热点之一,一些外源基因也被报道在乳酸菌中表达成功[6]。例如,Renye等[7]利用在乳酸菌表达中常用的Nisin (乳酸链球菌素)诱导表达系统NICE生产片球菌素;Abdullah等[8]用乳酸菌表达菌株中常用的乳酸乳球菌NZ9000表达了大肠杆菌的热休克蛋白DnaK。
荧光蛋白具有稳定性强、无物种专一性、易于在细胞内表达等特点[9-10],因此,已作为标记物而广泛应用于生命科学领域[11-15]。目前,国内已有荧光蛋白成功标记乳酸菌的报道,但相关研究成果并不多。2009年,陈晓雷等[16]构建了绿色荧光蛋白GFP标记穿梭表达载体pW425et-GFP,实现了绿色荧光蛋白GFP在嗜酸乳杆菌1.1878中的表达。2014年,徐一轲[17]成功构建了表达增强型绿色荧光蛋白eGFP的重组乳酸乳球菌BLCC02-0018。2017年寇田田等[18]构建了以红色荧光蛋白基因(dsred2)为标记,以α-淀粉酶(amy)为报告基因的表达载体,成功实现了融合基因dsred2-amy在干酪乳杆菌中的融合表达。前人报道表明不同荧光蛋白对不同宿主乳酸菌的标记能力有强弱,因此,更多的荧光蛋白标记需要被开发及应用于更多乳酸菌宿主菌株。
近年来,红色荧光蛋白mCherry用于荧光标记的研究报道逐渐增多,相对其他荧光蛋白,该荧光标记具有标记基因小、背景低等优点,且与已报道的dsred2红色荧光基因相比,其荧光强度及稳定性更好[19-25]。2015年van Zyl等[26]报道了利用mCherry荧光蛋白标记蒙氏肠球菌ST4SA和植物乳杆菌423的研究,这是mCherry荧光蛋白标记乳酸菌的首次报道,而国内关于mCherry荧光蛋白标记乳酸菌的研究尚未见报道。本文开展了mCherry荧光蛋白对植物乳杆菌的标记研究,选用的植物乳杆菌WCFS1菌株是全基因组序列率先公布的乳酸杆菌,不仅自身具有益生和肠道中高存活率特性[27],同时已有多个外源蛋白在该菌株中表达成功的报道[28-30],是公认的乳酸菌分子生物学研究中的模式菌株。胆盐水解酶BSH是本实验室研究多年的乳酸菌功能性酶,近期研究报道该酶可能和乳酸菌肠道定植相关[31-32]。因此,本文通过建立以mCherry荧光蛋白为标记、胆盐水解酶基因bsh为报告基因的融合蛋白表达系统,为更多外源活性蛋白在乳酸菌中进行功能性研究奠定基础,也为研究乳酸菌在生物体内的定位、示踪及益生性作用机制提供有效的技术手段。
1 材料与方法 1.1 材料 1.1.1 菌株和质粒本研究中涉及使用的菌株和质粒见表 1。
| Strains or plasmids | Relevant characteristicsa | References |
| Strains | ||
| Lactobacillus plantarum | ||
| WCFS1 | Wild type | Prof. Guangdong Shang |
| NB5462 | Derivative of WCFS1, bsh gene deleted (Δbsh1) | This lab |
| YE460 | Emr, derivative of WCFS1 containing pSIPH460 | This study |
| YeG471 | Emr, derivative of WCFS1 containing pSIPH471 with egfp gene | This study |
| YmC472 | Emr, derivative of WCFS1 containing pSIPH472 with mCherry gene | This study |
| YbE460 | Emr, NB5462 derivative containing pSIPH460 | This lab |
| YbB462 | Emr, NB5462 derivative containing pSIPH462 with bsh gene | This lab |
| YbBmC473 | Emr, NB5462 derivative, containing mCherry-BSH fusion expression vector pSIPH473 | This study |
| YbBmC673 | Emr, NB5462 derivative, containing mCherry-BSH fusion expression vector pLDHLH673 regulated by the promoter of ldhL gene | This study |
| YbBmC771 | Emr, NB5462 derivative, containing mCherry-BSH fusion expression vector pLDHLH673 regulated by P32 | This study |
| YbBmC871 | Emr, NB5462 derivative, containing mCherry-BSH fusion expression vector pLDHLH673 regulated by the double promoters of slpA gene | This study |
| Lactobacillus brevis | ||
| ATCC 367 | Containing S-layer protein and slpA gene | CGMCC |
| Escherichia coli | ||
| DH5α | Cloning host | This lab |
| Plasmids | ||
| pMD19-T | Ampr, TA cloning vector | TaKaRa |
| pMG36e | Emr, expression vector of lactic acid bacteria | This lab |
| pTracer-CMV3 | Ampr, containing egfp gene | Dr. Shunying Yang |
| pmCherry-C1 | Kanr, containing mCherry gene | This lab |
| pSIP403 | Emr, expression vector containing gusA gene regulated by PsppA | Prof. Lars Axelsson |
| pSIPH460 | Emr, pSIP403 derivative, gusA replaced by a multiple cloning site containing two his-tag fragments | This lab |
| pSIPH462 | Emr, pSIPH460 derivative containing bsh gene | This lab |
| pSIPH471 | Emr, pSIPH460 derivative containing egfp gene | This study |
| pSIPH472 | Emr, pSIPH460 derivative containing mCherry gene | This study |
| pSIPH473 | Emr, pSIPH472 derivative with bsh gene inserted | This study |
| pLDHLH673 | Emr, pSIPH472 derivative, PsppA replaced by PldhL | This study |
| pP32H771 | Emr, pSIPH472 derivative, PsppA replaced by P32 | This study |
| pSLPAH871 | Emr, pSIPH472 derivative, PsppA replaced by PslpA | This study |
| a Ampr: ampicillin resistant; Kanr: kanamycin resistant; Emr: erythromycin resistant; Cmr: chloromycetin resistant. | ||
大肠杆菌DH5α用LB培养基37 ℃振荡培养。植物乳杆菌用MRS培养基37 ℃静置培养。在大肠杆菌中,氨苄青霉素、卡那霉素和红霉素使用浓度分别为100 µg/mL、30 µg/mL和250 µg/mL。在乳酸菌中,红霉素使用浓度为10 µg/mL,诱导剂SppIP使用浓度为25 ng/mL。
1.1.3 引物mCherry和egfp基因序列的特殊性设计共用引物如下:P1和P2,此对引物引入了NdeⅠ和XbaⅠ两个酶切位点。根据质粒pSIPH462上bsh基因序列,设计引物:bsh1和bsh2,在目标片段两端皆引入XbaⅠ酶切位点。根据GenBank (登录号为AL935263.2)的WCFS1上ldhL基因的启动子序列、质粒pMG36e上P32启动子序列和GenBank (登录号为CP000416.1)的短乳杆菌ATCC 367上slpA基因的启动子序列,分别设计引物对如下:PldhL1-PldhL2、P32F-P32R以及PslpA1-PslpA2,上游引物皆引入BglⅡ酶切位点,下游引物皆引入NcoⅠ和NdeⅠ两个酶切位点。引物均由生工生物工程(上海)股份有限公司合成。本实验所需引物序列见表 2。
| Primer name | Primer sequence (5′-3′)a | Size (bp) |
| P1 | CATATGGTGAGCAAGGGCGAGGAG | 24 |
| P2 | TCTAGACTTGTACAGCTCGTCCATGC | 26 |
| bsh1 | TCTAGATGCACATCAGTAATTTATAC | 26 |
| bsh2 | TCTAGAATTTTGTTGGTTAATAATTTG | 27 |
| PldhL1 | AGATCTAATCTTCTCACCGTCTTG | 24 |
| PldhL2 | CATATGTCCCATGGTTGGCATGCTTGACAAAATAAG | 36 |
| P32F | AGATCTAGATTAATAGTTTTAGC | 23 |
| P32R | CATATGTCCCATGGTGAATTTTTCTGCTGAAACG | 34 |
| PslpA1 | AGATCTGGACGGGTGCTAGAGACG | 24 |
| PslpA2 | CATATGTCCCATGGTTAAACTTGATTGCATAATTTTTC | 38 |
| a Restriction sites are underlined. | ||
限制性内切酶NdeⅠ、XbaⅠ、EcoRⅠ、BglⅡ、HindⅢ、rTaq、Ex Taq、DNA marker、PrimerStar Max和DNA切胶回收试剂盒购自宝生物工程(大连)有限公司;氨苄青霉素、卡那霉素、红霉素、溶菌酶、SanPrep柱式质粒DNA小量抽提试剂盒、Bradford法蛋白浓度测定试剂盒、茚三酮等均购自生工生物工程(上海)股份有限公司;SppIP诱导肽(序列:MAGNSSNFIHKIKQIFTHR)也由生工生物工程(上海)股份有限公司合成;T4 DNA连接酶购自NEB(New England Biolabs)公司;His标签抗体等购自美国Cell Signaling公司。牛磺胆酸钠(Sodium Taurocholate,TCA)购自美国Sigma公司。
Personal PCR仪,AG 4309型高压脉冲电击转化仪,1 mm电击杯,CL-22M型高速冷冻离心机,德国Eppendorf公司;Gel Doc XR+型凝胶成像系统,美国Bio-Rad公司;HE-90型电泳仪,水平电泳槽,上海天能科技有限公司;生物样品匀质器,杭州奥盛仪器有限公司;IX73型倒置荧光显微镜,日本Olympus公司;Infinite 200 PRO酶标仪,瑞士TECAN公司。
1.2 方法 1.2.1 荧光蛋白eGFP和mCherry表达载体的构建及鉴定以载体pTracer-CMV3和pmCherry-C1质粒DNA为模板,利用引物P1和P2分别扩增egfp和mCherry基因,分别用NdeⅠ和XbaⅠ双酶切,与经相同酶切处理的pSIPH460载体片段连接,连接液转化大肠杆菌DH5α感受态细胞,在含红霉素的LB平板上筛选。构建获得的处于启动子PsppA调控下的egfp基因和mCherry基因的诱导型重组载体分别命名为pSIPH471和pSIPH472。电转化植物乳杆菌WCFS1感受态细胞后,获得的重组菌命名为YeG471和YmC472,含空载质粒pSIPH460的对照菌株命名为YE460。
1.2.2 荧光蛋白mCherry-BSH融合表达载体的构建及鉴定以本实验室保藏的含罗伊氏乳杆菌胆盐水解酶bsh基因的pSIPH462质粒为模板,利用引物对bsh1和bsh2通过PCR扩增bsh基因,用XbaⅠ单酶处理回收片段,与经相同酶处理的pSIPH472载体进行连接,含红霉素的LB平板上筛选,获得的阳性克隆命名为pSIPH473。将pSIPH460、pSIPH462、pSIPH472和pSIPH473先后电转化植物乳杆菌NB5462的感受态细胞中,获得的重组菌株相应命名为YbE460、YbB462、YbmC472和YbBmC473。
1.2.3 组成型荧光蛋白mCherry-BSH融合表达载体的构建及鉴定以植物乳杆菌WCFS1全基因组、质粒pMG36e及短乳杆菌ATCC 367全基因组为模板,利用PldhL1-PldhL2、P32F-P32R和PslpA1-PslpA2引物对,分别扩增PldhL、P32和PslpA 3种启动子DNA片段,然后进行TA克隆。测序正确的3种阳性克隆质粒经NdeⅠ和BglⅡ双酶切后,分别与经相同酶处理的pSIPH473连接,构建获得的含启动子PldhL、P32和PslpA的载体分别命名为pLDHLH673、pP32H771和pSLPAH871。电转化植物乳杆菌NB5462的感受态细胞后,获得的重组菌株相应命名为YbBmC673、YbBmC771和YbBmC871。
1.2.4 重组蛋白的表达及Western blotting鉴定诱导型重组菌株经25 ng/mL SppIP诱导表达过程按照Nguyen等所述方法进行[28, 33]。制备的粗酶液用Bradford法蛋白浓度测定试剂盒检测蛋白浓度,并通过调节蛋白浓度,使取液量中总蛋白量一致,12% SDS-PAGE检测。转PVDF膜,TBST洗涤10 min,浸于5%脱脂乳封闭液中,室温下封闭1 h。封闭结束后,经TBST振荡洗涤,加入稀释比例为1:2 500 His标签抗体孵育液,孵育1 h后洗涤显色。
1.2.5 重组蛋白表达的荧光活性观察与检测将诱导表达或连续培养的菌液离心收集沉淀,用PBS洗涤2次,取10 µL滴于载玻片上,盖上盖玻片,在倒置荧光显微镜下分别观察绿色和红色荧光。
为建立生长曲线和荧光强度变化趋势线,重组菌株于100 mL培养液中生长,每隔2 h取样,直至培养34 h停止。样品经PBS洗涤后,利用紫外分光光度计和酶标仪分别检测样品OD600值和荧光强度值,eGFP荧光蛋白于激发波长EX:488 nm和发射波长EM:511 nm下检测荧光强度,mCherry荧光蛋白于激发波长EX:587 nm和发射波长EM:620 nm下检测荧光强度[24]。
1.2.6 重组蛋白表达的胆盐水解酶活性测定重组蛋白表达方法见1.2.4,胆盐水解酶BSH活性测定方法参考文献[34]做少许修改。培养后的重组菌株,经离心,用1 mL 0.1 mol/L的PBS(pH 6.0)洗涤2次,沉淀重悬于500 µL PBS中。吸取100 µL菌悬液至1.5 mL的离心管中,反应体系200 µL,底物牛磺胆酸钠(TCA)终浓度20 mmol/L。水解反应于37 ℃水浴1 h,加入200 µL 15%三氯乙酸终止反应,立即12 000 r/min离心10 min,取100 µL上清液与900 µL茚三酮溶液混合,沸水浴14 min,冰浴5 min后测OD570数值计算酶活。根据牛磺酸-茚三酮标准曲线,得样本中游离氨基酸浓度,计算BSH酶活。一个酶活单位(U)定义为每分钟每毫升菌液与底物反应后释放出的游离氨基酸的量µmol。
2 结果与分析 2.1 eGFP和mCherry荧光蛋白重组表达质粒构建及酶切鉴定分别以质粒pmCherry-C1和pTracer-CMV3的质粒DNA为模板,PCR扩增egfp和mCherry两种荧光蛋白编码基因,目的片段均为约750 bp (图 1A),TA克隆后测序结果表明扩增到的egfp和mCherry基因与质粒公布的序列完全一致。依方法1.2.1构建的重组质粒经NdeⅠ和XbaⅠ双酶切鉴定,获得预期大小片段,进一步测序显示重组表达质粒pSIPH471 (含egfp基因)和pSIPH472 (含mCherry基因)构建正确(图 1B)。

|
| 图 1 eGFP和mCherry荧光蛋白重组表达质粒的鉴定 Fig. 1 Identification of recombinant plasmids containing fluorescent protein eGFP or mCherry. (A) Results of PCR amplification. M: DL2000 marker; 1: egfp gene; 2: mCherry gene. (B) Results of enzyme digestion. M: DL2000 marker; 1: egfp gene; 2: mCherry gene. |
| |
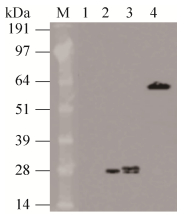
依方法1.2.4,在重组菌株诱导表达至3 h,取破碎后上清液做Western blotting检测,如图 2,结果显示重组蛋白表达出与预期eGFP (28.7 kDa)和mCherry (28.3 kDa)蛋白分子大小一致的条带,条带大小约为28 kDa。空载对照菌株则没有表达条带。
|
| 图 2 重组菌株表达蛋白的Western blotting鉴定 Fig. 2 Western blotting analysis of proteins expressed in recombinant strains. M: 14-191 kDa protein marker; 1: YE460 control; 2: GFP protein; 3: mCherry protein; 4: mCherry-BSH fusion protein. |
| |
将诱导表达3 h的重组乳酸菌YeG471和YmC472的菌悬液置于倒置荧光显微镜下观察。结果表明,在对应的荧光激发光条件下,重组eGFP菌株YeG471检测到绿色荧光,但是由于视野内背景绿色荧光也很强,因此菌株的荧光标记不明显(图 3A)。不同的是,重组mCherry菌株YmC472检测到红色荧光,但是由于视野内背景没有荧光,因此菌株的荧光标记清晰而显著(图 3B)。该结果说明相比eGFP荧光标记而言,mCherry红色荧光蛋白更适用于标记植物乳杆菌WCFS1,因此,本研究随后确定了mCherry蛋白作为植物乳杆菌WCFS1的荧光标记进行深入研究。

|
| 图 3 重组菌株的荧光观测结果 Fig. 3 Fluorescence detection results of recombinant strains. (A) Green fluorescence in the recombinant strain YeG471. (B) Red fluorescence in the recombinant cells YmC472. |
| |
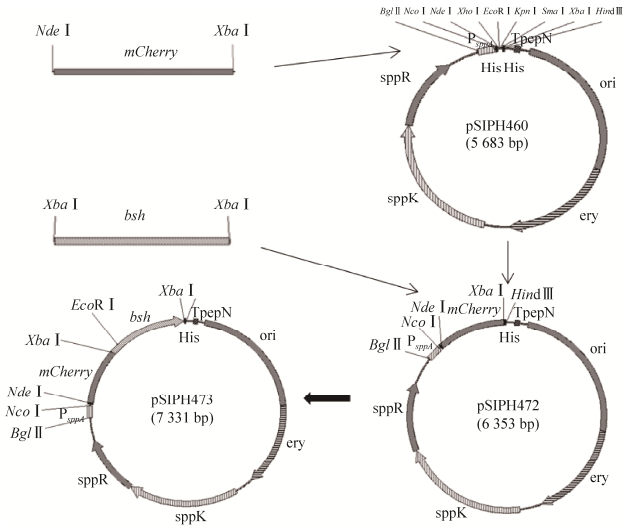
依方法1.2.2,将质粒pSIPH462上来源于罗伊氏乳杆菌的胆盐水解酶基因bsh克隆至pSIPH472质粒mCherry基因的C端,构建获得mCherry-BSH融合表达载体,命名为pSIPH473。将质粒pSIPH473电转化植物乳杆菌NB5462后获得的重组菌株命名为YbBmC473,构建策略见图 4。重组的融合蛋白mCherry-BSH编码基因是在启动子PsppA调节下表达的,由于该启动子是诱导型启动子,因此,融合蛋白mCherry-BSH的重组表达方式是诱导型的,需要向培养基中添加SppIP作为诱导剂从而开启基因的表达。
|
| 图 4 诱导型mCherry-BSH融合表达载体的构建示意图 Fig. 4 Construction diagram of inducible mCherry-BSH fusion expression vectors. |
| |
依方法1.2.4,在重组菌株经25 ng/mL SppIP诱导表达3 h后,取破碎后上清液进行Western blotting检测(图 2),结果显示重组蛋白表达出与预期mCherry-BSH融合蛋白(65.3 kDa)分子大小一致的条带,条带大小约为64 kDa。该结果说明了重组菌YbBmC473表达了mCherry-BSH融合蛋白。
将重组菌株YbBmC473活化转接至相应抗性培养基中,在OD600至0.3时加入诱导剂SppIP诱导蛋白表达。样品经测定后,建立了样品菌浓度、红色荧光强度以及胆盐水解酶BSH酶活性随时间变化曲线(图 5)。首先,从重组菌生长情况来看(图 5A),2个含mCherry基因的表达菌株YbBmC473和YbmC472与对照菌株之间无明显差别,表明mCherry荧光蛋白的表达对乳酸菌WCFS1无毒或低毒性,不影响菌株生长。其次,从重组菌荧光产生结果(图 5B)来看,重组菌YbmC472和YbBmC473均检测到荧光,2个菌株产生荧光的趋势相一致,即经SppIP诱导后荧光强度均是在菌体生长对数期中期(10 h)达到最高(37907 RFU和33707 RFU),随后逐渐下降,其中,融合了BSH蛋白的重组菌YbBmC473荧光强度略低。该结果说明单独表达的mCherry荧光蛋白可成功标记植物乳酸菌WCFS1,而融合目的蛋白后仍旧表达较高荧光强度。最后,从重组菌胆盐水解酶BSH活性表达情况来看(图 5C),融合了mCherry基因重组菌株YbBmC473检测到了BSH酶活性,酶活的产生趋势与该菌株荧光产生趋势相一致,在菌株发酵对数期中期(10 h) BSH活性最高,为0.603 U/mL,略低于bsh基因单独表达菌株YbB462。该结果说明mCherry-BSH蛋白融合表达成功,融合后的蛋白同时具有较高的荧光强度和BSH酶活性。

|
| 图 5 mCherry-BSH融合表达菌株活性鉴定结果 Fig. 5 The activity results of mCherry-BSH fusion expression recombinants. (A) The growth curve of recombinants. The strain NB5462 harboring pSIPH460, pSIPH462, pSIPH472 or pSIPH473 was cultured at 37 ℃ for 34 h and induced with 25 ng/mL of SppIP inducing peptides at ~0.3 OD600, respectively. (B) The fluorescent intensity curve of recombinant strains. (C) The BSH activity curve of recombinants. All data represent the average of three biological replicates. |
| |
综上所述,本研究实现了诱导型mCherry-BSH重组蛋白在植物乳杆菌WCFS1中的融合表达,该结果不仅成功建立了利用mCherry荧光蛋白标记植物乳杆菌WCFS1的有效方法,同时,对目标蛋白荧光标记的成功,也为在以植物乳杆菌WCFS1为例的乳酸菌宿主中异源表达重组蛋白的研究提供了有利条件。
2.4 组成型mCherry-BSH融合表达载体的构建、重组表达及活性鉴定在构建获得的质粒pSIPH473基础上,本研究另外构建了3种不同启动子调节下的mCherry-BSH融合蛋白表达载体,质粒的构建策略见图 6。本研究选择的另外3种启动子均来源于乳酸菌,且都是被报道在乳酸菌中成功表达外源基因的启动子。与PsppA不同的是,来源于植物乳杆菌的PldhL、乳酸乳球菌的P32和短乳乳杆菌的PslpA均是组成型启动子,在此类启动子调节下,重组蛋白的表达过程中无需添加诱导剂。

|
| 图 6 组成型表达载体构建策略图 Fig. 6 The construction strategy of constitutive expression vectors. The expression cassette was composed of three constitutive promoters (PldhL, P32 and PslpA) and the fusion protein mCherry-BSH encoding gene with the nucleotide and the corresponding amino acid sequence of the joint regions. The start codon and 4 or 8 amino acid residues from the original gene of these promoters were introduced into expressing vector. The amino acid residues marked in bold font were from the restriction enzyme recognition sites and first amino acid of the target gene. |
| |
以重组菌YbBmC473为对照,首先研究了3种组成型表达重组菌荧光表达情况,见图 7。与诱导型重组菌YbBmC473荧光产生的趋势不同,组成型重组菌的荧光强度在菌株生长稳定期(16 h)达到最高,且维持较长时间。在3种不同的启动子调节下,重组的mCherry-BSH融合蛋白表达的荧光强度也不同。其中,启动子PldhL调节下重组菌YbBmC673荧光强度最高,培养16 h后荧光值为23 897 RFU,启动子P32调节下重组菌YbBmC771荧光强度较低,为13 379 RFU,而启动子PslpA调节下重组菌YbBmC871没有检测到荧光。值得注意的是,荧光强度最高的组成型表达重组菌YbBmC673其荧光强度明显低于诱导型表达重组菌YbBmC473。

|
| 图 7 组成型融合表达菌株活性测定结果 Fig. 7 The activity detection of constitutive fusion expression recombinants. (A) The growth curve of constitutive fusion expression strains. The expression of fusion protein mCherry-BSH regulated by different promoters (PldhL, P32 and PslpA) in the host strain NB5462 were independent of inducing peptide. The inducing expression strain YbBmC473 was used as a control. (B) The fluorescence intensity of recombinants containing constitutive expression vectors. (C) The BSH activity results of recombinants. The strain YbE460 was used as a negative control, containing no BSH protein. In contrast, the positive control was YbB462. The different letters represented significant differences between two strains (P < 0.05); the same ones showed no significant differences. |
| |
随后,研究了最大荧光强度产生条件下的各个mCherry-BSH融合蛋白表达重组菌的BSH酶活性。结果显示(图 7),各个菌株的胆盐水解酶BSH活性高低与其融合的mCherry蛋白荧光强弱相对应,即诱导型表达重组菌BSH活性最高,为0.637 U/mL (发酵10 h),明显高于组成型表达重组菌,且启动子PldhL调节下重组菌YbBmC673的BSH酶活性最高,为0.416 RFU (发酵16 h),启动子P32调节下重组菌YbBmC771的BSH酶活性次之,而启动子PslpA调节下重组菌YbBmC871没有检测出BSH酶活性。
以上结果表明,在启动子PldhL调节下,组成型mCherry-BSH重组蛋白在植物乳杆菌WCFS1中也成功实现了融合表达,该表达过程中无需添加诱导剂,简化了表达过程。
3 讨论本研究利用mCherry红色荧光蛋白成功标记了植物乳杆菌WCFS1,是继2015年van Zyl等[26]报道之后mCherry荧光蛋白成功标记乳酸菌的又一例证,填补了国内空白。目前,已有一些文献报道了eGFP蛋白可标记乳酸菌产生绿色荧光[35-37],但是本研究结果表明,由于WCFS1菌体自身绿色荧光背景高,eGFP标记植物乳杆菌并不合适。相反,mCherry荧光蛋白标记植物乳杆菌则具有优势,由于WCFS1菌体自身无红色荧光背景,随着mCherry基因的诱导表达,重组菌检测到明显的红色荧光。本研究建立的采用mCherry蛋白标记植物乳酸菌的方法,可广泛应用于其他乳酸菌的荧光标记工作中,为研究乳酸菌在生物体内的分布、定植及存活情况从而揭示其益生功能的作用机理提供了有利条件。
目前,由于菌体自身安全、不分泌内毒素、表达外源蛋白无需纯化可直接同菌体一起进入胃肠道以及自身具有益生功能等优点,乳酸菌已然成为极具潜力的“新兴”基因工程菌,具有广阔的应用前景和研究价值。本研究选取的植物乳杆菌WCFS1菌株便是重要的基因工程模式菌株,已经有细胞因子、功能蛋白以及疫苗药物分子等在WCFS1中表达成功的报道[27-30, 38-39]。本研究构建了红色荧光蛋白mCherry基因和乳酸杆菌降胆固醇功能关键酶胆盐水解酶bsh基因的融合表达系统,转化植物乳杆菌WCFS1后,重组的融合蛋白mCherry-BSH同时检测到红色荧光和BSH酶活性,且两蛋白活性表达互不影响。可以预见,易于观察、便于检测的mCherry红色荧光蛋白将为更多活性蛋白在乳酸菌宿主中的表达、细胞定位、功能鉴定的研究奠定基础。
此外,本文还研究了3种不同组成型启动子调节下mCherry红色荧光融合蛋白表达情况,相较于PslpA调控的基因转录在WCFS1中无蛋白活性,在启动子PldhL调节下,融合蛋白mCherry-BSH具有较高的活性,且表达过程无需添加诱导剂。虽然实验结果显示组成型融合蛋白mCherry-BSH的蛋白活性不及诱导型,但是,无需添加诱导剂简化了操作过程,特别在诱导剂添加困难的目的蛋白表达研究中(如重组菌在实验动物胃肠道环境中表达蛋白)具有优势。值得注意的是,本研究构建获得的mCherry红色荧光融合蛋白表达系统也可作为植物乳杆菌启动子探针,为启动子的筛选提供有效工具。
| [1] | Wyszyńska A, Kobierecka P, Bardowski J, et al. Erratum to: lactic acid bacteria-20 years exploring their potential as live vectors for mucosal vaccination. Appl Microbiol Biotechnol, 2015, 99(10): 4531. DOI: 10.1007/s00253-015-6569-2 |
| [2] |
Bai FL, Zhang BL, Zhao HF. Progress in protein metabolism of lactic acid bacteria.
Food Sci, 2010, 31(19): 381–384.
(in Chinese). 白凤翎, 张柏林, 赵宏飞. 乳酸菌蛋白代谢研究进展. 食品科学, 2010, 31(19): 381-384. |
| [3] | van Pijkeren JP, Barrangou R. Genome editing of food-grade lactobacilli to develop therapeutic probiotics. Microbiol Spectr, 2017, 5(5): BAD-0013-2016. |
| [4] | Linares DM, Gómez C, Renes E, et al. Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front Microbiol, 2017, 8: 846. DOI: 10.3389/fmicb.2017.00846 |
| [5] | Saez-Lara MJ, Gomez-Llorente C, Plaza-Diaz J, et al. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. BioMed Res Int, 2015, 2015: 505878. |
| [6] |
Cui YQ, Wang JR, Wang YP. A review of research on lactic acid bacteria vectors for gene expression and their applications.
Food Sci, 2015, 36(9): 224–229.
(in Chinese). 崔月倩, 王菁蕊, 王艳萍. 乳酸菌基因表达载体及其应用研究进展. 食品科学, 2015, 36(9): 224-229. |
| [7] | Renye Jr JA, Somkuti GA. Nisin-induced expression of pediocin in dairy lactic acid bacteria. J Appl Microbiol, 2010, 108(6): 2142–2151. |
| [8] | Abdullah-Al-Mahin, Sugimoto S, Higashi C, et al. Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli DnaK. Appl Environ Microbiol, 2010, 76(13): 4277–4285. DOI: 10.1128/AEM.02878-09 |
| [9] | Enterina JR, Wu LS, Campbell RE. Emerging fluorescent protein technologies. Curr Opin Chem Biol, 2015, 27: 10–17. DOI: 10.1016/j.cbpa.2015.05.001 |
| [10] | Rodriguez EA, Campbell RE, Lin JY, et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem Sci, 2017, 42(2): 111–129. DOI: 10.1016/j.tibs.2016.09.010 |
| [11] | Piscitelli A, Pennacchio A, Cicatiello P, et al. Rapid and ultrasensitive detection of active thrombin based on the Vmh2 hydrophobin fused to a Green Fluorescent Protein. Biosens Bioelectron, 2017, 87: 816–822. DOI: 10.1016/j.bios.2016.09.052 |
| [12] | Kono J, Konno K, Talukder AH, et al. Distribution of corticotropin-releasing factor neurons in the mouse brain: a study using corticotropin-releasing factor-modified yellow fluorescent protein knock-in mouse. Brain Struct Funct, 2017, 222(4): 1705–1732. DOI: 10.1007/s00429-016-1303-0 |
| [13] | Vieira J, O'Hearn PM. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology, 2004, 325(2): 225–240. DOI: 10.1016/j.virol.2004.03.049 |
| [14] | Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science, 2003, 300(5616): 87–91. DOI: 10.1126/science.1082520 |
| [15] | Martinez-Jaramillo E, Garza-Morales R, Loera-Arias MJ, et al. Development of Lactococcus lactis encoding fluorescent proteins, GFP, mCherry and iRFP regulated by the nisin-controlled gene expression system. Biotech Histochem, 2017, 92(3): 167–174. DOI: 10.1080/10520295.2017.1289554 |
| [16] |
Chen XL, Yang GL, Wang CF. Construction of GFP labeling shuttle expression vector and preparation of restitution Lactobacillus.
J Jilin Agric Univ, 2009, 31(3): 325–329.
(in Chinese). 陈晓雷, 杨桂连, 王春凤. 绿色荧光蛋白标记穿梭表达载体构建及重组乳酸菌制备. 吉林农业大学学报, 2009, 31(3): 325-329. |
| [17] |
Xu YK. Construction of expressing enhanced green fluorescent protein in lactic acid bacteria.
Biotechworld, 2014(4): 4–6.
(in Chinese). 徐一轲. 表达增强绿色荧光蛋白重组乳酸菌的构建. 生物技术世界, 2014(4): 4-6. |
| [18] |
Kou TT, Bei TT, Li C, et al. Establishment of red fluorescent protein fusion expression system of lactic acid bacteria.
J Chin Inst Food Sci Technol, 2017, 17(3): 235–240.
(in Chinese). 寇田田, 北婷婷, 李晨, 等. 乳酸菌红色荧光蛋白融合表达系统的构建. 中国食品学报, 2017, 17(3): 235-240. |
| [19] | Fink D, Wohrer S, Pfeffer M, et al. Ubiquitous expression of the monomeric red fluorescent protein mCherry in transgenic mice. Genesis, 2010, 48(12): 723–729. DOI: 10.1002/dvg.v48.12 |
| [20] | Chapagain PP, Regmi CK, Castillo W. Fluorescent protein barrel fluctuations and oxygen diffusion pathways in mCherry. J Chem Phys, 2011, 135(23): 235101. DOI: 10.1063/1.3660197 |
| [21] | Ransom EM, Ellermeier CD, Weiss DS. Use of mCherry Red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol, 2015, 81(5): 1652–1660. DOI: 10.1128/AEM.03446-14 |
| [22] | Tauer C, Heinl S, Egger E, et al. Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb Cell Fact, 2014, 13: 150. DOI: 10.1186/s12934-014-0150-z |
| [23] | Russo P, Iturria I, Mohedano ML, et al. Zebrafish gut colonization by mCherry-labelled lactic acid bacteria. Appl Microbiol Biotechnol, 2015, 99(8): 3479–3490. DOI: 10.1007/s00253-014-6351-x |
| [24] | García-Cayuela T, de Cadiñanos LPZ, Mohedano ML, et al. Fluorescent protein vectors for promoter analysis in lactic acid bacteria and Escherichia coli. Appl Microbiol Biotechnol, 2012, 96(1): 171–181. |
| [25] | Mohedano ML, García-Cayuela T, Pérez-Ramos A, et al. Construction and validation of a mCherry protein vector for promoter analysis in Lactobacillus acidophilus. J Ind Microbiol Biotechnol, 2015, 42(2): 247–253. |
| [26] | van Zyl WF, Deane SM, Dicks LMT. Use of the mCherry fluorescent protein to study intestinal colonization by Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 in mice. Appl Environ Microbiol, 2015, 81(17): 5993–6002. DOI: 10.1128/AEM.01247-15 |
| [27] | van den Nieuwboer M, van Hemert S, Claassen E, et al. Lactobacillus plantarum WCFS1 and its host interaction: a dozen years after the genome. Microb Biotechnol, 2016, 9(4): 452–465. |
| [28] | Nguyen TT, Mathiesen G, Fredriksen L, et al. A food-grade system for inducible gene expression in Lactobacillus plantarum using an alanine racemase-encoding selection marker. J Agric Food Chem, 2011, 59(10): 5617–5624. DOI: 10.1021/jf104755r |
| [29] | Sak-Ubol S, Namvijitr P, Pechsrichuang P, et al. Secretory production of a beta-mannanase and a chitosanase using a Lactobacillus plantarum expression system. Microb Cell Fact, 2016, 15: 81. DOI: 10.1186/s12934-016-0481-z |
| [30] | Reverón I, Jiménez N, Curiel JA, et al. Differential gene expression by Lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in Tannin Degradation. Appl Environ Microbiol, 2017, 83(7): e03387–16. |
| [31] |
Huang Q, Huang L, Pan DD, et al. Expression in Escherichia coli, purification and functional characterization of recombination fusion protein MBP-BSH.
Food Sci, 2012, 33(7): 198–203.
(in Chinese). 黄茜, 黄璐, 潘道东, 等. 重组融合蛋白MBP-BSH在大肠杆菌中的表达及其纯化、功能鉴定. 食品科学, 2012, 33(7): 198-203. |
| [32] |
Huang Q, Huang L, Pan DD, et al. Molecular cloning and expression of the bile salt hydrolase gene (bsh) from Lactobacillus plantarum Y1.
J Nanjing Norm Univ: Nat Sci Ed, 2010, 33(3): 91–96.
(in Chinese). 黄茜, 黄璐, 潘道东, 等. 植物乳杆菌Lactobacillu plantarum Y1菌株胆盐水解酶基因(bsh)的克隆及重组表达. 南京师范大学学报:自然科学版, 2010, 33(3): 91-96. |
| [33] | Sørvig E, Grönqvist S, Naterstad K, et al. Construction of vectors for inducible gene expression in Lactobacillus sakei and L. plantarum. FEMS Microbiol Lett, 2003, 229(1): 119–126. DOI: 10.1016/S0378-1097(03)00798-5 |
| [34] | Moser SA, Savage DC. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl Environ Microbiol, 2001, 67(8): 3476–3480. DOI: 10.1128/AEM.67.8.3476-3480.2001 |
| [35] | Landete JM, Langa S, Revilla C, et al. Use of anaerobic green fluorescent protein versus green fluorescent protein as reporter in lactic acid bacteria. Appl Microbiol Biotechnol, 2015, 99(16): 6865–6877. DOI: 10.1007/s00253-015-6770-3 |
| [36] | Landete JM, Peirotén #193;, Rodríguez E, et al. Anaerobic green fluorescent protein as a marker of Bifidobacterium strains. Int Food Microbiol, 2014, 175: 6–13. DOI: 10.1016/j.ijfoodmicro.2014.01.008 |
| [37] | Geoffroy MC, Guyard C, Quatannens B, et al. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol, 2000, 66(1): 383–391. |
| [38] | Fredriksen L, Moen A, Adzhubei AA, et al. Lactobacillus plantarum WCFS1 O-linked protein glycosylation: an extended spectrum of target proteins and modification sites detected by mass spectrometry. Glycobiology, 2013, 23(12): 1439–1451. DOI: 10.1093/glycob/cwt071 |
| [39] | Fujii T, Ingham C, Nakayama J, et al. Two homologous Agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J Bacteriol, 2008, 190(23): 7655–7665. DOI: 10.1128/JB.01489-07 |
 2019, Vol. 35
2019, Vol. 35




