中国科学院微生物研究所、中国微生物学会主办
文章信息
- 徐超奕, 张婷, 蔡静晓, 余志良, 裘娟萍, 音建华
- Xu Chaoyi, Zhang Ting, Cai Jingxiao, Yu Zhiliang, Qiu Juanping, Yin Jianhua
- 革兰氏阴性菌中β-内酰胺酶诱导表达调控机制研究进展
- Progress in regulatory mechanism for inducing β-lactamase in Gram-negative bacteria
- 生物工程学报, 2018, 34(8): 1288-1296
- Chinese Journal of Biotechnology, 2018, 34(8): 1288-1296
- 10.13345/j.cjb.180187
-
文章历史
- Received: May 4, 2018
- Accepted: June 11, 2018
β-内酰胺类抗生素(简称β-内酰胺类,β-lactams)是临床治疗中应用最悠久且最广泛的一类抗菌药物。该类药物的化学结构中都含有β-内酰胺环,包括青霉素类、头孢菌素类、单环β-内酰胺类、碳青霉烯类和β-内酰胺酶抑制剂等[1]。青霉素结合蛋白(Penicillin binding protein,PBP)是细菌中重要的肽聚糖合成酶,负责肽聚糖糖链的聚合和链间的交联。β-内酰胺类能与青霉素结合蛋白不可逆共价结合,从而阻断肽聚糖的生物合成过程,致使细菌裂解死亡[2]。
近年来,由于抗生素的滥用和不合理使用导致细菌耐药性问题日益凸显。世界卫生组织公布了一份亟待研发新抗生素的12种耐药细菌清单,其中半数为β-内酰胺类耐药细菌,尤其是优先度最高的3种细菌均为耐碳青霉烯类的革兰氏阴性杆菌[3]。细菌对β-内酰胺类产生耐药的原因包括:一是修饰或改变抗生素的靶点,革兰氏阳性菌多采用该机制产生耐药性,如耐甲氧西林金黄色葡萄球菌(Methicillin resistant Staphylococcus aureus,MRSA)对β-内酰胺类的抗性即来源于此;二是编码β-内酰胺酶将抗生素水解,这是革兰氏阴性菌中最为普遍的耐药机制;三是减弱细胞外膜通透性,限制抗生素进入细胞内;四是提高药物外排泵的表达,将进入细胞内的抗生素外排[4]。
β-内酰胺酶广泛存在于各种临床致病菌和环境微生物中,根据氨基酸序列一致性可分为A、B、C和D四种类型[5]。其中,A、C和D型均为丝氨酸β-内酰胺酶,而B型为金属β-内酰胺酶。β-内酰胺酶的表达受β-内酰胺类诱导,但由于该类抗生素无法穿越细胞质膜,其如何诱导β-内酰胺酶的表达长期以来备受关注。目前研究最为深入的是部分革兰氏阴性杆菌中C型β-内酰胺酶AmpC诱导表达的调控,发现其与肽聚糖循环和LysR型转录因子AmpR密切相关。近年来,越来越多的研究表明双组分系统(Two component system,TCS)也能调控β-内酰胺酶的表达。本文将结合实验室研究工作,综述这两种β-内酰胺酶诱导表达调控机制,以期为新型抗生素的研发提供新靶点和新思路。
1 经典的ampR-ampC调控系统C型β-内酰胺酶AmpC (由ampC基因编码)是一种头孢菌素酶,能够水解青霉素类、头孢菌素类以及单环β-内酰胺类等抗生素[6-7]。AmpC的诱导表达是肠杆菌科细菌(如阴沟肠杆菌Enterobacter cloacae和弗氏柠檬酸杆菌Citrobacter freundii)以及非发酵革兰氏阴性杆菌(如铜绿假单胞菌Pseudomonas aeruginosa)对β-内酰胺类产生耐药性的主要原因。对这些细菌的研究表明,AmpC的诱导表达与肽聚糖循环密切相关,并且LysR型转录因子AmpR (由ampR基因编码)起核心调控作用,这种机制被称为经典的ampR-ampC调控系统[8-10]。
1.1 ampR-ampC divergon在含有ampR-ampC调控系统的细菌中,ampR与ampC基因在染色体上相邻排列,启动子区域相互重叠,但转录方向相反,即形成divergon结构。ampR基因编码LysR型调控蛋白AmpR,是ampC基因的转录激活因子。AmpR通常具有两个结构域:效应物结合结构域和DNA结合结构域。效应物结合结构域能与不同的效应物或配体结合,从而改变DNA结合结构域的构象,通过与ampC和ampR基因间隔区序列结合,调控两个基因的表达[11]。
1.2 肽聚糖循环过程细菌在正常生长过程中,肽聚糖始终处于一种动态平衡。在几十种肽聚糖合成酶和水解酶的共同作用下,不断地进行更新和循环[12]。由于β-内酰胺类的靶点是青霉素结合蛋白,抗生素处理时将不可避免地破坏肽聚糖循环的平衡。与β-内酰胺酶诱导表达密切相关的肽聚糖循环酶系主要包括AmpG、AmpD和NagZ[12]。AmpG是位于细胞质膜上的一种通透酶,含有10个跨膜结构域,负责肽聚糖水解产物的转运,如GlcNAc-anhMurNAc和GlcNAc-anhMurNAc-peptides等[13-14]。AmpD和NagZ均位于细胞质中,其中AmpD为N-乙酰胞壁酰-L-丙氨酸酰胺酶(N-acetylmuramyl-L-Ala amidase),能迅速水解anhMurNAc和L-Ala之间的酰胺键;而NagZ是β-乙酰氨基葡糖糖苷酶(β-N-acetylglucosaminidase),负责切开GlcNAc和anhMurNAc之间的β-1, 4糖苷键[15]。AmpD和NagZ的共同作用可将细胞质中的肽聚糖水解产物进一步生成GlcNAc、anhMurNAc以及anhMurNAc-peptides (图 1)。
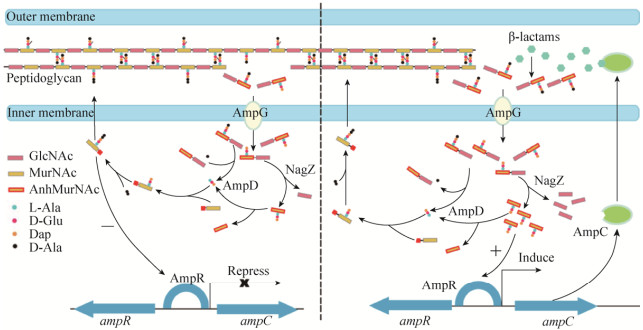
|
| 图 1 AmpR介导的β-内酰胺酶诱导表达调控模式图 Figure 1 Schematic representation of the induction of β-lactamase AmpC mediated by AmpR. In the absence of β-lactams (Left), UDP-MurNAc pentapeptides serves as a repressor ligand to mediate the repression of AmpC. In the presence of β-lactams (Right), the anhMurNAc peptides (usually tripeptide) act as an activator ligand for AmpC induction. |
| |
在缺乏β-内酰胺类诱导物时,由于AmpD和NagZ的作用使细胞内anhMurNac-peptides始终维持在较低的浓度,而三肽L-Ala-D-Glu-meso-Dap含量较高,进一步催化后生成UDP-MurNAc- pentapeptide。UDP-MurNAc-pentapeptide可作为阻遏信号分子与AmpR结合,从而抑制ampC基因的转录。当诱导物存在时,细胞周质空间中肽聚糖的损伤导致进入细胞质的肽聚糖水解产物大幅度增加,AmpD达到饱和状态,anhMurNAc-peptides大量积累,并作为诱导信号分子与UDP-MurNAc- pentapeptide竞争结合AmpR,引起AmpR蛋白构象的变化,从而激活ampC基因的转录(图 1)[11]。
AmpG-AmpD-NagZ-AmpR调控通路中任一基因的突变都有可能影响ampC基因的诱导表达。ampD基因的缺失阻断了anhMurNac-peptides的水解,使得诱导信号分子始终处于高浓度,引起AmpC高表达,致使很多临床菌株对β-内酰胺类产生耐药性[16-17]。P. aeruginosa中共有3个AmpD同源蛋白,它们的依次失活能引起AmpC表达水平和细菌耐药程度逐级增加[18-20]。AmpG的失活使诱导信号分子前体无法进入细胞质,导致ampC基因的表达不再被诱导[21-22]。NagZ可催化诱导信号分子的产生,因此nagZ基因突变与ampG效果一致,均能阻断ampC基因的表达。AmpG和NagZ的失活还能显著降低因ampD基因突变引起的AmpC高表达[23]。
这种经典的ampR-ampC调控系统同样适用于目前所研究的其他类型β-内酰胺酶的诱导表达,如霍乱弧菌Vibrio cholerae中B型β-内酰胺酶VanG[24]、野油菜黄单胞菌Xanthomonas campestris中A型β-内酰胺酶Blaxc[25-26]以及嗜麦芽窄食单胞菌Stenotrophomonas maltophilia中A型β-内酰胺酶L2 (BlaL2)和B型β-内酰胺酶L1 (BlaL1) [27-29]。S. maltophilia中blaL2基因和ampR基因构成divergon,但AmpR可同时激活L1/L2的表达。此外,该菌中mrcA基因(编码PBP1a)的失活致使这两个β-内酰胺酶呈组成型高表达,并且该过程依赖于AmpG-AmpD-AmpR,但不依赖于NagZ,表明L1/L2的表达除了遵循经典的ampR-ampC调控系统外,还存在一条不依赖于NagZ的调控通路[30-33]。
1.4 肽聚糖水解酶对β-内酰胺酶表达的影响正是由于肽聚糖循环与AmpC表达密切相关,细胞周质空间中肽聚糖的细微变化都有可能影响β-内酰胺酶的表达。目前已经发现部分肽聚糖水解酶与β-内酰胺酶的表达调控有关,如溶菌糖基转移酶(Lytic transglycosylases,LTs)和低分子量PBP等。通常,这些肽聚糖水解酶在细菌中数量较多,并且功能冗余,它们对β-内酰胺酶表达的影响在不同细菌中存在差异。LTs能将GlcNAc和MurNAc之间的β-1, 4糖苷键切开,释放ampR-ampC调控系统中的诱导信号分子前体GlcNAc-1, 6-anhMurNAc peptides。因此,LTs的失活势必影响β-内酰胺酶的表达。P. aeruginosa中含有11个LTs,但只有SltB1和MltB的失活增强AmpC的表达,而SltB的失活降低AmpC的表达[34-35]。S. maltophilia中含有6个LTs,MltD1的失活通过上调其他LTs (MltB1和MltD2)的表达增强L1/L2本底水平的表达,该过程依赖于AmpG-AmpD-NagZ-AmpR通路以及双组分系统CreBC[36]。低分子量PBP具有DD-羧肽酶和/或内肽酶活性,负责肽聚糖的修饰。P. aeruginosa中PBP4的突变使β-内酰胺酶AmpC以依赖于AmpR的方式过表达,显著增强对β-内酰胺类的耐药性[37-38]。
2 双组分系统介导的β-内酰胺酶诱导表达双组分系统是微生物中广泛存在的基因表达调控系统。该系统高度保守,一般由组氨酸激酶(Histidine kinase)和反应调节蛋白(Response regulator)二元组分构成。组氨酸激酶属跨膜蛋白,其N末端能够感知外界信号,催化C末端保守的组氨酸残基自磷酸化;然后将磷酰基转移至位于细胞内的反应调节蛋白,保守的天冬氨酸残基发生磷酸化,从而激活或抑制基因的表达,使微生物适应不同的环境变化[39]。组氨酸激酶感知的信号多种多样,如渗透压、细胞膜损伤、细胞壁损伤、金属离子和抗生素等;反应调节蛋白调控的生命过程也多种多样,如维持稳态、毒力响应、呼吸转换以及抗生素耐药等。
2.1 气单胞菌中β-内酰胺酶表达的调控气单胞菌属Aeromonas中的BlrAB是第一个被发现直接调控β-内酰胺酶表达的双组分系统。Aeromonas属细菌通常编码三种可诱导型β-内酰胺酶:B型CphA (也称Imi)、C型Cep和D型Amp[40-41]。对模式菌株嗜水气单胞菌A. hydrophila的研究发现,编码双组分系统的基因blrAB位于β-内酰胺酶基因ampH的上游,其中blrA编码反应调控蛋白BlrA,而blrB编码组氨酸激酶BlrB。β-内酰胺类诱导物抑制PBP的活性,致使肽聚糖平衡紊乱,二糖五肽单元(GlcNAc-MurNAc-pentapeptide)含量增加,很有可能作为信号分子通过直接或间接作用使BlrB自磷酸化;随后,磷酸基团转移至BlrA,激活的BlrA与特异的识别标签(cre/blr-tag:TTCACnnnnnnTTCAC)结合,从而募集RNA聚合酶启动β-内酰胺酶的表达[42] (图 2)。三个β-内酰胺酶基因的上游均存在该特异标签,并且标签的重复次数与β-内酰胺酶的表达水平呈正相关。ampH、cepH和imiH基因前的标签数分别为1、2、3个,β-内酰胺酶的表达水平为ImiH > CepH > AmpH[41, 43-44]。BlrB感知的信号源自于肽聚糖损伤,DD-羧肽酶/内肽酶(PBP4和BlrY)的失活显著提高二糖五肽单元含量,增强β-内酰胺酶的表达(图 2);而万古霉素能与D-Ala-D-Ala特异性结合,处理细菌后β-内酰胺酶的表达受到抑制。另外,blrAB基因下游含有一个编码内膜蛋白的基因blrD,其转录受BlrAB控制,但BlrD在β-内酰胺酶表达中的作用尚不清楚[44]。
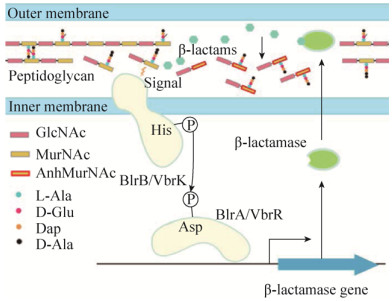
|
| 图 2 双组分系统介导的β-内酰胺酶表达调控示意图 Figure 2 Schematic representation of the induction of β-lactamase mediated by two component systems. Both BlrAB and VbrKR are involved in the regulation of β-lactamase production. The two component signal transduction pathway is activated by either β-lactams (for VbrKR) or GlcNAc-MurNAc-pentapeptide (for BlrAB). |
| |
大肠杆菌Escherchia coli中的双组分系统CreBC与BlrAB高度同源,最初发现CreBC是中间代谢的全局调控因子[43, 45]。反应调节蛋白CreB识别的特异序列与BlrA一致,当Aeromonas属来源的3个β-内酰胺酶基因连同上游序列一起转入E. coli DH5α菌株(CreB+)时,β-内酰胺酶可被成功表达[43]。
P. aeruginosa和S. maltophilia中的CreBC可能参与β-内酰胺酶表达的调控。P. aeruginosa中PBP4的突变特异性激活CreBC,然后上调CreD (与BlrD同源)的表达,间接参与β-内酰胺酶的表达[37, 46]。此外,P. aeruginosa CreBC还在β-内酰胺类胁迫、生物膜形成以及细菌适应度方面发挥重要的作用[46]。S. maltophilia中CreBC与细菌运动性密切相关[47],MltD1失活导致的β-内酰胺酶表达同时需要CreBC和AmpR的参与[36]。
2.2 弧菌中β-内酰胺酶表达的调控副溶血性弧菌Vibrio parahaemolyticus是一种重要的食源性致病菌,其对青霉素类的耐药性主要由A型β-内酰胺酶(blaAV100)介导[48]。该菌中含有32个可能的双组分系统,但只有VbrKR缺失后显著降低β-内酰胺酶的表达和β-内酰胺类耐药性。VbrK是一种能直接感知β-内酰胺类的组氨酸激酶,抗生素的结合使其空间构象发生变化,组氨酸激酶结构域与ATP酶结构域之间更加接近,随后VbrK自磷酸化并将磷酸基团转移至反应调节蛋白VbrR,从而触发β-内酰胺酶的表达[49] (图 2)。有趣的是,VbrKR还能调控PBP1a和PBP3的表达,但是否与β-内酰胺酶表达相关尚不清楚。
与BlrAB/CreBC明显不同的是,VbrK感知的信号并非来自于肽聚糖的损伤,而是直接与抗生素结合,然后迅速将信号传递至细胞内,通过诱导β-内酰胺酶的表达保证在细胞裂解之前将抗生素水解[50]。VbrK是目前革兰氏阴性菌中唯一发现能直接感知β-内酰胺类的受体蛋白。几乎所有弧菌属细菌都编码与VbrK同源的组氨酸激酶,因此弧菌属可能普遍存在VbrK介导的β-内酰胺酶表达调控机制。革兰氏阳性菌中β-内酰胺酶(如Bacillus licheniformis BlaP和Staphylococcus aureus BlaZ)的诱导表达也起始于抗生素与膜受体蛋白之间的直接相互作用,但受体蛋白感知抗生素后自身变成有活性的金属蛋白酶,通过水解阻遏蛋白诱导β-内酰胺酶的表达[51]。
2.3 希瓦氏菌中β-内酰胺酶表达的调控水环境中广泛存在的希瓦氏菌属Shewanella被认为是β-内酰胺类和喹诺酮类耐药基因的天然存储库,部分菌株也逐渐成为新兴致病菌[52-53]。该属的模式菌株奥奈达希瓦氏菌S. oneidensis编码7个假定的β-内酰胺酶,其中由染色体介导的D型β-内酰胺酶BlaA (也称为OXA-54)可能是碳青霉烯类水解酶Oxacillinase的祖先[54]。笔者近年来的研究发现,该酶的表达受氨苄青霉素诱导,但具体的诱导机制与AmpR介导的调控系统差异显著,主要表现在两个方面:一是基因组中blaA基因未与其他调控蛋白基因构成divergon,也不含有与AmpR高度同源的调控蛋白;二是主要的肽聚糖循环酶(如AmpG、NagZ和AmpD)对blaA表达的影响与经典的ampR-ampC调控系统相反[55-57]。ampG和nagZ基因缺失后blaA基因的表达仍可被诱导,表明该菌中存在一条不依赖于AmpG-NagZ-AmpR的β-内酰胺酶诱导表达通路。该通路与PBP1a及其外膜脂蛋白辅因子LpoA有关,这两个蛋白形成肽聚糖合成酶复合体PBP1a-LpoA,其失活导致blaA基因呈组成型高表达,暗示诱导blaA表达的信号分子位于周质空间。S. oneidensis中含有数目众多的双组分系统,虽不与CreBC或VbrKR高度同源,但有证据表明其他双组分系统参与blaA表达的调控。PBP1a与周质空间中的β-内酰胺类共价结合而失活,致使细胞被膜受损,激活双组分系统,从而诱导β-内酰胺酶的表达[58-60]。
3 结论与展望由此可见,革兰氏阴性菌中β-内酰胺酶诱导表达调控机制具有以下共同特点:LysR型转录因子或双组分系统在调控中起核心作用;如果β-内酰胺酶基因与LysR型转录因子编码基因组成divergon,那么该β-内酰胺酶的表达受LysR型转录因子调控;同一物种中可能同时包含两种调控通路(如P. aeruginosa和S. maltophilia),二者之间存在相互影响;肽聚糖循环在β-内酰胺酶诱导表达过程中发挥重要的作用,除V. parahaemolyticus VbrKR直接感知抗生素外,其他已经发现的调控蛋白均感知肽聚糖水解片段。
由于肽聚糖生物合成和循环过程非常复杂,目前我们对诱导β-内酰胺酶表达的信号分子仍不清晰,尤其是抗生素作用对肽聚糖的具体影响缺乏详实数据。未来的研究应该从以下几个方面展开:1)继续研究不同细菌中β-内酰胺酶诱导表达的调控机制,尤其是重要的临床致病菌和环境微生物。CRISPR-Cas9基因编辑技术的发展使得这些细菌的遗传改造变得方便快捷。2)利用生物信息学工具,从数据库中挖掘β-内酰胺酶诱导表达调控相关元件的分布规律,在进化层面上明确抗性基因的来源和转移规律。3)已有报道表明NagZ抑制剂能有效降低AmpC高产菌株对β-内酰胺类的耐药性[23],接下来应针对β-内酰胺酶诱导表达通路中的新靶点,加大筛选特异性药物的力度,从而有效解决革兰氏阴性菌中的β-内酰胺类耐药问题。
| [1] | Llarrull LI, Testero SA, Fisher JF, et al. The future of the β-lactams. Curr Opin Microbiol, 2010, 13(5): 551–557. DOI: 10.1016/j.mib.2010.09.008 |
| [2] | Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol Rev, 2008, 32(2): 361–385. DOI: 10.1111/j.1574-6976.2007.00095.x |
| [3] | Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature, 2017, 543(7643): 15. DOI: 10.1038/nature.2017.21550 |
| [4] | Blair JM, Webber MA, Baylay AJ, et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol, 2015, 13(1): 42–51. DOI: 10.1038/nrmicro3380 |
| [5] | Ambler RP. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci, 1980, 289(1036): 321–331. DOI: 10.1098/rstb.1980.0049 |
| [6] | Mark BL, Vocadlo DJ, Oliver A. Providing β-lactams a helping hand: targeting the AmpC β-lactamase induction pathway. Fut Microbiol, 2011, 6(12): 1415–1427. DOI: 10.2217/fmb.11.128 |
| [7] | Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother, 2010, 54(3): 969–976. DOI: 10.1128/AAC.01009-09 |
| [8] | Dietz H, Pfeifle D, Wiedemann B. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl- pentapeptide. Antimicrob Agents Chemother, 1997, 41(10): 2113–2120. |
| [9] | Jacobs C, Huang LJ, Bartowsky E, et al. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J, 1994, 13(19): 4684–4694. |
| [10] | Jacobs C, Frère JM, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell, 1997, 88(6): 823–832. DOI: 10.1016/S0092-8674(00)81928-5 |
| [11] | Zeng XM, Lin J. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol, 2013, 4: 128. |
| [12] | Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev, 2008, 72(2): 211–227. DOI: 10.1128/MMBR.00027-07 |
| [13] | Cheng QM, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol, 2002, 184(23): 6434–6436. DOI: 10.1128/JB.184.23.6434-6436.2002 |
| [14] | Chahboune A, Decaffmeyer M, Brasseur R, et al. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob Agents Chemother, 2005, 49(3): 1145–1149. DOI: 10.1128/AAC.49.3.1145-1149.2005 |
| [15] | Cheng QM, Li HS, Merdek K, et al. Molecular characterization of the β-N-Acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol, 2000, 182(17): 4836–4840. DOI: 10.1128/JB.182.17.4836-4840.2000 |
| [16] | Kopp U, Wiedemann B, Lindquist S, et al. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother, 1993, 37(2): 224–228. DOI: 10.1128/AAC.37.2.224 |
| [17] | Juan C, Maciá MD, Gutiérrez O, et al. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother, 2005, 49(11): 4733–4738. DOI: 10.1128/AAC.49.11.4733-4738.2005 |
| [18] | Langaee TY, Gagnon L, Huletsky A. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible ampC β-lactamase expression. Antimicrob Agents Chemother, 2000, 44(3): 583–589. DOI: 10.1128/AAC.44.3.583-589.2000 |
| [19] | Juan C, Moyá B, Pérez JL, et al. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob Agents Chemother, 2006, 50(5): 1780–1787. DOI: 10.1128/AAC.50.5.1780-1787.2006 |
| [20] | Schmidtke AJ, Hanson ND. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob Agents Chemother, 2006, 50(6): 2030–2037. DOI: 10.1128/AAC.01458-05 |
| [21] | Korfmann G, Sanders CC. ampG is essential for high-level expression of AmpC beta-lactamase in Enterobacter cloacae. Antimicrob Agents Chemother, 1989, 33(11): 1946–1951. DOI: 10.1128/AAC.33.11.1946 |
| [22] | Lindquist S, Weston-Hafer K, Schmidt H, et al. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol Microbiol, 1993, 9(4): 703–715. DOI: 10.1111/mmi.1993.9.issue-4 |
| [23] | Zamorano L, Reeve TM, Deng LH, et al. NagZ inactivation prevents and reverts β-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob Agents Chemother, 2010, 54(9): 3557–3563. DOI: 10.1128/AAC.00385-10 |
| [24] | Lin HV, Massam-Wu T, Lin CP, et al. The Vibrio cholerae var regulon encodes a metallo-β-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysR-type transcription factor. PLoS ONE, 2017, 12(9): e0184255. DOI: 10.1371/journal.pone.0184255 |
| [25] | Yang TC, Chen TF, Tsai JJP, et al. AmpG is required for BlaXc beta-lactamase expression in Xanthomonas campestris pv. campestris str. 17. FEMS Microbiol Lett, 2013, 340(2): 101–108. DOI: 10.1111/fml.2013.340.issue-2 |
| [26] | Yang TC, Chen TF, Tsai JJ, et al. NagZ is required for beta-lactamase expression and full pathogenicity in Xanthomonas campestris pv. campestris str. 17. Res Microbiol, 2014, 165(8): 612–619. DOI: 10.1016/j.resmic.2014.08.008 |
| [27] | Okazaki A, Avison MB. Induction of L1 and L2 β-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob Agents Chemother, 2008, 52(4): 1525–1528. DOI: 10.1128/AAC.01485-07 |
| [28] | Yang TC, Huang YW, Hu RM, et al. AmpDI is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother, 2009, 53(7): 2902–2907. DOI: 10.1128/AAC.01513-08 |
| [29] | Huang YW, Lin CW, Hu RM, et al. AmpN-AmpG operon is essential for expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Antimicrob Agents Chemother, 2010, 54(6): 2583–2589. DOI: 10.1128/AAC.01283-09 |
| [30] | Lin CW, Lin HC, Huang YW, et al. Inactivation of mrcA gene derepresses the basal-level expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. J Antimicrob Chemother, 2011, 66(9): 2033–2037. DOI: 10.1093/jac/dkr276 |
| [31] | Talfan A, Mounsey O, Charman M, et al. Involvement of mutation in ampD I, mrcA, and at least one additional gene in β-lactamase hyperproduction in Stenotrophomonas maltophilia. Antimicrob Agents Chemother, 2013, 57(11): 5486–5491. DOI: 10.1128/AAC.01446-13 |
| [32] | Huang YW, Hu RM, Lin CW, et al. NagZ-dependent and NagZ-independent mechanisms for β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob Agents Chemother, 2012, 56(4): 1936–1941. DOI: 10.1128/AAC.05645-11 |
| [33] | Juan C, Torrens G, González-Nicolau M, et al. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol Rev, 2017, 41(6): 781–815. DOI: 10.1093/femsre/fux043 |
| [34] | Cavallari JF, Lamers RP, Scheurwater EM, et al. Changes to its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother, 2013, 57(7): 3078–3084. DOI: 10.1128/AAC.00268-13 |
| [35] | Lamers RP, Nguyen UT, Nguyen Y, et al. Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. MicrobiologyOpen, 2015, 4(6): 879–895. DOI: 10.1002/mbo3.286 |
| [36] | Huang YW, Wu CJ, Hu RM, et al. Interplay among membrane-bound lytic transglycosylase D1, the CreBC two-component regulatory system, the AmpNG-AmpDI-NagZ-AmpR regulatory circuit, and L1/L2 β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob Agents Chemother, 2015, 59(11): 6866–6872. DOI: 10.1128/AAC.05179-14 |
| [37] | Moya B, D tsch A, Juan C, et al. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog, 2009, 5(3): e1000353. DOI: 10.1371/journal.ppat.1000353 |
| [38] | Ropy A, Cabot G, Sánchez-Diener I, et al. Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-lactam resistance, and peptidoglycan structure. Antimicrob Agents Chemother, 2015, 59(7): 3925–3934. DOI: 10.1128/AAC.05150-14 |
| [39] | Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem, 2000, 69(1): 183–215. DOI: 10.1146/annurev.biochem.69.1.183 |
| [40] | Avison MB, Niumsup P, Walsh TR, et al. Aeromonas hydrophila AmpH and CepH β-lactamases: derepressed expression in mutants of Escherichia coli lacking creB. J Antimicrob Chemother, 2000, 46(5): 695–702. DOI: 10.1093/jac/46.5.695 |
| [41] | Niumsup P, Simm AM, Nurmahomed K, et al. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of β-lactamase expression in Aeromonas spp. J Antimicrob Chemother, 2003, 51(6): 1351–1358. DOI: 10.1093/jac/dkg247 |
| [42] | Tayler AE, Ayala JA, Niumsup P, et al. Induction of β-lactamase production in Aeromonas hydrophila is responsive to β-lactam-mediated changes in peptidoglycan composition. Microbiology, 2010, 156(8): 2327–2335. DOI: 10.1099/mic.0.035220-0 |
| [43] | Avison MB, Horton RE, Walsh TR, et al. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem, 2001, 276(29): 26955–26961. DOI: 10.1074/jbc.M011186200 |
| [44] | Avison MB, Niumsup P, Nurmahomed K, et al. Role of the 'cre/blr-tag' DNA sequence in regulation of gene expression by the Aeromonas hydrophila β-lactamase regulator, BlrA. J Antimicrob Chemother, 2004, 53(2): 197–202. DOI: 10.1093/jac/dkh077 |
| [45] | Cariss SJL, Tayler AE, Avison MB. Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli. J Bacteriol, 2008, 190(11): 3930–3939. DOI: 10.1128/JB.00108-08 |
| [46] | Zamorano L, Moyà B, Juan C, et al. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother, 2014, 58(9): 5084–5095. DOI: 10.1128/AAC.02556-14 |
| [47] | Huang HH, Chen WC, Lin CW, et al. Relationship of the CreBC two-component regulatory system and inner membrane protein CreD with swimming motility in Stenotrophomonas maltophilia. PLoS ONE, 2017, 12(4): e0174704. DOI: 10.1371/journal.pone.0174704 |
| [48] | Chiou J, Li RC, Chen S. CARB-17 Family of β-lactamases mediates intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob Agents Chemother, 2015, 59(6): 3593–3595. DOI: 10.1128/AAC.00047-15 |
| [49] | Li L, Wang QY, Zhang H, et al. Sensor histidine kinase is a β-lactam receptor and induces resistance to β-lactam antibiotics. Proc Natl Acad Sci USA, 2016, 113(6): 1648–1653. DOI: 10.1073/pnas.1520300113 |
| [50] | Hofer U. Antimicrobials: β-lactam sensor discovered. Nat Rev Microbiol, 2016, 14(4): 195. DOI: 10.1038/nrmicro.2016.27 |
| [51] | Amoroso A, Boudet J, Berzigotti S, et al. A peptidoglycan fragment triggers β-lactam resistance in Bacillus licheniformis. PLoS Pathog, 2012, 8(3): e1002571. DOI: 10.1371/journal.ppat.1002571 |
| [52] | Ramírez MS, Merkier AK, Almuzara M, et al. Reservoir of antimicrobial resistance determinants associated with horizontal gene transfer in clinical isolates of the genus Shewanella. Antimicrob Agents Chemother, 2010, 54(10): 4516–4517. DOI: 10.1128/AAC.00570-10 |
| [53] | Janda JM, Abbott SL. The genus Shewanella: from the briny depths below to human pathogen. Crit Rev Microbiol, 2014, 40(4): 293–312. DOI: 10.3109/1040841X.2012.726209 |
| [54] | Poirel L, Héritier C, Nordmann P. Chromosome-encoded Ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob Agents Chemother, 2004, 48(1): 348–351. DOI: 10.1128/AAC.48.1.348-351.2004 |
| [55] | Yin JH, Sun LL, Dong YY, et al. Expression of blaA underlies unexpected ampicillin-induced cell lysis of Shewanella oneidensis. PLoS ONE, 2013, 8(3): e60460. DOI: 10.1371/journal.pone.0060460 |
| [56] | Yin JH, Mao YT, Ju LL, et al. Distinct roles of major peptidoglycan recycling enzymes in β-lactamase production in Shewanella oneidensis. Antimicrob Agents Chemother, 2014, 58(11): 6536–6543. DOI: 10.1128/AAC.03238-14 |
| [57] |
Wu GF, Yin JH. Peptidoglycan recycling and bacterial resistance to β-lactams.
Chin Pharm J, 2017, 52(3): 180–184.
(in Chinese). 吴根福, 音建华. 肽聚糖循环及细菌对β-内酰胺类抗生素的耐受性. 中国药学杂志, 2017, 52(3): 180-184. |
| [58] | Yin JH, Sun YY, Mao YT, et al. PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis. Antimicrob Agents Chemother, 2015, 59(6): 3357–3364. DOI: 10.1128/AAC.04669-14 |
| [59] | Yin JH, Sun YY, Sun YJ, et al. Deletion of Lytic transglycosylases increase β-lactam resistance in Shewanella oneidensis. Front Microbiol, 2018, 9: 13. DOI: 10.3389/fmicb.2018.00013 |
| [60] | Yin JH, Cai JX, Yuan Z, et al. Deletion of PBP1a/LpoA complex compromises cell envelope integrity in Shewanella oneidensis. FEMS Microbiol Lett, 2018. DOI: 10.1093/femsle/fny128 |
 2018, Vol. 34
2018, Vol. 34




