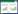中国科学院微生物研究所、中国微生物学会主办
文章信息
- 刘长命, 张显, 王永琦
- Changming Liu, Xian Zhang, Yongqi Wang
- 甜瓜S-腺苷甲硫氨酸脱羧酶基因的克隆及白粉病诱导表达分析
- Cloning and analysis of a powdery mildew responsive gene CmSAMDC from Cucumis melo L.
- 生物工程学报, 2018, 34(6): 928-936
- Chinese Journal of Biotechnology, 2018, 34(6): 928-936
- 10.13345/j.cjb.180038
-
文章历史
- Received: January 23, 2018
- Accepted: April 2, 2018
2 西北农林科技大学 园艺学院,陕西 杨凌 712100
2 College of Horticulture, Northwest A & F University, Yangling 712100, Shaanxi, China
Muskmelon is one of the most important cultivated plants worldwide. However, in China, melon was threatened by various abiotic and biotic stresses[1]. Powdery mildew, which is strictly biotrophic and completely dependent on the living host cells for survival[2], is a common disease of melon under field and greenhouse conditions worldwide. Many recent studies have shown that Podosphaera xanthii (P. xanthii) is predominant in most countries[3-5]. Based on our previous study[6], it could be preliminarily confirmed that the P. xanthii race 2F is the predominant physiological race within cucurbitaceous plants in Guanzhong areas, Shaanxi Province of China.
Currently, control of powdery mildew on melon is mainly achieved by widespread application of various fungicides. Although powdery mildew can be controlled by fungicides, the cost and the environmental impact of residues are undesirable consequences, and the long term use has led to fungicide resistance of powdery mildew races[7]. Furthermore, with the accumulation of toxic heavy metals in soil, the exceeded level of relevant metal ions will be found in melon, which seriously affects the melon food safety. Compared with the application of fungicides, using resistant cultivars is a more effective and environmentally safe approach to disease control.
Plants use various mechanisms to defend against pathogens, and these are often initiated by plant disease resistance genes, which may encode cell receptors that detect the presence of a specific pathogen and these initiate the activation of signal transduction pathways. Polyamines (PAs) are small ubiquitous compounds that have been implicated in the regulation of many physiological progresses and a variety of stress responses in plants[8]. The enhanced level of polyamines plays an important role in the protective response of plants to various abiotic stresses[9]. SAMDC is one of the key regulatory enzymes in the biosynthesis of polyamines. There are some reports about SAMDC genes responses to abiotic stresses in plants[10]. However, very few study about biotic stresses was conducted in melon.
In our previous study using cDNA-AFLP method, we observed an EST (expression sequence tag) that was induced in leaves of 'Yuantian930' upon infection with P. xanthii[6]. We analyzed the EST sequence and found that it was the only one completely consistent with SAMDC in melon genome sequence database (https://melonomics.net/). Since 'Yuantian930' has been shown to be highly resistant to powdery mildew, we undertook the present study to gain a better understanding of SAMDC in melon and verified its potential role in powdery mildew resistance. We cloned the full-length of CmSAMDC gene from 'Yuantian930' and further analyzed its expression in response to P. xanthii. The aim of this study was to determine whether or not there was a relationship between melon resistance and the response of CmSAMDC to P. xanthii infection. To our knowledge, this is the first report of SAMDC gene potentially related to powdery mildew resistance in melon.
1 Materials and methods 1.1 Plant material and inoculation methodThe powdery mildew (PM) resistant clone, Chinese wild melon C. melo 'Yuantian930' was used in this study, which was provided by the melon and watermelon research group of Horticulture Institute of Northwest A & F University, China. The P. xanthii isolate used in this work was collected from natural infections in cucurbit and through mono bacterial colony propagation. The melon seedlings were grown in the plant growth chamber at 30 ℃/18 ℃ (day/night) operating a 16 h/8 h (light/dark) photoperiod and 70%–85% relative humidity. PM inoculation were performed by spraying plants with a suspension of 106 sporangia/mL P. xanthii conidiophores, and then the plants were kept in the same conditions. The leaves from the infected plants (at the forth-leaf stage) were collected after 0 h, 12 h, 24 h, 48 h, 72 h and 120 h, stored at –80 ℃. As a control, leaves were collected from plants treated with sterile water only.
1.2 RNA extraction and cDNA synthesisTotal RNA was extracted from melon leaves using the Trizol reagent following the instructions of the Trizol kit (Tiangen, Beijing). Residual DNA was removed by DNase Ⅰ (Promega, Madison, WI, USA). Concentration of total RNA was measured with an ultraviolet spectrophotometer (V-550, JASCO, Japan) at 260 nm. RNA purity was checked by determining the A260/A280 ratio, and RNA integrity was examined by 1% agarose gel electrophoresis. SmartTM RACE cDNA Amplification Kit was used to synthesize cDNA, following the instructions of the Clontech kit (Clontech-TaKaRa Bio Inc., Japan).
1.3 Cloning of melon CmSAMDCOn the basis of the EST fragment of CmSAMDC from Chinese wild melon C. melo 'Yuntian-930' by cDNA-AFLP analysis and the data from melon genome sequence database, the gene-specific primers (Table 1) CmSAMDCs and CmSAMDCa containing full ORF (open reading frame) were designed using Primer Premier 5.0, and the BamH I and EcoR I restriction sites were added. PCR amplification of CmSAMDC with one microliter of cDNA from infected leaves was added to a PCR mix containing 2 U of Taq polymerase (MBI Fermentas), 1× PCR buffer, 2.5 mmol/L MgCl2, 0.1 mmol/L of each dNTP, and 0.5 mmol/L of the forward and reverse degenerate primers. The final volume of each reaction was 25 μL and PCR was performed using a Bio-Rad thermocycler following conditions: initial denaturation at 94 ℃ for 3 min; followed by 35 cycles of 94 ℃ for 30 s, 56 ℃ for 1 min, 72 ℃ for 1 min; and a final extension at 72 ℃ for 10 min. The PCR products were electrophoresed in 1.5% agarose gel. The fragments of expected size was isolated and purified using the Gel Extraction Kit (BioTake, USA). The pGEM-T Easy Vector System (Promega, Madison, USA) was used for DNA cloning. The positive clones identified by PCR and enzyme digestion were sequenced by the Beijing Aoke Biotech Co. Ltd. (Beijing, China).
| Primer name | Primer sequence (5'-3') |
| CmSAMDCs | GCGGATCCATGACGTGTCCAACCTCTG |
| CmSAMDCa | CGAATTCCTAATACTTCCCAACTTCCTCG |
| CmSAMDCf | ATCAAAACTTGCGGCACTAC |
| CmSAMDCr | AGCACCCTCACAATCAACTTAG |
| ActinS | TGCCCAGAAGTTCTATTCCAGC |
| ActinA | CATAGTTGAACCACCACTGAGGAC |
| The cutting sites of BamHⅠand EcoRⅠwere underlined. | |
The similarity analysis of nucleotide and protein sequences was carried out using the Blast tool at NCBI (http://www.ncbi.nlm.nih.gov/blast). The deduced amino acid sequence was analyzed with the Predict Protein Analysis System (http://www.predictprotein.org/). The parameters for the given protein were computed using the ProtParam tool at the ExPASy Proteomics Server (http://web.expasy.org/protparam/). The transmembrane regions and orientation of given protein was predicted using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The multiple sequences alignment was performed with Clustal X and Genedoc software, and a phylogenetic tree was constructed using the MEGA 5.1 software by the Neighbor-joining method. The reliability of each node on phylogenetic tree was tested by bootstrapping 1 000 times.
1.5 Quantitative real-time PCR and data analysisThe first-strand cDNA was synthesized using the Reverse Transcription System Kit (Promega, USA) according to the manufacturer's instructions. A pair of gene-specific primers (Table 1) CmSAMDCf and CmSAMDCf were designed following the recommended guidelines for qRT-PCR primer design, and primers specific for the melon Actin gene (GenBank Accession No. AY859055) were used as the internal control (Table 1). The product size was 147 bp.
Relative levels of gene expression were analyzed with the Themal Cycler Dice® Real Time System (Bio-Rad, Hercules, CA, USA) and normalized with the results for Actin. Each 20 μL PCR reaction contained 10 μL of SYBR® Premix ExTaqTMⅡ (2×), 0.8 μL of PCR Forward Primer (10 μmol/L), 0.8 μL of PCR Reverse Primer (10 μmol/L), 2 μL of cDNA (200 ng/μL), and 6.4 μL of ddH2O. The PCR cycling conditions consisted of an initial polymerase activation step at 95 ℃ for 30 s, followed by 40 cycles of 95 ℃ for 5 s and 60 ℃ for 30 s. Output data generated by the instrument on-board software iQ5 (Bio-Rad) were transferred to Sigmaplot software (v.10.0, Systat Inc., CA, USA) for analysis. Real-time quantitative RT-PCR was performed in three replicates for each sample. In addition, reverse transcription negative control was included to check for potential genomic DNA contamination. The relative expression of CmSAMDC gene was calculated according to the method of 2–△△Ct. The △△CT=(CT, Target–CT, Actin) Timex– (CT, Target–CT, Actin)Time0, CT, Target means the CT value of target gene, CT, Actin means the CT value of control gene, Time x means the post inoculation time (12 h, 24 h, 48 h, 72 h, 120 h), Time 0 means the time before inoculation (0 h).
1.6 Expression of CmSAMDC in E. coliAfter verifying the sequence, the recombinant plasmid of positive clone was digested with BamH Ⅰ/ EcoR Ⅰ and ligated directly into the prokaryotic expression vector pET-28a (Fermentas, USA), which was predigested with the same enzymes. A recombination reaction yielding a pET-28a and CmSAMDC fusion construct was conducted. After sequencing and confirming by restriction digestion, the positive recombinant prokaryotic expression plasmids, designated pET28a-CmSAMDC, were then transformed into competent E. coli strain BL21 (DE3) for expression of the fusion protein. Transformed E. coli cells were incubated at 37 ℃ overnight in Luria-Bertani medium with 50 mg/L Kan. 1 mL of the overnight cultures were added into 50 mL of fresh LB medium containing Kan and incubated with shaking at 37 ℃ until OD600 reached 0.6. Isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 1 mmol/L and the incubation was continued at 37 ℃. 2 mL of thallus were collected at 2 h, 4 h, 6 h and 8 h respectively after IPTG was added, followed by centrifuging at 12 000×g for 30 s, and then resuspended with 5× buffer solution [every 10 mL reaction contained: 0.6 mL Tris-HCl (1 mol/L, pH 6.8), 5 mL glycerol (50%), 2 mL SDS (10%), 0.5 mL mercaptoethanol, 1 mL bromophenol blue (1%), 0.9 mL ddH2O]. The resulting lysate was heated at 100 ℃ for 10 min, and then centrifuged at 12 000×g for 1 min. The supernatant was used as a sample for SDS-PAGE. After electrophoresis, the gel was stained with Coomassie Brilliant Blue.
2 Results and discussion 2.1 Isolation of CmSAMDC geneBased on the study conducted before, a length of 479 bp product was found highly identical to SAMDC in Cucumis sativus, Brassica juncea and Arabidopsis thaliana[11]. Given this, the full ORF of SAMDC gene with a length of 1 095 bp was amplified from 'Yuntian 930' and named CmSAMDC (GenBank Accession No. KF151861). Results from agarose gel electrophoresis showed that the PCR products had the expected molecular size of 1 095 bp, which suggested that the full length of CmSAMDC gene was successfully amplified by gene special primers (Fig. 1).

|
| 图 1 CmSAMDC基因的扩增 Figure 1 Amplification of CmSAMDC gene. M: DL2000 marker; 1: the CmSAMDC cDNA. |
| |
The results showed that the deduced peptide consists of 364 amino acid residues, with a predicted molecular weight of 40 kDa, and theoretical pI of 4.61. The transmembrane regions and orientation was predicted that the given protein probably has no transmembrane region (Fig. 2). Phylogenetic analysis comprising the SAMDC protein sequences from twenty plants revealed that the CmSAMDC protein had close distance with Cucumis sativus, Arabidopsis thaliana, Brassica juncea and Citrofortunella microcarpa, as they all belong to dicotyledon, and the lowest similarity was detected as to Saccharum officinarum, Triticum aestivum and Zea mays, since the last three belong to monocot (Fig. 3).

|
| 图 2 跨膜域与方向的预测 Figure 2 The transmembrane regions and orientation prediction. The line with solid dot and solid square means the polypeptide inside or outside the membrane respectively; the line with solid hexagon means the transmembrane region. |
| |

|
| 图 3 20种植物SAMDC蛋白氨基酸进化树分析 Figure 3 Phylogenetic tree analysis based on the alignment of deduced amino acid sequences of SAMDC protein in 20 plants. Two major classes were identified and classified to dicotyledon and monocot. The numbers below the branches refer to the percentage of 1 000 bootstrap replications supporting the nodes. |
| |
In this study, the expected size of SAMDC gene in melon was isolated, and the sequence analyses indicated that the CmSAMDC from 'Yuantian930' shares a high level of sequence homology with other plants of SAMDC genes [12-15]. Phylogenetic analysis and the alignment of the deduced amino acid sequences with other plants revealed a series of common features, which was consistent with former studies[16].
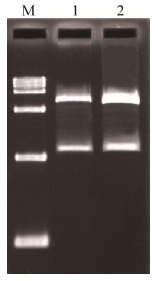
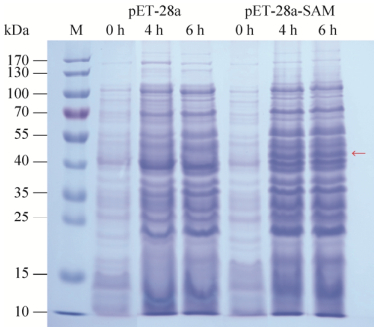
2.3 Expression of CmSAMDC in E. coli BL21 (DE3)After the array and orientation of the insert sequence were confirmed by restriction digestion with enzyme EcoRⅠ/BamHⅠ and sequencing, the PCR product was directly cloned into pET-28a. Sequencing results showed that CmSAMDC gene has been inserted correctly into pET-28a vector (Fig. 4). Upon IPTG induction, fusion proteins containing the desired protein were expressed in E. coli BL21 (DE3). As illustrated by the SDS-PAGE (Fig. 5), the fusion protein was detected as a broad and a predominant band at 43 kDa (including 3 kDa His-tag) in the induced cell lysate with pET28a-SAMDC (lane 4 h and 6 h), and six hours of IPTG induction would be suitable. The molecular mass was consistent with the molecular weight calculated from the amino acid sequence. No reaction was observed with no-loading vector and 0 h as well.
|
| 图 4 融合表达载体的酶切鉴定 Figure 4 Validation of fusion expression vector by digestion. M: DNA marker (250, 1 000, 2 500, 5 000, 10 000, 12 500, 15 000 bp); 1: pGEM-T-SAMDC digested with EcoRⅠand BamHⅠ; 2: pET28a-SAMDC digested with EcoRⅠand BamHⅠ. |
| |
|
| 图 5 pET-28a-CmSAMDC重组体的原核表达分析 Figure 5 CmSAMDC proteins expressed in E. coli and separated by SDS-PAGE. The arrows showed the molecular weight of the target protein agreed well with the predicted. |
| |
The expression level of CmSAMDC in response to P. xanthii in PM-resistant clone 'Yuntian930' was investigated at different time by qRT-PCR. As shown in Fig. 6, CmSAMDC was constitutively expressed at low levels in leaves of 'Yuantian930' before inoculation, then it was peaked at 48 h as almost 7 times high as observed in control. These results suggested that transcription of CmSAMDC is indeed upregulated in Chinese wild PM-resistant melon C. melo 'Yuntian-930' upon infection of P. xanthii pathogens.

|
| 图 6 中国野生甜瓜白粉病菌侵染后CmSAMDC基因表达分析 Figure 6 Expression profiles of CmSAMDC gene in Chinese wild melon after P. xanthii infection. Each experiment was repeated three times, and the standard error bars were indicated. Total RNA was extracted from leaves of C. melo clone 'Yuntian930', and analyzed at 0 h, 12 h, 24 h, 48 h, 72 h and 120 h after inoculation with P. xanthii. The leaves of the plants on which sterile water was sprayed were collected as the control leaves (CK). |
| |
For coping with biotic and abiotic stresses, plants have evolved a wide range of defense mechanisms. These defense mechanisms depend on a series of proteins, including many involved in regulating cellular response to stress and signaling crosstalk[17]. Many of these proteins involved the production of phytohormones, pathogenesis-related proteins and transcription factors[18]. The pathogen defense mechanism in plants has been reported to be associated with early and strong gene expression[19]. Most plant resistance genes are transcriptionally regulated in response to pathogen attack. In rice, the transcription of resistance gene Xa1 appears to increase following pathogen inoculation[20], which indicates that the transcription of the resistance gene depends on the type of plant-pathogen interaction.
During this experiment, we have examined the expression pattern of SAMDC in PM-resistant clone 'Yuantian930' in response to P. xanthii and it showed higher levels and stronger expression, which was almost 7 times as high as observed in no pathogen inoculation. This trend is similar to SAMDC expressing in sugarcane[13], tall fescue[14] and tomato under abiotic stress[21]. In addition, the author's further research also shows that SAMDC gene plays protective roles in melon resistance to powdery mildew infection by transgenic arabidopsis plants expressing CmSAMDC[22]. Thus, SAMDC was involved in P. xanthii-induced defense response besides the important role in response to abiotic stress[23-26]. However, the close relationship of SAMDC and powdery mildew stress need to be further verified.
3 ConclusionIn summary, we have demonstrated that CmSAMDC gene was induced by P. xanthii in PM-resistant melon leaves and likely plays a positive role in defense mechanisms that confer resistance to powdery mildew in melon. Further investigations of CmSAMDC may aid us in understanding the mechanism(s) of PM resistance in melon.
| [1] |
Akkurt M, Welter L, Maul E, et al. Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L. and Vitis sp.). Mol Breed, 2007, 19(2): 103-111. DOI:10.1007/s11032-006-9047-9 |
| [2] |
Cheng H, Kun WP, Liu DS, et al. Molecular cloning and expression analysis of CmMlo1 in melon. Mol Bio Rep, 2012, 39(2): 1903-1907. DOI:10.1007/s11033-011-0936-6 |
| [3] |
Fekete C, Fung RWM, Szabó Z, et al. Up-regulated transcripts in a compatible powdery mildew-grapevine interaction. Plant Physiol Biochem, 2009, 47(8): 732-738. DOI:10.1016/j.plaphy.2009.03.006 |
| [4] |
Kim M, Panstruga R, Elliott C, et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature, 2002, 416(6879): 447-451. DOI:10.1038/416447a |
| [5] |
Consonni C, Humphry ME, Hartmann HA, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet, 2006, 38(6): 716-720. DOI:10.1038/ng1806 |
| [6] |
Xian F. Genetic mechanisms, hormonal changes and cDNA-AFLP analysis of resistance to powdery mildew in wild melon[sp. Agrestis (NAUD. ) Greb. ][D]. Yangling: Northwest A & F University, 2012 (in Chinese). 咸丰. 野生甜瓜[sp. Agrestis (Naud. ) Greb. ]抗白粉病的遗传机制和激素变化及其cDNA-AFLP分析[D]. 杨凌: 西北农林科技大学, 2012. |
| [7] |
Fekete C, Fung RWM, Szabó Z, et al. Up-regulated transcripts in a compatible powdery mildew-grapevine interaction. Plant Physiol Biochem, 2009, 47(8): 732-738. DOI:10.1016/j.plaphy.2009.03.006 |
| [8] |
Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature, 2001, 411(6839): 826-833. DOI:10.1038/35081161 |
| [9] |
Kubiś J. Exogenous spermidine differentially alters activities of some scavenging system enzymes, H2O2 and superoxide radical levels in water-stressed cucumber leaves. J Plant Physiol, 2008, 165(4): 397-406. DOI:10.1016/j.jplph.2007.02.005 |
| [10] |
Walters DR. Polyamines and plant disease. Phytochemistry, 2003, 64(1): 97-107. DOI:10.1016/S0031-9422(03)00329-7 |
| [11] |
Liu CM, Xian F, Tian ZG, et al. Analysis of gene expression profiling in the incompatible interaction between Chinese wild melon 'Yuntian930' and Podosphaera xanthii. Acta Phytopathol Sin, 2014, 44(1): 65-73. 刘长命, 咸丰, 田治国, 等. 野生甜瓜'云甜930'与白粉病菌互作的基因表达特征. 植物病理学报, 2014, 44(1): 65-73. |
| [12] |
Roy M, Wu R. Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Sci, 2002, 163(5): 987-992. DOI:10.1016/S0168-9452(02)00272-8 |
| [13] |
Liu JX, Que YX, Guo JL, et al. Molecular cloning of sugarcane S-adenosylmethionine decarboxylase gene (Sc-SAMDC) and its expression analysis. Sci Agric Sin, 2010, 43(7): 1448-1457. 刘金仙, 阙友雄, 郭晋隆, 等. 甘蔗S-腺苷蛋氨酸脱羧酶基因Sc-SAMDC的克隆和表达分析. 中国农业科学, 2010, 43(7): 1448-1457. |
| [14] |
Wang XL, Liu XX, Wang SY, et al. Cloning and differential expression analysis of S-adenosylmethionine decarboxylase gene FaSAMDC in tall fescue. Acta Pratacult Sin, 2011, 20(4): 169-179. 王小利, 刘晓霞, 王舒颖, 等. 高羊茅腺苷甲硫氨酸脱羧酶基因FaSAMDC的克隆与差异表达分析. 草业学报, 2011, 20(4): 169-179. DOI:10.11686/cyxb20110421 |
| [15] |
Wi SJ, Kim WT, Park KY. Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep, 2006, 25(10): 1111-1121. DOI:10.1007/s00299-006-0160-3 |
| [16] |
Li CQ, Shao JF, Wang YJ, et al. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genomics, 2013, 14: 851. DOI:10.1186/1471-2164-14-851 |
| [17] |
Alcázar R, Altabella T, Marco F, et al. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta, 2010, 231(6): 1237-1249. DOI:10.1007/s00425-010-1130-0 |
| [18] |
Koh S, André A, Edwards H, et al. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J, 2005, 44(3): 516-529. DOI:10.1111/tpj.2005.44.issue-3 |
| [19] |
Wen XP, Pang XM, Matsuda N, et al. Over-expression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Res, 2008, 17(2): 251-263. DOI:10.1007/s11248-007-9098-7 |
| [20] |
Kim ST, Kim SG, Hwang DH, et al. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus Magnaporthe grisea. Proteomics, 2004, 4(11): 3569-3578. DOI:10.1002/(ISSN)1615-9861 |
| [21] |
Liu ZY, Wang XX, Gao JC, et al. Cloning and sequence analysis of a S-adenosylmethionine decarboxylase gene (SlSAMDC1) in tomato. Acta Horticult Sin, 2008, 35(8): 1137-1146. 刘志勇, 王孝宣, 高建昌, 等. 番茄S-腺苷蛋氨酸脱羧酶基因SlSAMDC1的克隆与序列分析. 园艺学报, 2008, 35(8): 1137-1146. |
| [22] |
Liu CM, Li XL, Yang RP, et al. The protective roles of S-adenosylmethionine decarboxylase (SAMDC) gene in melon resistance to powdery mildew infection. Hort Environ Biotechnol, 2014, 55(6): 557-567. DOI:10.1007/s13580-014-0026-5 |
| [23] |
Perchepied L, Bardin M, Dogimont C, et al. Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology, 2005, 95(5): 556-565. DOI:10.1094/PHYTO-95-0556 |
| [24] |
Sabelli PA, Larkins BA. Regulation and function of retinoblastoma related plant genes. Plant Sci, 2009, 177(6): 540-548. DOI:10.1016/j.plantsci.2009.09.012 |
| [25] |
Zhang SP, Xiao YN, Zhao JR, et al. Digital gene expression analysis of early root infection resistance to Sporisorium reilianum f. sp. zeae in maize. Mol Genet Genomics, 2013, 288(1/2): 21-37. |
| [26] |
Crowley T, Walters DR. Polyamine metabolism in an incompatible interaction between barley and the powdery mildew fungus, Blumeria graminis f. sp. hordei. J Phytopathol, 2002, 150(11/12): 581-586. |
 2018, Vol. 34
2018, Vol. 34