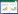中国科学院微生物研究所、中国微生物学会主办
文章信息
- 惠希武, 陈虹, 黄秉仁
- Xiwu Hui, Hong Chen, Bingren Huang
- N-糖基化位点突变的人IFN-λ1在毕赤酵母中的表达、纯化及表征
- Expression, purification and characterization of N-glycosylation mutant human IFN-λ1 in Pichia pastoris
- 生物工程学报, 2018, 34(4): 613-624
- Chinese Journal of Biotechnology, 2018, 34(4): 613-624
- 10.13345/j.cjb.170390
-
文章历史
- Received: October 9, 2017
- Accepted: December 15, 2017
2 石药集团中奇制药技术 (石家庄) 有限公司,河北 石家庄 050035
2 CSPC Zhongqi Pharmaceutical Technology (Shijiazhuang) Co. Ltd., Shijiazhuang 050035, Hebei, China
IFN-λ1, also known as interleukin-29 (IL-29), is a member of a new family of interferons called type Ⅲ IFNs, which have similar functions to type Ⅰ IFNs, IFN-α and IFN-β. IFN-λ1 interacts with a heterodimeric IFN-λ1 receptor complex composed of a unique IFN-λR1 chain and the IL-10R2 chain[1-2]. IFN-λ1 and type Ⅰ IFNs bind to unrelated functional receptors, but they share the same signal transduction pathway (Janus kinase/signal transducer and activator of transcription and JAK-STAT), and activate the transcription of a similar set of effectors. IFN-λ1 activates IFN-stimulated response elements (ISRE) and mediates a number of biological effects, including inhibition of viral replication, cellular growth inhibition, and immunoregulatory[2-6]. rhIFN-λ1 has been developed as biopharmaceutical regent for hepatitis C virus therapy. Results from a phase 1b study of PEG-IFN-λ1 treatment of patients with chronic HCV infection showed that PEG-IFN-λ1 is safe, effective, and has fewer side effects compared with PEG-IFN-α2a[7-8].
P. pastoris glycosylate foreign proteins at N-linked glycosylation sites, although to lesser extent than in mammalian cells[9-13]. It is known that the high-mannose type glycosylation can change the protein structure, influence the protein characteristics and functions, and bring immunogenicity[14]. In human body, the high-mannose type oligosaccharide chains bind mannose receptor, leading to poor pharmacokinetic properties and immune response[15-16], and these effects limit the applications of P. pastoris expression system in producing recombinant proteins. Therefore, many studies have focused on the humanization of P. pastoris N-glycosylated protein, in the hope of generating human-like oligosaccharide chain structure. However, no significant progress achieved to date. So far, the main method to avoid hyper-glycosylation is site-directed mutations of N-glycosylation sites of recombinant proteins.
IFN-λ1 contains a potential N-linked (Asn-X-Ser/Thr) glycosylation sites at Asn46 (NWS, Fig. 1), and expressed hyper-glycosylated IFN-λ1 (HG-IFN-λ1) and low-glycosylated IFN-λ1 (LG-IFN-λ) in P. pastoris[17]. To reduce immunogenicity and increase the expression and purification efficiency of recombinant proteins, we used N-glycosylation site-directed mutagenesis approach to express mutant IFN-λ1 in P. pastoris. Here we described the construction of P. pastoris strains capable of secreting biologically active mutant IFN-λ1, rhIFN-λ1-Nm, and the expression, purification, physicochemical properties characterization and biological activity analysis of mutant rhIFN-λ1. Our results indicated that the mutant rhIFN-λ1 could be legitimate substitutes for IFN-λ1 in developing into a more promising therapeutic reagent.
|
| 图 1 人IFN-λ1的核苷酸及氨基酸序列(方框内为潜在N-糖基化位点(NWS); 下划线为信号肽; 核苷酸及氨基酸序列均来自NCBI; 核苷酸及氨基酸序列号分别为NM_172140.1;NP_742152.1) Figure 1 The nucleotide and amino acid sequence of human IFN-λ1. The potential N-glycosylation site(NWS) show in box, and the signal peptide is underlined prior to the mature protein sequence in bold. The nucleotide and amino acid sequence were both from NCBI. The number of nucleotide and amino acid sequence are NM_172140.1 and NP_742152.1 separately. |
| |
E. coli DH5α (New England Biolabs) was used to construct the recombinant plasmids. Yeast P. pastoris GS115 (his4) (Invitrogen) was used as a host strain for expressing rhIFN-λ1 and rhIFN-λ1-Nm.
The human embryonic kidney cells, HEK293 cells were cultured in DMEM medium supplemented with 10% FBS (heat-inactivated fetal bovine serum) in presence of 100 U/mL penicillin and 0.1 mg/mL streptomycin maintained at 37 ℃, 5% CO2 incubator. The human hepatocellular cancer cell lines HepG2 and Hep3b were maintained in MEM medium supplemented with 10% FBS, 100 U/mL penicillin and 0.1 mg/mL streptomycin maintained at 37 ℃, 5% CO2 incubator. Cells were transfected with transfection reagents (Vigorous) according to the manufacturer's protocol.
1.2 Construction of multicopies N-glycosylation mutant expression plasmidIFN-λ1 mutated at Asn-46(IFN-λ1-Nm) was generated by overlapping PCR with Pfu-DNA polymerase using pAO-1×IFNλ1 as template. A first round of PCR were performed respectively with primers, α Factor-F and IFN-λ1-Nm-R or IFN-λ1-Nm-F and IFN-λ1-R1, for 5 cycles of 94 ℃ for 1 min, 48 ℃ for 1 min, and 72 ℃ for 1 min, followed by 25 cycles of 94 ℃ for 40 s, 55 ℃ for 40 s, and 72 ℃ for 1 min. Both reaction products, which contain codon CAA encoding Gln instead of AAC for Aln, were mixed together, and then re-amplified with primers α Factor-F and IFN-λ1-R1. Thus, the DNA fragment of α Factor-IFN-λ1-Nm was obtained. The sequences of the primers used in this study are shown in Table 1.
| Primer name | Sequences (5′–3′) |
| α Factor-F | GGAATTCACCATGAGATTTCCTTCAATTTTT |
| IFN-λ1-Nm-R | TGCAACTCCATTGTTTCAGCTTG |
| IFN-λ1-Nm-F | CAAGCTGAAACAATGGAGTTGCA |
| IFN-λ1-R1 | CGGAATTCGTGGGGTGTCAGGTGGACTCAG |
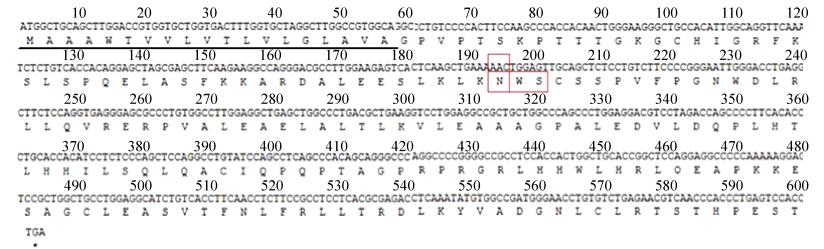
The α Factor-IFN-λ1-Nm fragment was digested with EcoR Ⅰ and cloned into plasmid pAO815 (Invitrogen) (Fig. 2A) at the unique EcoR Ⅰ site. After proving the correct insertion orientation of α Factor-IFN-λ1-Nm in pAO815, the plasmid pAO-1×IFN-λ1-Nm was used as a source of IFN-λ1-Nm expression cassette (5′AOX1-α Factor-IFN-λ1-Nm-3′AOX1TT) for further cloning. IFN-λ1-Nm expression cassette was released by digestion of Bgl Ⅱ and BamH Ⅰ. To achieve two tandem copies of the IFN-λ1-Nm expression cassette in a vector, pAO-1×IFN-λ1-Nm was linearized at the unique BamH Ⅰ site which lies immediately downstream of the 3′AOX1TT. Insertion of the second expression cassette in correct orientation involved a Bgl Ⅱ/BamH Ⅰ cohesive end ligation, thereby retained a unique Bgl Ⅱ site at the 5′ end of the promoter for the first cassette and a single BamH Ⅰ site at the 3′ end of the terminator for the second cassette in pAO-2×IFN-λ1-Nm. Similarly, pAO-6×IFN-λ1-Nm was constructed by ligating a 2×IFN-λ1-Nm cassette to pAO-4×IFN-λ1-Nm at the unique BamH Ⅰ site (Fig. 2B).
|
| 图 2 表达质粒pAO-n×IFN-λ1-Nm的构建. (A)表达载体pAO815图谱. (B) 1×, 2×, 4×, and 6×IFN-λ1-Nm表达质粒构建图. (C)质粒酶切电泳图. M: λDNA/Hind Ⅲ分子量标准:23 kb, 9.4 kb, 6.5 kb, 4.3 kb, 2.3 kb, 2.0 kb; 1: pAO-IFN-λ1-Nm; 2: pAO-2×IFN-λ1-Nm; 3: pAO-4×IFN-λ1-Nm; 4: pAO-6× IFN-λ1-Nm的Bgl Ⅱ和BamH Ⅰ双酶切图谱, 箭头指示不同拷贝数表达盒. Figure 2 Construction of plasmid pAO-n×IFN-λ1-Nm. (A) Integration and intracellular expression vector pAO815. (B) Sketch map of the IFN-λ1-Nm expression cassette contained in the 1×, 2×, 4×, and 6×IFN-λ1-Nm expression plasmids. (C) Plasmid digested with different restriction enzyme. M: λDNA/Hind Ⅲ and their bp (base pairs) numbers are: 23 kb, 9.4 kb, 6.5 kb, 4.3 kb, 2.3 kb, 2.0 kb; 1: pAO-IFN-λ1-Nm; 2: pAO-2×IFN-λ1-Nm; 3: pAO-4×IFN-λ1-Nm; 4: pAO-6× IFN-λ1-Nm digested with Bgl Ⅱ and BamH Ⅰ. Arrows showed different copies of expression cassette. |
| |
The pAO-6×IFN-λ1-Nm expression plasmid described above (10 μg) was linearized with Sal Ⅰ restriction enzyme and used to transform P. pastoris strains GS115 by electroporation using a Gene Pulser Ⅱ (Bio-Rad) (0.2 cm cuvette, 1.8 kV, 25 μF, 200 Ω). P. pastoris cells were spreaded on minimal dextrose(MD) plate and incubated for 4–6 d at 30 ℃. His+ transformants (GS115/ IFN-λ1-Nm) were then simultaneously patched both on a minimal methanol (MM) plate and a MD plate to screen Mut+ or Muts phenotype. To induce expression of rhIFN-λ1-Nm, the Mut+ clones were grown in 10 mL BMG medium at 30 ℃ in shaking tubes. After 48 h of cultivation, the yeast cells were spun down, resuspended in 5 mL of BMMY medium to an OD600 of 2.5–5 and incubated at 30 ℃ with shaking[18]. Methanol was added every 24 h at 1% (V/V).
The supernatants of GS115/IFN-λ1-Nm cultures were diluted with 5 volumes of 50 mmol/L sodium phosphate, pH 7.0. The rhIFN-λ1-Nm was eluted from the column with a linear gradient to 50 mmol/L sodium phosphate, 1 mol/L NaCl, pH 7.0. rhIFN-λ1-Nm proteins were further separated on a size exclusion chromatography (SEC)-Superdex 75 prepacked column (1.25 cm×60 cm, GE) with 20 mmol/L sodium phosphate, 0.15 mol/L NaCl, pH 7.2.
1.4 Coomassie brilliant blue staining, PAS staining and Western blottingSecreted rhIFN-λ1-Nm in the media was evaluated by SDS-PAGE using 15% gel and visualized by Coomassie brilliant blue staining or Periodic Acid-Schiff (PAS) staining. rhIFN-λ1-Nm was further identified by Western blotting with a polyclonal antibody against human IFN-λ1 (R & D Systems). PAS staining of glycosylated protein was performed as described previously[19]. Briefly, the gels were fixed in 25% isopropyl alcohol/10% acetic acid, overnight; and then in 10% isopropyl alcohol/10% acetic acid, 2 h; 0.5% periodic acid, 2 h; 0.5% sodium arsenate/5% acetic acid, 30–60 min; 0.1% sodium arsenate/5% acetic acid, 20 min, repeated twice; acetic acid, 10–20 min. The gels were then stained by one hundred milliliter Schiff reagent overnight, followed by 0.1% sodium metabisulfite/0.01 mol/L HCl for several hours, and repeated until the rinse solution failed to turn pink.
1.5 Analytical methods 1.5.1 N-terminal sequencingThe proteins were concentrated and separated by SDS-PAGE as above, and then electro-transferred onto PVDF membrane in 10 mmol/L Caps/10% methanol, with 200 mA for 2 h at 4 ℃. After staining with 0.1% Coomassie brilliant blue R-250/50% methanol, the membrane was distained with 50% methanol/10% acetic acid and washed adequately with distilled water. The corresponding bands were analyzed by automated Edman degradation using a PPSQ-33A (SHIMADZU).
1.5.2 MALDI-TOF mass spectrometry analysisMALDI-TOF-MS spectra were performed on a reflex time of flight instrument (5800 plus MALDI-TOF/TOF, Ab Sciex, Massachusetts, USA) equipped with a scout ion source operating in positive linear mode. Ions generated by a pulsed UV laser beam (nitrogen laser, λ 337 nm) were accelerated to 25 kV. A saturated solution of sinapinic acid in water/ACN (1:1) was used as matrix and mixed with the samples dissolved in 0.1% TFA aqueous solution at a V/V ratio of 1:1.
1.5.3 Circular dichrosim analysisCircular dichroism (CD) analysis were performed with A J-810 spectropolarimetry (Jasco, Easton, MD, USA) and in 0.1 cm patch length quartz cells at 20 ℃. The analysis was carried out using 0.1 mg/mL protein in 10 mmol/L NaAC. Both near and far UV spectra (in the range 195–250 nm and 250–330 nm respectively, with a data pitch of 0.1 nm) were recorded by averaging five individual scans. The distribution of main secondary structures was calculated according to a described unsupervised learning neural network method[20].
1.6 Biological activity of rhIFN-λ1-Nm 1.6.1 Activity detection used dual-luciferase reporter gene assay (RGA)2×105 HEK293 cells per well were plated in 6-well plate and transfected with pISRE-luc (5 μg) and pRL-SV40 (0.1 μg) plasmids. At 24 h after transfection, cells were stimulated with rhIFN-λ1 for 16 h. Cells were harvested and lysed using Promega 1×passive lysis buffer. Relative ISRE-Luciferase activity was measured using the dual luciferase reporter assay system (Promega). The mixture of cell lysates and buffer was tested on a SPECTRA MAX XPS (Molecular Devices).
1.6.2 Expression of HLA-ABC induced by rhIFN-λ1-Nm5×105 HepG2 and Hep3B cells per well were plated in 6-well plate and stimulated with rhIFN-λ1 in DMEM containing 0.5% FBS for 72 h, respectively. After treated with trypsin and percussion into a single cell suspension, and washed twice with PBS, the HepG2 and Hep3B cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-HLA-A, B, C antibody (BD Biosciences, San Jose, CA, USA). The fluorescence intensity was analyzed with a BD Accuri C6 Flow Cytometer (BD Biosciences).
1.7 Statistical analysisThe data were analyzed by SPSS software. Results were expressed as x±s standard deviations and the differences were considered significant as P < 0.05.
2 Result 2.1 Construction of plasmid pAO-6×IFNλ1-Nm and P. pastoris recombinant strainsThe N-glycosylation site mutagenesis of human IFN-λ1 at Asn46Gln was described as "rhIFN-λ1-Nm". As described above, DNA fragment α F-IFN-λ1-Nm containing the correct sequence of N-glycosylation site mutant was successfully cloned into plasmid pAO815, a plasmid for generating multicopies expression cassettes in vitro. It will remain two Bgl Ⅱsites and one BamH Ⅰ site in every recombinant plasmid if expression cassettes are ligated in correct orientation. This makes it feasible to identify the number of gene expression cassette by Bgl Ⅱ and BamH Ⅰ double digestion (Fig. 2C). The section of ~2.1 kb between the restriction sites of Bgl Ⅱ and BamH Ⅰ is the expression cassette consisting of the AOX1 promoter (5′AOX1) and the gene of α F-IFN-λ1-Nm followed by AOX1 transcription termination (TT). Via a series of cloning procedure, pAO815-6×IFN-λ1-Nm containing six copies of IFN-λ1-Nm expression cassette was successfully constructed in vitro (Fig. 2C). The expression cassettes were integrated at the His4 or AOX1 locus of GS115 genome by homologous integration, giving rise to recombinant strains GS115/IFN-λ1-Nm. Plasmids linearized with Sal Ⅰ favored their insertion at His4 to generate His+Mut+ phenotype.
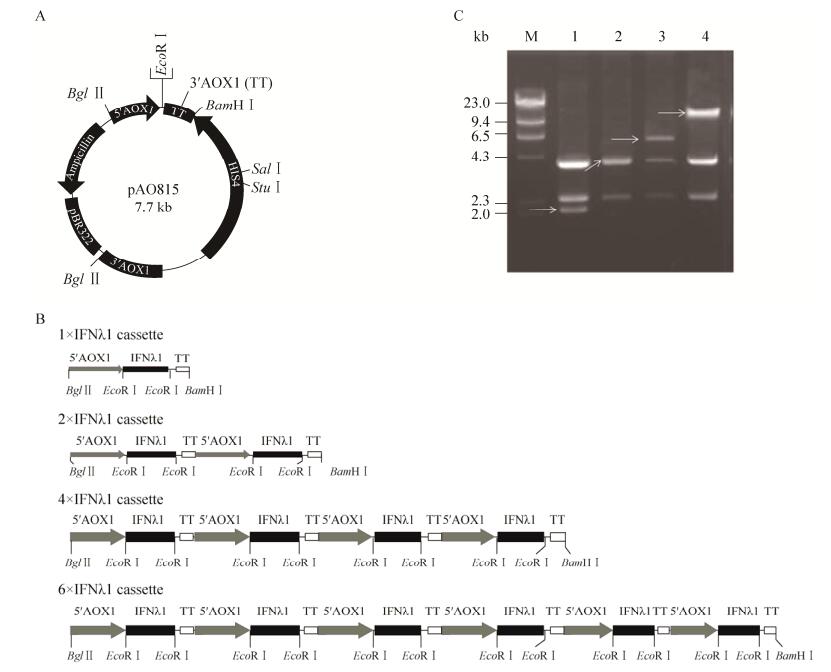
2.2 Expression and purification of rhIFNλ1-Nm in P. pastorisSmall-scale cultures of the His+ Mut+ clones were subjected to methanol induction to identify clones capable of secreted rhIFN-λ1-Nm. At different time points (24 h, 48 h, 72 h, 96 h) after methanol induction, the medium supernatants were harvested, and expression products were visualized by SDS-PAGE. Compared with rhIFN-λ1, only one major protein band at 21 kDa was showed in the supernatant of rhIFN-λ-Nm by Coomassie brilliant blue staining, Western blotting and PAS staining (Fig. 3C). After N-glycosylation site mutagenesis, the 35 kDa protein band appeared in the sample of rhIFN-λ1 was not detected in the product of rhIFN-λ1-Nm, indicating the absence of hyper-glycosylation IFN-λ1.
|
| 图 3 毕赤酵母表达rhIFN-λ1和rhIFN-λ1-Nm的纯化及SDS-PAGE鉴定. (A) rhIFN-λ1的SPFF及Superdex 75层析图谱; 圆圈标出为HG-IFN-λ1, 箭头指示IFN-λ1. (B) rhIFN-λ1-Nm的SPFF及Superdex 75层析图谱; 箭头指示IFN-λ1, 无HG-IFN-λ1. (C) rhIFN-λ1及rhIFN-λ1-Nm考马斯亮蓝染色, Western boltting及PAS糖染色分析. M:蛋白分子量标准86, 43, 34, 26, 19 kDa; 1, 2: rhIFN-λ1及rhIFN-λ1-Nm表达上清; 3, 4: SPFF洗脱峰; 5, 6: SEC洗脱峰. Figure 3 Purification and SDS-PAGE determination of rhIFN-λ1 and rhIFN-λ1-Nm expressed in Pichia pastoris. (A) SPFF and Superdex 75 chromatography of rhIFN-λ1; the peak of HG-IFN-λ1 with a circle around it and the arrow shows IFN-λ1. (B) SPFF and Superdex 75 chromatography of rhIFN-λ1-Nm. The arrow shows IFN-λ1-Nm, and there is no HG-IFN-λ1 peak. (C) Coomassie brilliant blue staining, Western blotting and PAS staining analysis of rhIFN-λ1 and rhIFN-λ1-Nm after expression and purification. M: the standard proteins and their sizes were 86, 43, 34, 26, 19 kDa; 1, 2: the induced culture medium supernatant of rhIFN-λ1 and rhIFN-λ1-Nm; 3, 4: the elution fraction of SPFF after loading culture medium supernatant; 5, 6: the protein eluted with the SEC after SPFF chromatography. |
| |
The fermentation supernatant of GS115/IFN-λ1-Nm or GS115/IFN-λ1 fermentation was purified by a two-step process. Cation exchange chromatography was a crucial step to remove the most of the HG-IFN-λ1 of the fermentation supernatant of GS115/IFN-λ1 and native secreted proteins of P. pastoris. The crudely purified proteins were further purified on a Superdex 75 size exclusion column. Coomassie brilliant blue staining, PAS staining and Western blotting showed that both rhIFN-λ1-Nm and rhIFN-λ1 was eluted with a purity of > 98% (Fig. 3C). But we have to give up the purified rhIFN-λ1 because it is hard to separate the HG-IFN-λ1 from the LG-IFN-λ1 completely (Fig. 3A and 3B), and this led to reduced purification recovery efficiency. Therefore, we turned to the purification of rhIFN-λ1-Nm.
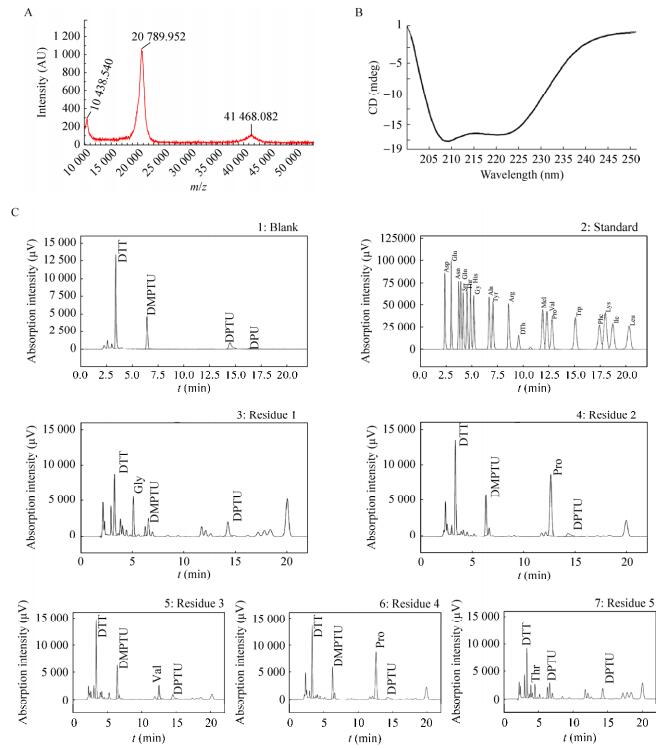
2.3 Characterization of rhIFN-λ1-NmMALDI-TOF-MS, N-terminal amino acid sequence analyzer and circular dichroism were used to analyze the molecular weight, N-terminal amino acid sequence and secondary structure of N-glycosylation mutant recombinant protein to detect possible changes. The MS reported in Fig. 4A shows the major signal with mass value of molecular weight 20 789.95 Da, which is consistent with the theoretical Mw of rhIFN-λ1. The other two signals with mass of 10 438.54 and 41 468.08 Da correspond to the doubly-charged IFN-λ1 and a dimeric of IFN-λ1, respectively. As showed in Fig. 4B, N-terminal amino acid sequence of the protein is G-P-V-P-T-, which is consistent with rhIFN-λ1. The circular dichroism spectrum result showed that there is no significantly difference of secondary structure before and after the mutation. Both of them have a predominantly α-helical structure with two characteristic strong negative shoulder peaks at 208 nm and 222 nm as revealed by the far UV-CD spectrum shown in Fig. 4C. Together, these results suggest that N-glycosylation site mutation does not affect the second structure of rhIFN-λ1.
|
| 图 4 rhIFN-λ1-Nm表征结果(A:MALDI-TOF-MS检测rhIFN-λ1-Nm分子量大小; B:rhIFN-λ1-Nm圆二色谱结; C:rhIFN-λ1-Nm N末端测序结果) Figure 4 The characteristics of rhIFN-λ1-Nm. (A) The molecular weight of rhIFN-λ1-Nm detected by MALDI-TOF-MS. (B) Circular dichroism spectra of rhIFN-λ1-Nm. (C) N-terminal sequence of rhIFN-λ1-Nm. |
| |
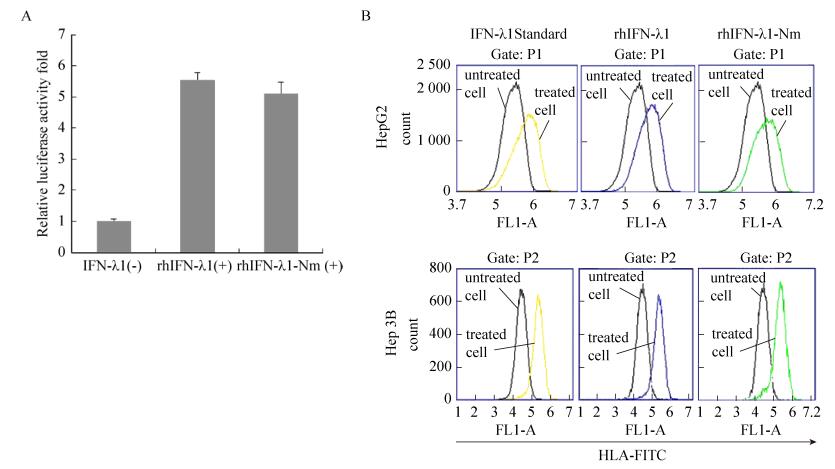
To determine whether the mutant N-glycosylation site of IFN-λ1 could influence the biological activity of rhIFN-λ1, a pISRE luciferase reporter assay was used to assess the transactivation of ISRE-modulated gene. HEK293 cells were co-transfected with both of the plasmids pISRE-Luc and pRL-SV40, and then stimulated with purified rhIFN-λ1 or rhIFN-λ1-Nm. As shown in Fig. 5A, cells stimulated with rhIFN-λ1-Nm induced 5-fold higher luciferase activity of ISRE than media alone, which is similar to standard IFN-λ1[2], and there was no significant difference compared with rhIFN-λ1.
|
| 图 5 rhIFN-λ1及rhIFN-λ1-Nm的生物学活性检测(A:用100 ng/mL的rhIFN-λ1及rhIFN-λ1-Nm处理ISRE荧光素酶报告基因质粒转染的HEK 293细胞, 检测相对荧光素酶活性; B:肝细胞中MHC Ⅰ分子表达的流式细胞仪分析) Figure 5 Bioactivity of rhIFN-λ1 and rhIFN-λ1-Nm. (A) HEK 293 cells were transfected with an ISRE reporter gene plasmid and luciferase activity was analyzed after treatment with 100 ng/mL rhIFN-λ1 and rhIFN-λ1-Nm. Relative luciferase activity was determined by dividing the relative light unit (RLU) value of each experimental sample by the RLU value of medium alone. Values are means sd (n=3). (B) Flow cytometric analysis of MHC class Ⅰ molecule expression in hepatic cells. IFN-λs-treated or untreated cells were stained with anti-HLA antibody. |
| |
To further detect the bioactivity of rhIFN-λ1-Nm, we examined whether rhIFN-λ1-Nm could increase the expression of MHC Ⅰ. The major histocompatibility complex (MHC) is a set of cell surface molecules encoded by a large gene family in all vertebrates. MHC is not only involved in transplant rejection and T cell differentiation and development, but also plays an important role in regulating the immune start-up and immune response. Interferon can improve the expression level of MHC Ⅰ molecules in cell surface, leading to enhanced killing effect of cytotoxic T cells to target cells; increase the cracking potential of NK cells, and finally activate the effective antiviral and antitumor immune response. After stimulation of rhIFN-λ1 and rhIFN-λ1-Nm, the expressions of HLA-ABC in HepG2 and Hep3B cell surface were significantly unregulated, and no noticeable difference was observed between rhIFN-λ1 and rhIFN-λ1-Nm (Fig. 5B).
3 ConclusionWe recently purified recombinant human IFN-λ1 (rhIFN-λ1) in a soluble form from P. pastoris. However, the N-glycosylation site in IFN-λ1 at Asn46 (NWS), bring immunogenicity and affects the purification efficiency. In this study, we successfully generated and expressed the mutant IFN-λ1-Nm, which contains the Asn46Gln mutation, in P. pastoris. A higher yield of pure protein of mutant IFN-λ1 was obtained in flask culture as compared with rhIFN-λ1. Our results showed that there is no highly glycosylated portion in the rhIFN-λ1-Nm preparation, and the oligosaccharide content of the mutant IFN-λ1 is significantly reduced as compared with the rhIFN-λ1. After purified by SPFF and Superdex 75 chromatography, we obtained > 98% pure recombinant mutant protein that had high recovery rate. The MALDI-TOF mass spectrometry, N-terminal amino acid sequencing and CD spectrum analysis showed that both purified rhIFN-λ1-Nm and rhIFN-λ1 have standard molecule weight, the same N-terminal amino acid sequence, and the similar second structure. These results indicate that the N-glycosylation mutation does not affect the physicochemical properties of IFN-λ1.
It is important to establish a method to determine the biological activity of the recombinant protein in the pharmaceutical quality control. The potency of IFN has been determined traditionally by the antiviral assay (AVA)[21-22], in which the activity of IFN is measured based on its inhibitory effects on viral replication. However, AVA is inherently disadvantageous because of higher assay variations, the requirement for working with virus in Biosafety level-2 (BSL-2) laboratories, and the need to titrate the virus[23-26]. It is highly desirable to establish new bioassay methods. Here we used a RGA based on ISRE-driven luciferase activity measurement[22, 27] to detect the activity of rhIFN-λ1 and rhIFN-λ1-Nm. And, also we examined whether rhIFN-λ1-Nm could increase the expression of MHC Ⅰ to detect the immunoregulatory activity of IFN-λs. Both results indicated that there is no significant difference between rhIFN-λ1 and rhIFN-λ1-Nm in bioactivity.
In conclusion, we have shown that elimination of the N-glycosylation sites in human IFN-λ has no effect on characteristics or bioactivity in IFN-λ1 proteins. Therefore, the mutation legitimately substitutes for wild-type in developing into a more promising therapeutic reagent in the future, that reduced immunogenicity and improved efficiency of rhIFN-λ1 purification from P. pastoris.
| [1] |
Kotenko SV, Gallagher G, Baurin VV, et al. IFN-λs mediate antiviral protection through a distinct class Ⅱ cytokine receptor complex. Nat Immunol, 2003, 4(1): 69-77. DOI:10.1038/ni875 |
| [2] |
Sheppard P, Kindsvogel W, Xu WF, et al. IL-28, IL-29 and their class Ⅱ cytokine receptor IL-28R. Nat Immunol, 2003, 4: 63-68. DOI:10.1038/ni873 |
| [3] |
Lasfar A, Lewis-Antes A, Smirnov SV, et al. Characterization of the mouse IFN-λ ligand-receptor system: IFN-λs exhibit antitumor activity against B16 melanoma. Cancer Res, 2006, 66(8): 4468-4477. DOI:10.1158/0008-5472.CAN-05-3653 |
| [4] |
Vilcek J. Novel interferons. Nat Immunol, 2003, 4(1): 8-9. DOI:10.1038/ni0103-8 |
| [5] |
Dumoutier L, Tounsi A, Michiels T, et al. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-λ1: similarities with type Ⅰ interferon signaling. J Biol Chem, 2004, 279(31): 32269-2274. DOI:10.1074/jbc.M404789200 |
| [6] |
Dumoutier L, Lejeune D, Hor S, et al. Cloning of a new type Ⅱ cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem J, 2003, 370(2): 391-396. DOI:10.1042/bj20021935 |
| [7] |
Miller DM, Klucher KM, Freeman JA, et al. Interferon lambda as a potential new therapeutic for hepatitis C. Ann NY Acad Sci, 2009, 1182: 80-87. DOI:10.1111/j.1749-6632.2009.05241.x |
| [8] |
Muir AJ, Shiffman ML, Zaman A, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology, 2010, 52(3): 822-832. DOI:10.1002/hep.23743 |
| [9] |
Grinna LS, Tschopp JF. Size distribution and general structural features of N-linked oligosaccharides from the methylotrophic yeast, Pichia pastoris. Yeast, 1989, 5(2): 107-115. DOI:10.1002/(ISSN)1097-0061 |
| [10] |
Trimble RB, Atkinson PH, Tschopp JF, et al. Structure of oligosaccharides on Saccharomyces SUC2 invertase secreted by the methylotrophic yeast Pichia pastoris. J Biol Chem, 1991, 266(34): 22807-22817. |
| [11] |
Kang HA, Sohn JH, Choi ES, et al. Glycosylation of human α1-antitrypsin in Saccharomyces cerevisiae and methylotrophic yeasts. Yeast, 1998, 14(4): 371-381. DOI:10.1002/(ISSN)1097-0061 |
| [12] |
Montesino R, García R, Quintero O, et al. Variation in N-linked oligosaccharide structures on heterologous proteins secreted by the methylotrophic yeast Pichia pastoris. Prot Expr Purif, 1998, 14(2): 197-207. DOI:10.1006/prep.1998.0933 |
| [13] |
Urbatsch IL, Wilke-Mounts S, Gimi K, et al. Purification and characterization of N-glycosylation mutant mouse and human P-glycoproteins expressed in Pichia pastoris cells. Arch Biochem Biophys, 2001, 388(1): 171-177. DOI:10.1006/abbi.2001.2299 |
| [14] |
Guo MJ, Hang HF, Zhu TC, et al. Effect of glycosylation on biochemical characterization of recombinant phytase expressed in Pichia pastoris. Enzyme Microb Technol, 2008, 42(4): 340-345. DOI:10.1016/j.enzmictec.2007.10.013 |
| [15] |
Walsh G. Biopharmaceutical benchmarks-2003. Nat Biotechnol, 2003, 21(8): 865-870. DOI:10.1038/nbt0803-865 |
| [16] |
Khandekar SS, Silverman C, Wells-Marani J, et al. Determination of carbohydrate structures N-linked to soluble CD154 and characterization of the interactions of CD40 with CD154 expressed in Pichia pastoris and Chinese hamster ovary cells. Prot Expr Purif, 2001, 23(2): 301-310. DOI:10.1006/prep.2001.1501 |
| [17] |
Xie YF, Chen H, Huang BR. Expression, purification and characterization of human IFN-λ1 in Pichia pastoris. J Biotechnol, 2007, 129(3): 472-480. DOI:10.1016/j.jbiotec.2007.01.018 |
| [18] |
Clare JJ, Romanes MA, Rayment FB, et al. Production of mouse epidermal growth factor in yeast: high-level secretion using Pichia pastoris strains containing multiple gene copies. Gene, 1991, 105(2): 205-212. DOI:10.1016/0378-1119(91)90152-2 |
| [19] |
Fairbanks G, Steck TL, Wallach DFH. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry, 1971, 10(13): 2606-2617. DOI:10.1021/bi00789a030 |
| [20] |
Andrade MA, Chacón P, Merelo JJ, et al. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Prot Eng, 1993, 6(4): 383-390. DOI:10.1093/protein/6.4.383 |
| [21] |
Meager A, Gaines DR, Zoon K, et al. Establishment of new and replacement World Health Organization International Biological Standards for human interferon alpha and omega. J Immunol Methods, 2001, 257(1/2): 17-33. |
| [22] |
Meager A. Biological assays for interferons. J Immunol Methods, 2002, 261(1/2): 21-36. |
| [23] |
Canosi U, Mascia M, Gazza L, et al. A highly precise reporter gene bioassay for type Ⅰ interferon. J Immunol Methods, 1996, 199(1): 69-76. DOI:10.1016/S0022-1759(96)00168-8 |
| [24] |
Giard DJ, Fleischaker RJ. A study showing a high degree of interlaboratory variation in the assay of human interferon. J Biol Stand, 1984, 13(3): 265. DOI:10.1016/S0092-1157(84)80005-0 |
| [25] |
Fray MD, Mann GE, Charleston B. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-Ⅰ interferon. J Immunol Methods, 2001, 249(1/2): 235-244. |
| [26] |
Lleonart R, Näf D, Browning H, et al. A novel, quantitative bioassay for type Ⅰ interferon using a recombinant indicator cell line. Nat Biotechnol, 1990, 8(12): 1263-1267. DOI:10.1038/nbt1290-1263 |
| [27] |
Meager A, Das RG. Biological standardization of human interferon beta: establishment of a replacement world health organization international biological standard for human glycosylated interferon beta. J Immunol Methods, 2005, 306(1/2): 1-15. |
 2018, Vol. 34
2018, Vol. 34