中国科学院微生物研究所、中国微生物学会主办
文章信息
- 李晓姣, 赵圣国, 郑楠, 程建波, 王加启
- Li Xiaojiao, Zhao Shengguo, Zheng Nan, Cheng Jianbo, Wang Jiaqi
- 细菌脲酶蛋白复合物及其活化机制
- Progress in bacterial urease complexes and their activation mechanisms
- 生物工程学报, 2019, 35(2): 204-215
- Chinese Journal of Biotechnology, 2019, 35(2): 204-215
- 10.13345/j.cjb.180239
-
文章历史
- Received: June 10, 2018
- Accepted: October 11, 2018
- Published: November 21, 2018
2. 中国农业科学院北京畜牧兽医研究所 农业农村部奶产品质量安全风险评估实验室,北京 100193;
3. 安徽农业大学 动物科技学院,安徽 合肥 230036
2. Laboratory of Quality & Safety Risk Assessment for Dairy Products of China Ministry of Agriculture and Rural Affairs, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China;
3. College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, Anhui, China
脲酶(尿素酶,EC 3.5.1.5)广泛存在于植物、动物、细菌、真菌和人体中,可以催化尿素水解形成氨和碳酸氢盐[1]。尿素在自然界中大量存在,是生命过程主要的代谢物之一,通常情况下,尿素非常稳定,极难水解。脲酶存在情况下,尿素的水解速率是无催化反应速率的1014倍。脲酶的主要作用是将尿素变成可供有机体使用的氮源,在自然界氮的新陈代谢过程中扮演着关键性的角色。
植物脲酶的主要作用是催化被植物体吸收的尿素的水解,使其作为可供吸收的氮源参与植物的生命过程,还可以参与植物防御系统中蛋白氮的转运通路,履行对植物病原体的防御功能。在反刍动物生产中,饲喂尿素可以节约蛋白质资源,降低饲料成本,尿素在瘤胃内被瘤胃壁和瘤胃内容物中主要来自于链球菌属Streptococcus、埃希氏菌属Escherichia、乳酸杆菌属Lactobacillus等的微生物分解并释放氨[2],氨与酮酸结合被用于合成微生物蛋白,为机体提供优质氮源[3-4]。尽管脲酶的存在有着积极的意义,但也会造成不利影响。尿素水解过快会造成氨的非生产性挥发,当氨超出瘤胃微生物的利用能力,则易导致动物氨中毒[5]。此外,幽门螺杆菌Helicobacter pylori、结核分枝杆菌Mycobacterium tuberculosis、奇异变形杆菌Proteus mirabilis等细菌脲酶与人和动物的许多发病机制相关,如幽门螺杆菌水解尿素产生的氨可以毒害胃粘膜的上皮细胞,因此医学上通常使用脲酶活性测试实验来确诊幽门螺杆菌的感染。
由于脲酶独特的生物学功能,其相关研究一直受到广泛关注,包括不同来源脲酶的蛋白质晶体结构及活化机制、脲酶的催化水解机理、脲酶抑制剂的研发应用等。脲酶的活性是影响尿素分解的主要因素,具有活性的脲酶属于蛋白复合物,其活化需要各蛋白(结构蛋白UreABC、辅助蛋白UreD/UreH、UreE、UreF和UreG等)相互配合共同发挥作用将Ni2+传递至脲酶活性中心。此外,脲酶的活化还需要二氧化碳和GTP等的共同参与。文中重点介绍细菌脲酶蛋白质的结构特性、促进脲酶金属中心组装的相关辅助蛋白的功能以及脲酶蛋白的活化路径,以期为调控脲酶活性提供理论指导。
1 细菌脲酶基因簇结构不同细菌来源的脲酶基因簇的结构有所不同,它们在基因的数量和顺序上存在差异,包含结构基因(ureA、ureB和ureC)、辅助基因(ureD、ureE、ureH、ureF、ureG和ureI等)和调节基因(ureR等)[6]。如图 1,编码产气克雷伯氏菌Klebsiella aerogenes脲酶的3个结构基因(ureABC)的侧翼是编码辅助蛋白的4个基因(ureD位于上游,ureE、ureF和ureG位于下游)。豆科根瘤菌Rhizobium leguminosarum的脲酶基因顺序与产气克雷伯氏菌相同,但有几个未被注释的开放阅读框(ORF)插入其中[7]。除此之外,许多细菌ureD位于ureG之后,如嗜热性芽孢杆菌Bacillus sp. TB-90[8]。在胸膜肺炎放线杆菌Actinobacillus pleuropneumoniae中,ureABCEFGD簇前面含有编码镍转运(cbiK、cbiL、cbiM、cbiQ、cbiO)和尿素转运的基因(utp)[9]。幽门螺杆菌脲酶基因簇由ureA、ureB以及5个下游基因ureI、ureE、ureF、ureG和ureH (与其他细菌的ureD同源)组成。还有一些微生物具有独特的脲酶基因,如幽门螺杆菌除了含有完整的脲酶基因簇以外还具有第二组结构基因[10]。枯草芽孢杆菌Bacillus subtilis缺乏可识别的辅助蛋白基因[11],这表明辅助蛋白不是体内脲酶活化的必需因素。
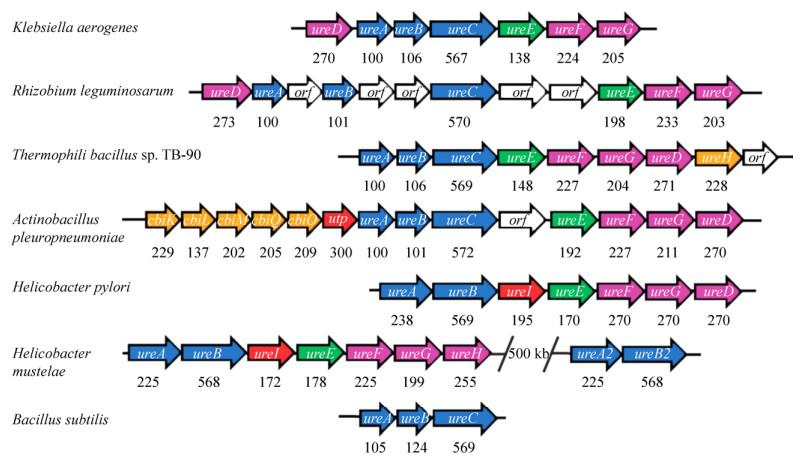
|
| 图 1 细菌脲酶的基因簇组成[12] Fig. 1 Organization of representative urease gene clusters[12]. Genes encoding urease subunits are shown in blue, the ureE gene encoding a metallochaperone is green, other urease accessory genes are purple, genes encoding proteins involved in nickel uptake are orange, those encoding proteins related to urea transport are red, and unknown genes are white. The sizes of the arrows do not accurately reflect the sizes of the genes, and the number below the arrow indicates the number of amino acid residues contained in the protein encoded by the gene |
| |
细菌脲酶蛋白包括结构蛋白和辅助蛋白。由结构基因ureA、ureB和ureC编码的γ、β和α亚基所形成的三聚体结构为脱辅基脲酶(Apourease),不具有脲酶活性。大多数细菌脲酶的活化需要至少4种辅助蛋白UreD/UreH、UreF、UreG和UreE的协助,分别由辅助基因ureD/H、ureF、ureG和ureE编码。此外,一些细菌在脲酶基因簇内还包含有其他辅助基因,这些辅助基因编码的蛋白质可能具有ATP结合、尿素转运、镍离子转运等功能,如HypA、HypB和UreI等。
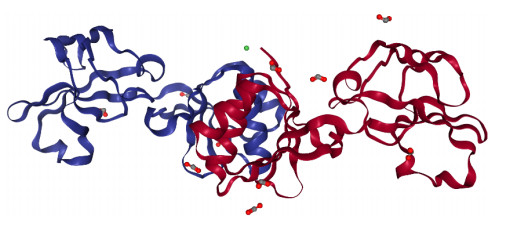
2.1 结构蛋白虽然不同来源的脲酶亚基数目、单体结构均有不同,但这些单体通常具有保守的氨基酸序列(相似性超过50%)[12]。单体的活性区域均位于α亚基,每个亚基含有被氨基甲酰化赖氨酸桥接的双核镍离子活性位点[13-14]。细菌脲酶通常由ureC编码的α亚基(分子量在60–76 kDa)、ureB编码的β亚基(分子量8–21 kDa)和ureA编码的γ亚基(分子量6–14 kDa)组成(αβγ)3三聚体结构,如产气克雷伯氏菌脲酶(图 2A)和巴氏芽孢杆菌Sporosarcina pasteurii脲酶。幽门螺杆菌脲酶的单体α亚基(ureB编码)和β亚基(ureA编码)组成((αβ)3)4杂聚体结构(图 2B),其UreA与产气克雷伯氏菌的UreA-UreB融合体同源,UreB与产气克雷伯氏菌的UreC同源。
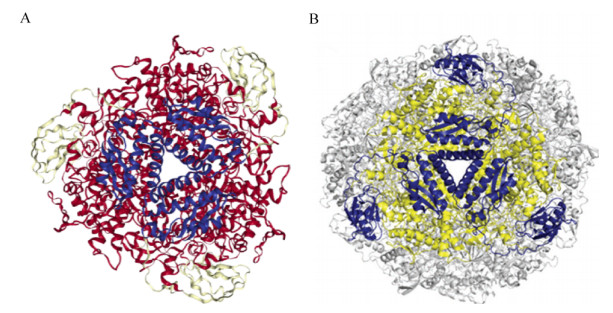
|
| 图 2 产气克雷伯氏菌和幽门螺旋杆菌的脲酶结构蛋白[12] Fig. 2 Urease structural protein of K. aerogenes and H. pylori[12]. (A) K. aerogenes urease (PDB code: 1fwj) with UreA depicted in blue, UreB in white, and UreC in red, together forming a (UreABC)3 structure. (B) H. pylori urease (1e9z) with UreA (corresponding to a fusion of the two small subunits in the K. aerogenes enzyme) depicted in blue and UreB (analogous to UreC in the K. aerogenes protein) shown in yellow for one (UreAB)3 unit, with three more (UreAB)3 units shown in gray included in the biologically relevant [(UreAB)3]4 structure |
| |
在大肠杆菌中外源表达产生的UreD/UreH是不溶的,与麦芽糖结合蛋白融合时(MBP-UreD),产气克雷伯氏菌的UreD是可溶的,并结合2当量的镍离子[14]。在脲酶金属中心组装的过程中,UreD/UreH可能作为募集其他辅助蛋白的支架和镍插入活性部位的直接促进剂,但其具体的镍转移方式仍不清楚。目前对于UreD的镍转移方式有2种推测,一种是UreD通过表面残基转移镍离子,另外一种是镍离子穿过UreD蛋白内部通道进入脲酶活性中心[15]。此外,UreD/UreH对脲酶的活性具有重要意义,单独添加镍离子也可以在不存在UreE、UreF或UreG的情况下使脱辅基脲酶产生部分活性,但UreD/UreH不存在时无法激活脲酶[16]。
2.2.2 UreF产气克雷伯氏菌外源表达的UreF蛋白也是不溶的,幽门螺杆菌UreF的天然形式是可溶的并且在单体和二聚体之间呈现平衡[17]。研究发现,UreF高度保守的C末端残基在UreF:UreH复合物的形成中起重要作用,UreF:UreH复合物形成后,UreF的C末端残基的排序发生改变,形成α螺旋(残基236–243),其与UreH链15–17位的氨基酸残基结合。在UreF不存在的情况下,UreH链15–17位上暴露的疏水性残基可能会使蛋白质不稳定,导致包涵体形成[18]。但是单独的UreF的C末端残基不足以与UreH形成可溶性复合物,还可能涉及UreF的其他残基。此外,UreF和UreH之间的界面保守性较差,这可能是由于UreF和UreD/UreH序列内的相似性较低的缘故。
2.2.3 UreGUreG是一种可溶性蛋白,其金属结合特性已经从多种微生物中得到鉴定,包括产气克雷伯氏菌[19]、巴氏芽孢杆菌[20]、结核分枝杆菌[21]和幽门螺杆菌[22]。产气克雷伯氏菌的UreG是单体,结合1当量的锌离子或镍离子,巴氏芽孢杆菌和结核分枝杆菌的UreG蛋白质是二聚体,2个单体通过二硫键连接,每个二聚体可以结合3个锌离子和4个镍离子[20]。幽门螺旋杆菌UreG蛋白的单体结合0.5当量锌离子和2当量镍离子[22]。
UreG是参与调控镍供应的SIMIBI (信号识别颗粒MinD和BioD)类的GTP酶[23-24]。SIMIBI GTP酶的生物功能通常由二聚化调控[25]。在GTP、镍和碳酸氢盐存在的条件下,UreG在单体和二聚体形式之间的转换使其能够动态组装和拆卸金属结合位点以将镍递送至目标脲酶。UreG的GTP酶活性还可通过调控UreF来提高脲酶金属中心组装的保真度[26]。研究表明,2个UreG的单体间的二聚体界面含有Cys66-Pro67-His68金属结合基序,锌的配位层是由来自UreG二聚体的每个单体的残基Cys66和His68形成的,而UreG的Cys66和His68突变会导致其镍离子结合能力丧失[27]。
2.2.4 UreEUreE是金属伴侣蛋白,在产气克雷伯氏菌中,每个UreE同源二聚体结合6个镍离子[28]。大多数镍与UreE富含组氨酸的C末端(HGHHHAHHDH HAHSH)结合,但缺乏该区域的截短形式(H144* UreE)形成的同源二聚体仍然可以结合2个镍离子[29]。幽门螺杆菌UreE蛋白在其C末端缺少富含组氨酸的区域,每个二聚体结合1个镍离子[30]。
UreE可以与UreG相互作用成UreG:UreE复合物[31-33]。对于幽门螺旋杆菌,UreE二聚体与2个UreG单体结合,其结构可以通过锌离子稳定[34]。而来自产气克雷伯氏菌的UreE二聚体与1个UreG单体结合后的结构,锌离子或镍离子均可使其达到稳定状态[35]。
2.2.5 其他辅助蛋白HypA和HypB与细菌中[Ni, Fe]-氢化酶生物合成相关,它们对幽门螺杆菌和肝螺旋杆菌H. hepaticus的脲酶活性具有重要作用[36]。氢化酶是一种与细菌能量来源相关的酶,HypA是一种镍结合蛋白,被认为是氢化酶成熟的镍伴侣蛋白[37]。UreE可与HypA形成三元复合物,由1个UreE二聚体和1个HypA单体组成。这种相互作用促进镍从HypA向UreE的传递并增强脲酶活性[38]。幽门螺杆菌HypB不结合镍,但其与UreG同源,具有氢化酶激活所需的GTP酶活性[39]。通常认为HypA和HypB通过形成HypA:HypB复合物将镍插入到氢化酶的大亚基中,化学交联研究表明HypA和HypB可形成异二聚体。在上述两种螺杆菌中,hypA和hypB的突变几乎消除了脲酶和氢化酶的活性[40]。
ureI存在于胃和口腔细菌中,例如幽门螺杆菌和唾液链球菌Streptococcus salivarius,该基因编码的蛋白被称为尿素的转运蛋白。UreI转运蛋白是分子量为21.7 kDa的内膜蛋白,其结构由6个亚基组成,每个亚基各自独立运作并对应单一的功能通道[41]。UreI可以将尿素递送至胞内,随后脲酶将尿素转化为氨,UreI蛋白的活性对pH呈现很强的依赖性,其转运作用在pH 6.0左右时关闭,在pH 5.0或更低时完全打开,使尿素迅速转运至细胞质内[42]。
胸膜肺炎放线杆菌的cbiK、L、M、Q基因编码的蛋白可能具有镍转运功能,用大肠杆菌表达cbiKLMQ基因及脲酶基因(ureABC-DFGE),在普通LB培养基的培养条件下即可具有完整脲酶活性,而仅含脲酶基因(ureABC-DFGE)的细胞需要补充镍离子以实现完整的脲酶活性[9]。假结核耶尔森菌Yersinia pseudotuberculosis脲酶基因簇中的5个基因(yntA、B、C、D和E)也具有镍转运的功能,该区域的缺失会消除脲酶活性并降低微生物吸收镍的速率[43]。唾液链球菌Streptococcus salivarius 57.I的脲酶基因簇的下游含有3个基因(ureM、Q和O),ureM的插入失活会导致脲酶蛋白无法组装镍离子并丧失活性[44]。
3 细菌脲酶的活化过程脲酶的活化实质上是镍离子与脲酶蛋白的金属组装过程,UreE将镍离子传递给UreG[27, 45]之后,携带镍离子的UreG需要将镍离子转移给UreF,然后再穿过UreD/H通道进入脲酶,才能完成金属中心的装配[14, 46-47]。化学交联试验表明,UreF与UreH的结合可以诱导脲酶的构象变化[48],使镍离子和二氧化碳进入活性位点[49]。
通过确定不同物种的脱辅基脲酶蛋白结构,发现脲酶的活性中心含有2个催化所必需的镍离子,还有1个羧酸化的赖氨酸作为桥键,其羧基的2个氧原子分别与镍离子配位[11]。在活化过程中,UreB是促进辅助蛋白与脲酶脱辅基蛋白相互作用的主要参与者,并指导镍离子高效掺入(UreC)新生活性位点[50]。有研究显示,用含碳酸氢盐与镍离子的缓冲液孵育脲酶脱辅基蛋白,可产生约15%的活性脲酶蛋白;而先用镍离子孵育脲酶脱辅基蛋白再加入碳酸氢盐,则没有检测到脲酶活性,这一发现表明,CO2必须先于镍与脲酶蛋白结合才能使其激活[51]。
目前提出的脲酶活化路径有3种(图 3),差异主要体现在辅助蛋白与结构蛋白的结合方式以及辅助蛋白之间的结合顺序上,但最后均需形成UreABC:UreD:UreF:UreG复合物,该复合物的形成被认为是镍插入脲酶活性位点的关键步骤。此外,UreE-UreG的结合在脲酶的活化过程中也具有重要作用,将UreABC:UreD:UreF:UreG与纯化的UreE混合,用生理浓度下的碳酸氢盐、镍和GTP孵育可将脲酶蛋白激活至野生型水平,但它们之间具体的分子机制还不完全清楚。
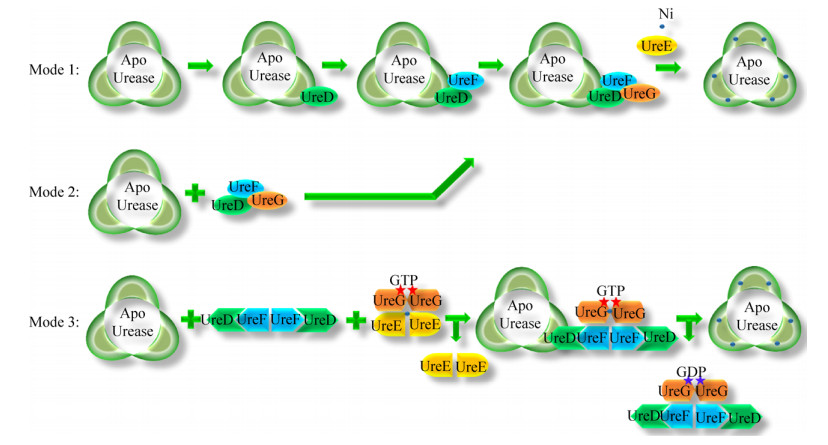
|
| 图 3 脲酶活化机制与模式示意图 Fig. 3 Schematic representation of the proposed mechanisms for urease activation. The urease activation mechanisms proposed so far involve either the sequential binding of UreD, UreF and UreG to apourease (mode 1)[1] or the direct interaction of a preformed UreDFG protein complex (mode 2) to the inactive form of the enzyme, to build a preactivation complex that prepares apourease for nickel binding. Ni(Ⅱ) ions were suggested to be directly delivered by UreE to apourease in the final step of the process[52]. The mode 3 shows that UreE/Ni is first combined with UreG to form UreG2UreE2/Ni to transfer Ni to UreG, then UreG/Ni is separated from UreG2UreE2/Ni and combined with UreD:UreF and apourease[27]. GTP (red five-pointed star) plays an important role in the activation of urease, which hydrolyzes to GDP (blue five-pointed star) |
| |
UreD是产气克雷伯氏菌脲酶体内活化所必需的,UreD与编码脲酶结构蛋白的基因过度表达时,可以得到纯化的UreABC:UreD蛋白复合物[51, 53]。MBP-UreD可以在细胞内与UreABC联合,但不能与UreAC (即脲酶缺失UreB)联合。化学交联研究结果表明,UreD紧邻UreB和UreC[48],而小角度X射线散射(SAXS)研究也证实了UreABC: UreD复合物中,UreD与UreABC复合物的UreB结合[49]。
UreF和UreD以及UreABC的高度表达可以形成UreABC:UreD:UreF复合物,但是仅表达UreF与UreABC无法形成复合物。将纯化得到的UreABC:UreD:UreF复合物用天然凝胶的蛋白免疫印迹进行分析,发现每个(UreABC)3脱辅基蛋白含有0–3个UreDF异二聚体[17]。小角度X射线散射实验显示UreD和UreF之间紧密接触,2种辅助蛋白都结合在UreB附近[49]。化学交联和蛋白水解片段的基质辅助激光解吸电离质谱进一步证明了UreF与UreD之间的关系,研究显示,UreD抗体不能检测到UreABC:UreD:UreF复合物,但是能检测到UreABC:UreD复合物,这表明UreF与UreD抗原表位结合[17]。
将UreG与UreABC:UreD:UreF混合可以得到UreABC:UreD:UreF:UreG复合物,在标准激活条件下,该复合物可以得到60%的活性脲酶,使用高于生理水平的碳酸氢盐和镍离子孵育脲酶蛋白,会降低脲酶活性,但提高GTP的浓度会极大地促进脲酶活化[54]。将UreG的与GTP相关的作用位点进行突变,会极大地降低UreABC:UreD:UreF: UreG复合物脲酶的活性,说明UreG的GTP酶的作用对脲酶的活化具有重要意义。
3.2 模式2:UreD:UreF:UreG直接与UreABC结合产气克雷伯氏菌可以在体内单独表达UreD: UreF:UreG复合物,但其溶解性很差[19]。幽门螺杆菌的UreH:UreF:UreG复合物的结构显示(图 4),UreF需要与UreH结合形成复合物引发构象变化后,UreF二聚体才具有结合2个UreG的能力。UreG先前被认为是一种特定的锌离子结合伴侣,没有镍结合能力[40],而UreG:UreF:UreH复合物的结构清楚地表明UreG可以结合镍离子[55]。
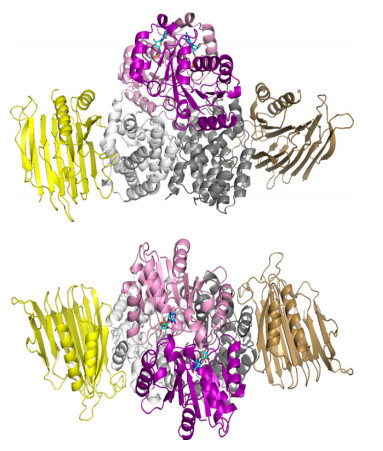
|
| 图 4 幽门螺杆菌(UreH:UreF:UreG)2复合物的结构[53] Fig. 4 Crystal structure of H. pylori urease accessory protein UreF/H/G complex (PDB code: 4HI0)[53]. Shown are two views of the (UreH:UreF:UreG)2 complex from H. pylori (UreH, UreF, and UreG in shades of yellow, gray, and magenta, respectively). A GDP molecule (cyan) is located in each UreG |
| |
MBP-UreD:UreF:UreG复合物具有很好的溶解性,并且可以与脱辅基脲酶结合[51]。在含有高浓度镍和碳酸氢盐的标准缓冲液中,异源三聚体UreG:UreF:UreH可以与脱辅基脲酶结合并促进脲酶活化,说明UreG:UreF:UreH复合物作为脲酶特异性分子伴侣发挥功能[56-57]。
3.3 模式3:UreE:UreG与UreABC、UreD: UreF的结合UreE被认为是脲酶活化过程中的镍来源,UreE与镍结合的结构位于2个UreE单体之间的界面处(图 5)[30]。拉下实验(Pull-down assay)表明,产气螺杆菌UreG和UreE在镍存在下形成复合物,它们之间通过蛋白质相互作用进行镍的转移[35]。有研究提出,UreG与UreE之间的相互作用位置可能位于UreG镍结合位点附近的蛋白质表面,而这个位置被埋在(UreH:UreF:UreG)2复合物中[27, 58]。GTP可以诱导UreG的构象变化,使(UreH:UreF:UreG)2复合物不稳定,UreG从复合物中解离并与携带镍荷电的UreE二聚体形成(UreE:UreG)2复合物[59]。
幽门螺杆菌中,UreG与UreE在GDP存在的条件下形成UreE2:UreG复合物,但在GTP存在的条件下则会形成(UreE:UreG)2复合物,并接受来自UreE的镍[27]。携带镍离子的UreG二聚体可以与脱辅基蛋白和(UreH:UreF)2形成具有活性的脲酶复合物[49]。这可能是由于GTP水解为GDP导致UreG的CPH基序的构象发生变化,破坏了由Cys66/His68组成的正方形平面构型,使(UreE:UreG)2复合物释放了携带镍离子的UreG。而此时的UreG处于CDP约束状态更倾向与(UreH:UreF)2形成(UreH:UreF:UreG)2复合物,该复合物继续转运镍离子进一步促进脲酶活化[59]。
4 总结与展望综上所述,细菌脲酶的活化需要结构蛋白、辅助蛋白(UreD/H、UreE、UreF和UreG)、镍离子、GTP水解及碳酸氢盐共同发挥作用。在活化过程中,UreD是第一个与结构蛋白结合的辅助蛋白;UreF可以与UreD结合增加UreD的可溶性,也可作为GTP酶的活化蛋白来调节UreG的功能。UreG是一种GTP酶,在脲酶活化中,将GTP替换为不可被水解的GTP类似物会抑制脲酶活化,表明UreG的GTP酶活性是脲酶活化所必需的。最后,UreE在GTP存在的条件下,将其募集的镍离子转移给UreG或结构蛋白与UreF2H2形成的复合物。脲酶活化是一个复杂的过程,目前研究较为广泛的是产气克雷伯氏菌和幽门螺旋杆菌细菌脲酶蛋白的功能性质以及活化方式,但是还有许多基本问题未得到解答,各脲酶蛋白在活化过程中的确切功能仍然未知。此外,自然界中的微生物大部分都是未培养微生物,无法通过分离培养获得菌株及其生理代谢特征,因此未培养微生物脲酶的基因结构与功能是否与可培养微生物类似值得深入研究。基于目前的研究现状,笔者认为,未来还应在以下几个方面加强研究。
1) 本实验室从事瘤胃尿素分解菌研究多年,包括瘤胃尿素分解菌的体外分离与培养、脲酶抑制剂的开发应用以及通过高通量测序确定了瘤胃中的脲酶优势分解菌群等。在尿素分解菌的体外培养方面,我们通过厌氧滚管和调整培养基等技术,从瘤胃液分离得到了溶糊精琥珀酸弧菌、布氏密螺旋体和白色瘤胃球菌,从瘤胃壁上分离获得了地衣芽孢杆菌和奇异变形菌,共5种尿素分解菌[60]。为了抑制瘤胃脲酶活性,我们实验室还对脲酶抑制剂乙酰氧肟酸的合成工艺进行了优化,通过奶牛饲养实验验证了该产品显著抑制了瘤胃尿素的分解。还运用自主研发的人工瘤胃模拟系统进行了体外发酵实验,尿素和乙酰氧肟酸分别作为激活剂和抑制剂,利用高通量测序技术研究尿素分解菌16S rRNA基因变化,揭示了瘤胃优势尿素分解菌为假单胞菌属Pseudomonas、链球菌属Streptococcus、嗜血杆菌属Haemophilus、芽孢杆菌属Bacillus、奈瑟氏菌属Neisseria、放线菌属Actinomyces和琥珀酸弧菌科Succinivibrionaceae[5]。
但是我们对抑制剂抑制尿素分解菌的作用原理并不清楚,因此将研究方向进一步扩展为:找到瘤胃优势脲酶分解菌,研究该细菌脲酶的活化模式,继而为开发新的脲酶抑制剂垫定基础。由于目前对瘤胃中尿素分解菌的分布和多样性知之甚少,而脲酶基因(ureC)是分析各种环境中的尿素分解菌的目标基因。我们通过尿素饲喂奶牛,收集瘤胃液相、固相和瘤胃壁的细菌样品,使用Miseq进行ureC基因的扩增和测序,多样性分析表明,甲基球菌科Methylococcaceae、梭菌科Clostridiaceae、类芽孢杆菌科Paenibacillaceae、螺杆菌科Helicobacteraceae和嗜甲基菌科Methylophilaceae的ureC基因丰度较高[4]。
随着宏基因组学的快速发展,为研究未培养微生物种类与功能提供了重要研究方法,因此通过宏基因组技术获得未培养微生物脲酶基因簇,开展结构分析、活化过程和影响因素研究,将是未来重点开展的研究方向。所以我们利用宏基因组测序技术将得到的基因片段进行拼接、Binning (分箱分析)、绘制基因草图,得到了多个基因簇。再将这些基因簇与之前得到的优势细菌ureC序列进行比对,最终获得优势脲酶细菌的基因簇。将脲酶基因全长和结构基因在体外诱导表达,发现脲酶全长基因表达的蛋白的活性远高于结构蛋白的活性,还需进一步研究该细菌脲酶蛋白的生物功能。
2) 辅助蛋白的功能研究:已有研究证明,带镍的UreG二聚体可以与UreF2H2和结构蛋白形成复合物,并且UreG和UreF2H2之间的相互作用对于脲酶活化至关重要,但是尚不清楚UreG二聚体是如何与结构蛋白和UreF2H2相互作用的。并且UreG可以与UreE形成UreE2G2复合物,该复合物中UreG是以单体的形式还是二聚体的形式与UreE2结合也未知。目前比较缺乏对UreD/UreH与UreF的研究,我们在实验过程中也发现UreD与UreF相对于其他辅助蛋白稳定性和可溶性均较差,而UreD/UreH与UreF是脲酶活化方面的关键辅助蛋白,所以UreF与UreD/UreH之间如何转移镍离子并传递至结构蛋白将是今后研究的重点。
3) 脲酶抑制剂的开发:先通过将具有活性的脲酶蛋白与抑制剂进行体外实验,通过IC50等指标筛选出具有活性抑制作用的产品。之后利用等温量热或表面等离子共振等技术研究该抑制剂所抑制的关键功能蛋白,对新型脲酶抑制剂的开发应用具有重要意义。
| [1] | Zambelli B, Musiani F, Benini S, et al. Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc Chem Res, 2011, 44(7): 520–530. DOI: 10.1021/ar200041k |
| [2] | Konieczna I, Żarnowiec P, Kwinkowski M, et al. Bacterial urease and its role in long-lasting human diseases. Curr Protein Pept Sci, 2012, 13(8): 789–806. DOI: 10.2174/138920312804871094 |
| [3] |
Zhao SG, Wang JQ, Liu KL, et al. Analysis for the diversity of ureolytic bacterium from dairy rumen based on metagenomics.
J China Agric Univ, 2010, 15(1): 55–61.
(in Chinese). 赵圣国, 王加启, 刘开朗, 等. 宏基因组学方法分析奶牛瘤胃尿素分解菌的多样性. 中国农业大学学报, 2010, 15(1): 55-61. DOI:10.3969/j.issn.1009-508X.2010.01.006 |
| [4] | Jin D, Zhao SG, Zheng N, et al. Differences in ureolytic bacterial composition between the rumen digesta and rumen wall based on ureC Gene Classification. Front Microbiol, 2017, 8: 385. |
| [5] | Jin D, Zhao SG, Wang PP, et al. Insights into abundant rumen ureolytic bacterial community using rumen simulation system. Front Microbiol, 2016, 7: 1006. |
| [6] |
Zhao SG, Wang JQ, Bu DP, et al. Biochemistry and molecular biology of bacterial ureases.
Microbiology, 2008, 35(7): 1146–1152.
(in Chinese). 赵圣国, 王加启, 卜登攀, 等. 细菌尿素酶的生化和分子生物学特点. 微生物学通报, 2008, 35(7): 1146-1152. DOI:10.3969/j.issn.0253-2654.2008.07.028 |
| [7] | Toffanin A, Cadahia E, Imperial J, et al. Characterization of the urease gene cluster from Rhizobium leguminosarum cv. viciae. Arch Microbiol, 2002, 177(4): 290–298. DOI: 10.1007/s00203-001-0392-0 |
| [8] | Maeda M, Hidaka M, Nakamura A, et al. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J Bacteriol, 1994, 176(2): 432–442. DOI: 10.1128/jb.176.2.432-442.1994 |
| [9] | Bossé JT, Gilmour HD, MacInnes JI. Novel genes affecting urease activity in Actinobacillus pleuropneumoniae. J Bacteriol, 2001, 183(4): 1242–1247. DOI: 10.1128/JB.183.4.1242-1247.2001 |
| [10] | Stoof J, Breijer S, Pot RGJ, et al. Inverse nickel-responsive regulation of two urease enzymes in the gastric pathogen Helicobacter mustelae. Environ Microbiol, 2010, 10(10): 2586–2597. |
| [11] | Kim JK, Mulrooney SB, Hausinger RP. Biosynthesis of active Bacillus subtilis urease in the absence of known urease accessory proteins. J Bacteriol, 2005, 187(20): 7150–7154. DOI: 10.1128/JB.187.20.7150-7154.2005 |
| [12] | Carter EL, Flugga N, Boer JL, et al. Interplay of metal ions and urease. Metallomics, 2009, 1(3): 207–221. DOI: 10.1039/b903311d |
| [13] | Benini S, Rypniewski WR, Wilson KS, et al. A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure, 1999, 7(2): 205–216. DOI: 10.1016/S0969-2126(99)80026-4 |
| [14] | Carter EL, Hausinger RP. Characterization of the Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J Bacteriol, 2010, 192(9): 2294–2304. DOI: 10.1128/JB.01426-09 |
| [15] | Farrugia MA, Wang BB, Feig M, et al. Mutational and computational evidence that a nickel-transfer tunnel in UreD is used for activation of Klebsiella aerogenes urease. Biochemistry, 2015, 54(41): 6392–6401. DOI: 10.1021/acs.biochem.5b00942 |
| [16] | Park IS, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc Natl Acad Sci USA, 1994, 91(8): 3233–3237. DOI: 10.1073/pnas.91.8.3233 |
| [17] | Moncrief MB, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol, 1996, 178(18): 5417–5421. DOI: 10.1128/jb.178.18.5417-5421.1996 |
| [18] | Fong YH, Wong HC, Chuck CP, et al. Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF-UreH protein complex. J Biol Chem, 2011, 286(50): 43241–43249. DOI: 10.1074/jbc.M111.296830 |
| [19] | Moncrief MB, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J Bacteriol, 1997, 179(13): 4081–4086. DOI: 10.1128/jb.179.13.4081-4086.1997 |
| [20] | Zambelli B, Stola M, Musiani F, et al. UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+. J Biol Chem, 2005, 280(6): 4684–4695. DOI: 10.1074/jbc.M408483200 |
| [21] | Zambelli B, Musiani F, Savini M, et al. Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry, 2007, 46(11): 3171–3182. DOI: 10.1021/bi6024676 |
| [22] | Zambelli B, Turano P, Musiani F, et al. Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins, 2010, 74(1): 222–239. |
| [23] | Leipe DD, Wolf YI, Koonin EV, et al. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol, 2002, 317(1): 41–72. |
| [24] | Bange G, Sinning I. SIMIBI twins in protein targeting and localization. Nat Struct Mol Biol, 2013, 20(7): 776–780. DOI: 10.1038/nsmb.2605 |
| [25] | Gasper R, Meyer S, Gotthardt K, et al. It takes two to tango: regulation of G proteins by dimerization. Nat Rev Mol Cell Biol, 2009, 10(6): 423–429. |
| [26] | Boer JL, Hausinger RP. Klebsiella aerogenes UreF: identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry, 2012, 51(11): 2298–2308. DOI: 10.1021/bi3000897 |
| [27] | Yang XM, Li HY, Lai TP, et al. UreE-UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori. J Biol Chem, 2015, 290(20): 12474–12485. DOI: 10.1074/jbc.M114.632364 |
| [28] | Lee MH, Pankratz HS, Wang SK, et al. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci, 2010, 2(6): 1042–1052. |
| [29] | Brayman TG, Hausinger RP. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J Bacteriol, 1996, 178(18): 5410–5416. DOI: 10.1128/jb.178.18.5410-5416.1996 |
| [30] | Banaszak K, Martin-Diaconescu V, Bellucci M, et al. Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni+ and Zn+ reveals a role for the disordered C-terminal arm in metal trafficking. Biochem J, 2012, 441(3): 1017–1035. |
| [31] | Shi R, Munger C, Asinas A, et al. Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry, 2010, 49(33): 7080–7088. DOI: 10.1021/bi100372h |
| [32] | Song HK, Mulrooney SB, Huber R, et al. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J Biol Chem, 2001, 276(52): 49359–49364. DOI: 10.1074/jbc.M108619200 |
| [33] | Remaut H, Safarov N, Ciurli S, et al. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J Biol Chem, 2001, 276(52): 49365–49370. DOI: 10.1074/jbc.M108304200 |
| [34] | Bellucci M, Zambelli B, Musiani F, et al. Helicobacter pylori UreE, a urease accessory protein: specific Ni2+ and Zn2+ binding properties and interaction with its cognate UreG. Biochem J, 2009, 422(1): 91–100. DOI: 10.1042/BJ20090434 |
| [35] | Boer JL, Quiroz-Valenzuela S, Anderson KL, et al. Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry, 2010, 49(28): 5859–5869. DOI: 10.1021/bi1004987 |
| [36] | Chan KH, Lee KM, Wong KB. Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS ONE, 2012, 7(2): e32592. DOI: 10.1371/journal.pone.0032592 |
| [37] | Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol, 2003, 185(3): 726–734. DOI: 10.1128/JB.185.3.726-734.2003 |
| [38] | Yang XM, Li HY, Cheng TF, et al. Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics, 2014, 6(9): 1731–1736. DOI: 10.1039/C4MT00134F |
| [39] | Sydor AM, Lebrette H, Ariyakumaran R, et al. Relationship between Ni(Ⅱ) and Zn(Ⅱ) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB. J Biol Chem, 2014, 289(7): 3828–3841. DOI: 10.1074/jbc.M113.502781 |
| [40] | Benoit SL, Zbell AL, Maier RJ. Nickel enzyme maturation in Helicobacter hepaticus: roles of accessory proteins in hydrogenase and urease activities. Microbiology, 2007, 153(11): 3748–3756. DOI: 10.1099/mic.0.2007/010520-0 |
| [41] | Strugatsky D, McNulty R, Munson K, et al. Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature, 2013, 493(7431): 255–258. |
| [42] | Cáceres-Delpiano J, Teneb J, Mansilla R, et al. Variations in periplasmic loop interactions determine the pH-dependent activity of the hexameric urea transporter UreI from Helicobacter pylori: a molecular dynamics study. BMC Struct Biol, 2015, 15(1): 1–12. DOI: 10.1186/s12900-014-0028-7 |
| [43] | Sebbane F, Mandrand-Berthelot MA, Simonet M. Genes encoding specific nickel transport systems flank the chromosomal urease locus of Pathogenic yersiniae. J Bacteriol, 2002, 184(20): 5706–5713. DOI: 10.1128/JB.184.20.5706-5713.2002 |
| [44] | Chen YYM, Burne RA. Identification and characterization of the nickel uptake system for urease biogenesis in Streptococcus salivarius 57.I. Virology, 2003, 183(23): 6773–6779. |
| [45] | Sigel A, Sigel H, Sigel RKO. Nickel and Its Surprising Impact in Nature, Volume 2. Hoboken, NJ: John Wiley & Sons, Ltd, 2007: 519-544. |
| [46] | Musiani F, Gioia D, Masetti M, et al. Protein tunnels: the case of urease accessory proteins. J Chem Theory Comput, 2017, 13(5): 2322–2331. DOI: 10.1021/acs.jctc.7b00042 |
| [47] | Zambelli B, Berardi A, Martin-Diaconescu V, et al. Nickel binding properties of Helicobacter pylori UreF, an accessory protein in the nickel-based activation of urease. J Biol Inorg Chem, 2014, 19(3): 319–334. DOI: 10.1007/s00775-013-1068-3 |
| [48] | Chang ZZ, Kuchar J, Hausinger RP. Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J Biol Chem, 2004, 279(15): 15305–15313. DOI: 10.1074/jbc.M312979200 |
| [49] | Quiroz-Valenzuela S, Sukuru SCK, Hausinger RP, et al. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch Biochem Biophys, 2008, 480(1): 51–57. |
| [50] | Park IS, Hausinger RP. Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science, 1995, 267(5201): 1156–1158. DOI: 10.1126/science.7855593 |
| [51] | Carter EL, Boer JL, Farrugia MA, et al. Function of UreB in Klebsiella aerogenes urease. Biochemistry, 2011, 50(43): 9296–9308. DOI: 10.1021/bi2011064 |
| [52] | Soriano A, Colpas GJ, Hausinger RP. UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry, 2000, 39(40): 12435–12440. DOI: 10.1021/bi001296o |
| [53] | Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter. J Biol Chem, 2013, 288(19): 13178–13185. DOI: 10.1074/jbc.R112.446526 |
| [54] | Soriano A, Hausinger RP. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc Natl Acad Sci USA, 1999, 96(20): 11140–11144. DOI: 10.1073/pnas.96.20.11140 |
| [55] | Fong YH, Wong HC, Yuen MH, et al. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol, 2013, 11(10): e1001678. DOI: 10.1371/journal.pbio.1001678 |
| [56] | Stingl K, Schauer K, Ecobichon C, et al. In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol Cell Proteomics, 2008, 7(12): 2429–2441. DOI: 10.1074/mcp.M800160-MCP200 |
| [57] | Farrugia MA, Han LJ, Zhong YY, et al. Analysis of a soluble (UreD:UreF:UreG)2 accessory protein complex and its interactions with Klebsiella aerogenes urease by mass spectrometry. J Am Soc Mass Spectrom, 2013, 24(9): 1328–1337. DOI: 10.1007/s13361-013-0677-y |
| [58] | Merloni A, Dobrovolska O, Zambelli B, et al. Molecular landscape of the interaction between the urease accessory proteins UreE and UreG. Biochim Biophys Acta, 2014, 1844(9): 1662–1674. DOI: 10.1016/j.bbapap.2014.06.016 |
| [59] | Yuen MH, Fong YH, Nim YS, et al. Structural insights into how GTP-dependent conformational changes in a metallochaperone UreG facilitate urease maturation. Proc Natl Acad Sci USA, 2017, 114(51): E10890–E10898. DOI: 10.1073/pnas.1712658114 |
| [60] |
Xing L, Zhao SG, Zheng N, et al. Advance in isolation and culture techniques of uncultured microbes: a review.
Microbiol China, 2017, 44(12): 3053–3066.
(in Chinese). 邢磊, 赵圣国, 郑楠, 等. 未培养微生物分离培养技术研究进展. 微生物学通报, 2017, 44(12): 3053-3066. |
 2019, Vol. 35
2019, Vol. 35