中国科学院微生物研究所、中国微生物学会主办
文章信息
- 苟艳, 刘忠川, 王刚刚
- Gou Yan, Liu Zhongchuan, Wang Ganggang
- 异戊二烯合成酶研究进展
- Advances in isoprene synthase research
- 生物工程学报, 2017, 33(11): 1802-1813
- Chinese Journal of Biotechnology, 2017, 33(11): 1802-1813
- 10.13345/j.cjb.170040
-
文章历史
- Received: February 13, 2017
- Accepted: April 19, 2017
2 中国科学院成都生物研究所 环境微生物四川省重点实验室,四川 成都 610041;
3 中国科学院大学,北京 100049
2 Key Laboratory of Environmental Microbiology of Sichuan Province, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu 610041, Sichuan, China;
3 University of Chinese Academy of Sciences, Beijing 100049, China
异戊二烯(2-甲基-1, 3-丁二烯,isoprene)是一种挥发性的五碳萜类化合物。1957年,Sanadze等第一次发现了木本植物排放异戊二烯,此后,人们发现其他生物也具有这种能力,包括动物、真菌、细菌等[1-3]。异戊二烯的排放是大气中碳氢化合物最主要的生物来源,据估计,全球异戊二烯在大气中的年均自然排放量约几百兆克,与大气中各种来源的甲烷排放量相当[4]。生物排放异戊二烯的过程对其自身具有一定的保护作用[5-7],但同时会对大气带来不良影响[8-9],另外,异戊二烯还是一种具有重要用途的化工原料,被广泛应用于化工、医药、能源等领域[5]。
异戊二烯是最简单的类异戊二烯家族成员, 所以人们推断它可能来源于类异戊二烯的生物合成途径中共有的五碳中间代谢物。Sanadze等认为异戊二烯的形成是一个非酶过程,它是由酸催化烯丙基二磷酸(Dimethylallyl diphosphate,DMAPP),离子化DMAPP中的焦磷酸基团,产生一个过渡态的碳正离子而形成异戊二烯[10]。直到1991年,Sliver等在山杨Populus tremuloides michx.的树叶中发现了异戊二烯合成酶,该酶能够在中性pH条件下发挥作用[11],后来,在其他植物中也分离纯化到了Isps,在一些细菌的粗酶液中也检测到该酶的活性,并证明了在这些生物中,异戊二烯是由Isps催化形成的[12]。
异戊二烯合成酶能够催化类异戊二烯中间体——烯丙基二磷酸(Dimethylallyl diphosphate,DMAPP),生成异戊二烯(Isoprene)和焦磷酸(Pyrophosphate),该反应依赖二价阳离子[13]。异戊二烯合成酶是自然条件下异戊二烯排放过程的关键酶,其基因表达量和酶活性能够调控异戊二烯的排放量,因此,Isps在异戊二烯的排放过程中具有重要的作用。本文就Isps最新研究进展进行综述,比较分析不同生物来源的Isps的生化特征、结构特征,探讨其催化机制,并对该酶在生物工程领域的应用进行展望。
1 异戊二烯合成酶的生化特征到目前为止,人们已经对多种不同来源的Isps进行了研究,并对它们的生化特征进行分析,包括分子量大小、最适pH、最适温度、催化反应需要的二价阳离子等。从表 1可以看出,它们具有一些相似的生化特性,但也有一定的差别:第一,Isps的活性对温度有很强的依赖性, 最适温度一般为35-40 ℃,在55-60 ℃的范围快速失活,目前,只有英国栎Pedunculate oak Isps的最适温度达到了50 ℃[14];第二,大多数植物来源的Isps最适pH在7.5-8.0之间,例如橡树、杨树等[11, 13-15],而苔藓植物Isps的最适pH在8.6左右[16],褪色柳salix discolor Isps的最适pH约为10[17-18],另外,与植物Isps相比,细菌Isps的最适pH较低,如枯草芽孢杆菌Isps粗酶液的最适pH为6.2[19];第三,大多数异戊二烯合成酶催化活性主要依赖于Mg2+和Mn2+[13, 15], 但对于苔藓植物,如曲柄藓属植物Campylopus introflexus,它的Isps催化活性相较于Mg2+更加优先利用Mn2+[16];第四,Isps的米氏常数通常达到mmol/L水平,与单萜合酶、倍半萜合酶的米氏常数相比要高2-3个数量级[20]。
| Source of enzyme | Mr (kDa) | pHopt | Topt (℃) | Metal ion requirement | Km for DMAPP (mmol/L) |
| Populus tremuloides | 120 | 8.0 | 45 | Mg2+﹥Mn2+ | 8[11, 13] |
| Mucuna sp. | N/A | 7.8-8.5 | N/A | Mg2+﹥Mn2+﹥﹥Ca2+ | 1-9[15] |
| Salix discolor | 73 | 10.0 | N/A | Mg2+, Mn2+ | 8[17-18] |
| Quercus robur | 95 | 7.3-7.7 | 50 | N/A | 0.53[14] |
| Populus alba | 62 | 8 | 40 | Mg2+﹥Mn2+﹥Co2+ | 8.7[21] |
| Populus×canescens | 51 | 7.5-8.5 | 40 | Mg2+ | 2.5±0.1[22] |
| Pueraria montana | N/A | N/A | N/A | N/A | 7.7[23] |
| Heath star moss | N/A | 8.1-9.1 | 37-43 | Mn2+﹥Mg2+ | 0.37±0.28[16] |
| Bacillus subtilis | N/A | 6.2 | 35-40 | Mn2+﹥Mg2+﹥﹥Ca2+ | N/A[19] |
| Eucalyptus globulus | 66 | N/A | 40 | N/A | 0.16[24-25] |
| Arachis hypogaea | N/A | 7.6 | 34 | Mg2+ | 18.3±6.538[26] |
| N/A: data not available. | |||||
另外,已知的所有植物异戊二烯合成酶均位于叶绿体中,但具有两种不同的存在形式,一种结合在内囊体膜上,另一种处于溶解状态。Silver和Kuzma分别在山杨和绒毛豆树叶的叶绿体中提取出了溶解状态的异戊二烯合成酶[13, 15],后来,Wildermuth和Fall又在褪色柳Salix discolor等植物中发现了紧密结合在内囊体膜上的异戊二烯合成酶,他们使用了多种溶解方法均不能使该酶从膜上脱落下来而保持活性[17-18]。这两种不同形式的异戊二烯合成酶的性质基本类似,只是结合于膜上的酶具有较高的最适pH (pH 10) [15]。
2 异戊二烯合成酶的结构特征在GenBank数据库中查找不同来源的Isps及其他萜类合成酶的序列,利用clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/)进行序列比对,比对结果再用Aline软件编辑[27] (图 1)。序列比对结果表明,不同来源的Isps之间,以及Isps与其他的萜类合成酶之间均具有较高的序列相似性。例如,来源于不同种类杨树的异戊二烯合成酶的氨基酸序列具有97%-100%的相似性,而来源于杂交灰杨Populus×canescens树叶的异戊二烯合成酶(PcIsps)与植物单萜合成酶的氨基酸序列具有约50%的相似性。

|
| 图 1 异戊二烯合成酶以及其他的萜类合成酶之间的氨基酸序列比对 Figure 1 Multiple sequence alignment of isoprene synthase and other terpene synthases. PcIsps: isoprene synthase(Populus×canadensis)[28]; PaIsps: isoprene synthase 3(Populus alba)[29]; FsIsps: isoprene synthase (Ficus septic)[6]; MnIsps: Isoprene synthase (Morus notabilis)[30]; FvIsps: isoprene synthase (Ficus virgata)[6]; TwTps22: terpene synthase 22 (Tripterygium wilfordii)[31]; PlBos: beta-ocimene synthase (Phaseolus lunatus)[32]; EgMts: monoterpene synthase (Eucalyptus globulus)[33]; MtEBos: (E)-beta-ocimene/myrcene synthase (Medicago truncatula)[34]; OeTps3: terpene synthase 3 (Olea europaea)[35]. |
| |
萜类合成酶(Terpene synthases,Tps)是指能够将含异戊二烯基焦磷酸的非环状结构前体物质转化成萜类分子的一类酶[36]。根据反应机理和产物形式可以将该酶分成两种类型,分别为Tps-Ⅰ和Tps-Ⅱ[37-38]。已知的大多数萜类合成酶都是Tps-Ⅰ类型的,也有少数萜类合成酶同时具有Tps-Ⅰ和Tps-Ⅱ两种类型的功能域。另外,根据Tps的生化功能、亚细胞定位、底物专一性以及产物的信息和定位,可以将Tps的基因分为7个进化分支,分别为tps-a、b、c、d、e/f、g、h[39],这7个进化分支中均具有Ⅰ型酶,而Ⅱ型酶仅限于Tps-c中[38]。其中Isps属于Ⅰ型萜类合成酶,基因属于Tps-b进化分支[25]。
目前仅有PcIsps和PtIsps的晶体结构已被解析,它们分别来源于杂交灰杨Populus×canescens和美洲山杨Populus tremuloides的树叶,且这两种Isps的氨基酸序列具有98.5%的一致性[5, 40]。通过比较这两种酶的结构发现,它们具有同型结构,都是由两个α螺旋结构域组成。它们的N末端为α-桶状的Ⅱ型萜类合酶折叠模式,没有催化活性;而包含活性位点的C末端α-螺旋结构域具有Ⅰ型萜类合酶折叠模式,并且两个单体能够通过C端催化结构域的相互作用形成二聚体形式。分析PcIsps结构发现,在该酶C端的催化结构域中具有两个保守的金属离子结合基序,一个是富含天冬氨酸的DDXXD序列,另一个是“NSE/DTE”序列(NDXXSXXXE),这两个基序在植物、细菌、真菌来源的萜类环化酶中保守,异戊二烯合成酶是发现的第一个具有这两种基序的非环化萜类合成酶[5],而一般的非环化萜类合成酶具有两个富含天冬氨酸的序列,如催化类异戊二烯的链延伸反应的法尼基焦磷酸合酶[41]。
另外,在异戊二烯合成酶C末端的活性位点一般具有两个保守的苯丙氨酸,它们分别位于活性位点的两端,通过与底物产生范德华力而使活性口袋减小,从而阻止比DMAPP更大的底物与之结合。而这些氨基酸残基并不存在于Tps-b家族的单萜合酶或者其他的萜类合成酶中,因为它们的底物更大,是十碳或者具有更多碳原子的含异戊二烯基的焦磷酸。因此,推测这些氨基酸残基的作用与底物特异性有关[5, 23, 41]。
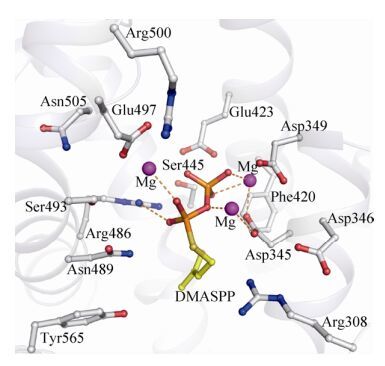
3 异戊二烯合成酶的催化机制早在1995年,Silver等就已提出异戊二烯的生物合成是一个由金属离子(如Mg2+、Mn2+等)触发的DMAPP去焦磷酸过程[13]。通过分析PcIsps与底物类似物DMASPP (DMAPP中磷酸酯键的氧原子被替代为硫原子)以及3个Mg2+形成的复合体(PcIsps-(Mg2+)3-DMASPP)的晶体结构,发现DMASPP与PcIsps活性位点上的氨基酸形成氢键,其焦磷酸基团上的氧原子能与Mg2+形成配位键;Mg2+通过与金属离子结合基序中的氨基酸形成配位键而结合在活性位点上[5, 42-43](图 2)。一般在Ⅰ型萜类合成酶中,当底物与3个金属离子结合到活性位点形成复合体后,该酶的活性口袋将由开放状态转变为闭合状态,推测这一构象变化是为了防止催化过程中形成的碳正离子中间体过早淬灭,从而避免对萜类化合物的形成造成影响[5, 44-46]。
|
| 图 2 杂交银灰杨异戊二烯合成酶(PcIsps)活性中心结合底物类似物(DMASPP)和Mg2+的结构 Figure 2 The structure of active center in PcIsps-DMASPP-Mg2+complex. Atoms are color coded as follows: carbon: gray; nitrogen: blue; oxygen: red. Mg2+ appears as magenta sphere. |
| |
通过对结构的进一步分析表明,异戊二烯的生物合成是通过烯丙基二磷酸中间体经过两步反应得到的(图 3)。在Isps的催化作用下, 底物DMAPP形成烯丙基的碳正离子中间体,通过顺叠消除机制脱去无机焦磷酸基团(PPi),然后烯丙基的甲基基团C31再发生去质子化,最终形成产物,这一过程与许多单萜合成酶的起始步骤是一致的。在该催化反应过程中,底物甲基基团C31的去质子化还需要合适的广义碱帮助,而有趣的是,在这里提供广义碱的是离开底物DMAPP的焦磷酸基团。另外,对PcIsps-Mg2+-DMASPP复合体结构的研究发现,DMASPP以七元环椅式结构结合到PcIsps上,这个构象使DMASPP的α-磷酸基团的氧原子更加接近C31原子,从而表明底物DMASPP对顺叠消除机制的帮助[5, 47]。另外,Isps在催化过程中具有正协同作用。Isps单体能够通过C端催化域的相互作用而形成同型二聚体,在PcIsps研究中发现,当底物与该酶结合后,二聚体界面发生了一定的构象变化,在两个单体之间新形成了2个氢键和4个盐桥。这些结构特点也许正是Isps具有正协同作用的结构基础[5, 48-50]。

|
| 图 3 Isps催化机制(Isps催化生成异戊二烯是通过烯丙基二磷酸中间体经过两步反应得到,首先是DMAPP通过顺叠消除机制而脱去焦磷酸基团,然后是C31去质子化形成异戊二烯) Figure 3 Catalytic mechanism for Isps. The generation of isoprene by Isps occurs in two steps via an allylic carbo cation intermediate. First, iPP is removed from DMAPP by syn-periplanar elimination reaction. Then, the C31is deprotonated in the assistance of the removed diphosphate group serving as the general base. Eventually, the isoprene is formed. |
| |
生物向大气排放大量的异戊二烯,据估计每年植物的排放量大约为600兆克/年,相当于生物来源的挥发性有机化合物(Biogenic volatile organic compound,BVOC)排放总量的1/3[51]。异戊二烯的排放对这些生物具有一定的作用,但同时也对大气环境产生较大的影响,从异戊二烯合成酶发现以来,科学家们在这方面进行了大量的研究工作。
其一,研究发现,并不是所有的生物都具有异戊二烯的排放能力,对于研究的主要对象——陆生植物来说,也并不是都具有此能力[52],据统计,超过1/3的被子植物和裸子植物,约一半的蕨类植物以及94%的苔藓植物能够排放异戊二烯,但它们的排放能力具有差异[53],这说明异戊二烯的排放对生物来说并不是必需的,或者说具有可替代的途径。而关于异戊二烯排放对生物的作用,目前还不是很清楚。对于研究最多的植物而言,也仅仅部分了解,例如,有研究发现异戊二烯可以与受紫外激发形成的单线态氧在体内反应产生异丁烯醛和甲基乙烯基酮,这两种产物的热稳定差,可以从植物体内排出,从而防止单线态氧进一步形成其他的活性氧而损伤植物[54]。另外,也有文献表明排放异戊二烯可以保护这些植物不受高温、辐射、臭氧等环境压力的破坏;可以稳定脂膜以避免热诱导引起的相变;可以直接或间接地防卫草食动物危害植物等[5-6, 55]。通过对异戊二烯合成酶基因进行系统进化分析等研究,了解异戊二烯排放能力的演变,从而有望了解生物排放异戊二烯的具体作用。
其二,每年大量异戊二烯的排放对大气化学产生巨大的影响,它通过与羟自由基、氮氧化物等反应而提高大气中的臭氧、有机酸、有机硝酸盐浓度,延长了温室气体甲烷在大气中的寿命,形成二次有机气溶胶等[8-9]。通过对异戊二烯合成酶的研究发现,Isps催化形成异戊二烯的过程受到复杂的调控,涉及酶的热动力学、代谢流及其反馈作用、细胞器内外的代谢物交换等[56-57],Isps的活性受到光照、温度、细胞内二氧化碳浓度等因素的影响[52, 58-60]。只有对这些调控因素进行深入研究,研究人员才能更准确地估计异戊二烯的排放率,以及伴随着异戊二烯排放而给大气环境造成的影响[61]。
5 异戊二烯合成酶与人工合成异戊二烯异戊二烯是一种重要的化工原料,被广泛应用于多个领域,包括形成以异戊二烯为基础的燃料,作为石油派生聚合物的合成前体,应用于合成橡胶、农药、医药、香料及黏结剂生产等[62-65]。目前,异戊二烯主要是通过石油基原料异戊烷、异戊烯脱氢法、化学合成法(包括异丁烯-甲醛法、乙炔-丙酮法、丙烯二聚法)和裂解C5馏分萃取蒸馏法生产。然而,随着化石资源的日益枯竭,开发新的异戊二烯合成技术已成为当前发展的必然趋势[65],而利用微生物发酵生产异戊二烯具有经济、清洁、可持续等得天独厚的优势[66-67],因而受到了广泛关注和深入研究。
微生物发酵方法生产异戊二烯可以分为两个部分,首先是通过2-甲基-D-赤藓糖醇-4-磷酸途径(Methylerythritol-phosphate pathway,MEP)或者甲羟戊酸途径(Mevalonate pathway,MVA)合成前体物质DMAPP,DMAPP经Isps催化形成异戊二烯。如果合成的DMAPP不能及时转化而过量积累,会对宿主细胞造成毒性。因此,如何提高Isps的催化效率,加快DMAPP转化成异戊二烯的速率成了提高微生物发酵生产异戊二烯产量的关键。
因此,通过蛋白质工程改造异戊二烯合成酶是目前研究的重点之一,即对Isps进行修饰改造提高其活性,从而实现在微生物中高效合成异戊二烯[16, 68]。目前对杨树(Poplar)和葛藤(Kudzu vine)两个植物家族来源的Isps研究较为深入,且已经解析了PcIsps和PtIsps的晶体结构。在这些工作的基础上,通过蛋白质工程改造异戊二烯合成酶方面的研究工作已经取得了一些成果。如对来源于银白杨Populus alba的Isps进行改造,截掉氨基酸序列N末端的L90R突变体的活性明显提升[69];柳树Salix alba、Salix babylonica来源的Isps的S288C突变体也具有比野生型更高的稳定性[40]。另外,有研究结果显示,将银白杨Populus alba来源的Isps在酿酒酵母Saccharomyces cerevisiae中表达生产异戊二烯,经过一系列代谢调控优化后,在摇瓶条件下培养24 h异戊二烯产量能达到23.6 mg/L。而在此基础上,根据Isps的催化机制和结构信息对其进行定向进化改造,发现A570T/ F340L突变株生产异戊二烯的产量可以达到50.2 mg/L[70-71]。
除了改造已知的Isps,从自然环境或者基因库中发现新的isps也是目前的研究热点之一。例如,发现豆类中的isps可用于发酵生产异戊二烯;在大肠杆菌Escherichia coli中异源表达花生Arachis hypogaea isps能使异戊二烯的产量提高到35 mg/(L·h·OD)[72];通过同源性克隆和在基因库搜索符合杨树Isps活性位点特征的萜类合酶等方法发现了9种新的被子植物isps,并且这些isps在大肠杆菌中表达后具有活性,其中蓝桉树Eucalyptus globules Isps的Km比其他已知的Isps都低[25];新的研究发现,甘薯Ipomoea batatas、芒果Mangifera indica、杜英Elaeocarpus photiniifolius的Isps都能在大肠杆菌中表达并具有催化活性,且甘薯来源的Isps产异戊二烯的能力优于已知的杨树、葛藤属的Isps [73]。另外,除了植物来源的Isps外,人们也致力于发掘微生物来源的Isps,例如枯草芽孢杆菌B. subtilis的粗酶液具有Isps活性,它的最适pH为6.2,并且所需要的二价阳离子水平很低(100 mmol/L)[19],这样的催化条件与宿主微生物的培养条件更接近。如果能够鉴定枯草芽孢杆菌的Isps编码基因,将其表达用于发酵生产异戊二烯可能会获得更高的产量。
另外,选择合适的宿主构建微生物细胞工厂对于人工生产异戊二烯具有重要意义。构建产异戊二烯的工程菌株首先就要使外源的isps能够在该微生物中表达,植物来源的isps在导入微生物中需要经过一些修饰,包括切除定位到叶绿体的信号肽、优化密码子等[20]。目前常用的表达宿主有枯草芽孢杆菌B. subtilis、大肠杆菌E. coli、蓝细菌Cyanobacterium synechocystis以及酿酒酵母S. cerevisiae[20]。在这些宿主细胞中已经存在异戊二烯前体化合物的合成途径,通过优化前体合成途径和异源表达Isps便可实现异戊二烯合成。
6 结语从发现异戊二烯合成酶到现在已经有30多年的历史,通过研究人员的广泛研究,使我们对该酶的基本生化特征和结构功能都有了一定的认识。这些研究成果不仅对人们认识大气中的异戊二烯排放有重要意义,而且对人工合成异戊二烯具有重要的指导作用。由于不同来源的异戊二烯合成酶在结构、生化特征等方面具有多样性,因此,今后仍需要对异戊二烯合成酶进行深入研究,主要包括:1)异戊二烯合成酶的结构功能关系,基于结构和功能关系对酶进一步改造,以提高Isps的催化效率;2)发掘新的异戊二烯合成酶,扩大异戊二烯合成酶的基因库,为微生物发酵产异戊二烯提供更多的选择。只有系统深入地研究异戊二烯合成酶的生化性质和结构功能关系,才能更好地改造和利用该酶。
| [1] | Karl T, Prazeller P, Mayr D, et al. Human breath isoprene and its relation to blood cholesterol levels: new measurements and modeling. J Appl Physiol, 2001, 91(2): 762–770. |
| [2] | Kuzma J, Nemecek-Marshall M, Pollock WH, et al. Bacteria produce the volatile hydrocarbon isoprene. Curr Microbiol, 1995, 30(2): 97–103. DOI: 10.1007/BF00294190 |
| [3] | Sharkey TD, Yeh SS. Isoprene emission from plants. Plant Biol, 2001, 52: 407–436. |
| [4] | Guenther A, Hewitt CN, Erickson D, et al. A global model of natural volatile organic compound emissions. J Geophys Res, 1995, 100(D5): 8873–8892. DOI: 10.1029/94JD02950 |
| [5] | Köksal M, Zimmer I, Schnitzler JP, et al. Structure of isoprene synthase illuminates the chemical mechanism of teragram atmospheric carbon emission. J Mol Biol, 2010, 402(2): 363–373. DOI: 10.1016/j.jmb.2010.07.009 |
| [6] | Oku H, Inafuku M, Ishikawa T, et al. Molecular cloning and biochemical characterization of isoprene synthases from the tropical trees Ficus virgata, Ficus septica, and Casuarina equisetifolia. J Plant Res, 2015, 128(5): 849–861. DOI: 10.1007/s10265-015-0740-9 |
| [7] | Sharkey TD, Wiberley AE, Donohue AR. Isoprene emission from plants: why and how. Ann Bot, 2008, 101: 5–18. |
| [8] | Fehsenfeld F, Calvert J, Fall R, et al. Emissions of volatile organic compounds from vegetation and the implications for atmospheric chemistry. Glob Biogeochem Cycl, 1992, 6(4): 389–430. DOI: 10.1029/92GB02125 |
| [9] | Kiendler-Scharr A, Wildt J, dal Maso M, et al. New particle formation in forests inhibited by isoprene emissions. Nature, 2009, 461(7262): 381–384. DOI: 10.1038/nature08292 |
| [10] | Deneris ES, Stein RA, Mead JF. Acid-catalyzed formation of isoprene from a mevalonate-derived product using a rat liver cytosolic fraction. J Biol Chem, 1985, 260(3): 1382–1385. |
| [11] | Silver GM, Fall R. Enzymatic synthesis of isoprene from dimethylallyl diphosphate in aspen leaf extracts. Plant Physiol, 1991, 97(4): 1588–1591. DOI: 10.1104/pp.97.4.1588 |
| [12] | Sharkey TD. Isoprene synthesis by plants and animals. Endeavour, 1996, 20(2): 74–78. DOI: 10.1016/0160-9327(96)10014-4 |
| [13] | Silver GM, Fall R. Characterization of aspen isoprene synthase, an enzyme responsible for leaf isoprene emission to the atmosphere. J Biol Chem, 1995, 270(22): 13010–13016. DOI: 10.1074/jbc.270.22.13010 |
| [14] | Lehning A, Zimmer I, Steinbrecher R, et al. Isoprene synthase activity and its relation to isoprene emission in Quercus robur L. leaves. Plant Cell Environ, 1999, 22(5): 495–504. DOI: 10.1046/j.1365-3040.1999.00425.x |
| [15] | Kuzma J, Fall R. Leaf isoprene emission rate is dependent on leaf development and the level of isoprene synthase. Plant Physiol, 1993, 101(2): 435–440. DOI: 10.1104/pp.101.2.435 |
| [16] | Lantz AT, Cardiello JF, Gee TA, et al. Biochemical characterization of an isoprene synthase from Campylopus introflexus (heath star moss). Plant Physiol Biochem, 2015, 94: 209–215. DOI: 10.1016/j.plaphy.2015.06.008 |
| [17] | Wildermuth MC, Fall R. Light-dependent isoprene emission (characterization of a thylakoid-bound isoprene synthase in Salix discolor chloroplasts). Plant Physiol, 1996, 112(1): 171–182. DOI: 10.1104/pp.112.1.171 |
| [18] | Wildermuth MC, Fall R. Biochemical characterization of stromal and thylakoid-bound isoforms of isoprene synthase in willow leaves. Plant Physiol, 1998, 116(3): 1111–1123. DOI: 10.1104/pp.116.3.1111 |
| [19] | Sivy TL, Shirk MC, Fall R. Isoprene synthase activity parallels fluctuations of isoprene release during growth of Bacillus subtilis. Biochem Biophys Res Commun, 2002, 294(1): 71–75. DOI: 10.1016/S0006-291X(02)00435-7 |
| [20] | Ye LD, Lv XM, Yu HW. Engineering microbes for isoprene production. Metab Eng, 2016, 38: 125–138. DOI: 10.1016/j.ymben.2016.07.005 |
| [21] | Sasaki K, Ohara K, Yazaki K. Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett, 2005, 579(11): 2514–2518. DOI: 10.1016/j.febslet.2005.03.066 |
| [22] | Schnitzler JP, Zimmer I, Bachl A, et al. Biochemical properties of isoprene synthase in poplar (Populus×canescens). Planta, 2005, 222(5): 777–786. DOI: 10.1007/s00425-005-0022-1 |
| [23] | Sharkey TD, Yeh S, Wiberley AE, et al. Evolution of the isoprene biosynthetic pathway in kudzu. Plant Physiol, 2005, 137(2): 700–712. DOI: 10.1104/pp.104.054445 |
| [24] | Loreto F, Delfine S. Emission of isoprene from salt-stressed Eucalyptus globulus leaves. Plant Physiol, 2000, 123(4): 1605–1610. DOI: 10.1104/pp.123.4.1605 |
| [25] | Sharkey TD, Gray DW, Pell HK, et al. Isoprene synthase genes form a monophyletic clade of acyclic terpene synthases in the TPS-B terpene synthase family. Evolution, 2013, 67(4): 1026–1040. DOI: 10.1111/evo.2013.67.issue-4 |
| [26] | Beatty MK, Hayes K, Hou ZL, et al. Legume isoprene synthase for production of isoprene: US, 8895277B2. 2014-11-25. |
| [27] | Bond CS, Schüttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr, 2009, 65(Pt 5): 510–512. |
| [28] | Hu BQ, Wang B, Wang CG, et al. Microarray analysis of gene expression in triploid black poplar. Silv Genet, 2012, 61(4/5): 148–157. |
| [29] | Vickers CE, Possell M, Hewitt CN, et al. Genetic structure and regulation of isoprene synthase in Poplar (Populus spp.). Plant Mol Biol, 2010, 73(4/5): 547–558. |
| [30] | He NJ, Zhang C, Qi XW, et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun, 2013, 4: 2445. |
| [31] | Arimura GI, Köpke S, Kunert M, et al. Effects of feeding Spodoptera littoralis on Lima bean leaves: Ⅳ. Diurnal and nocturnal damage differentially initiate plant volatile emission. Plant Physiol, 2008, 146(3): 965–973. DOI: 10.1104/pp.107.111088 |
| [32] | Hansen NL, Heskes AM, Hamberger B, et al. The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J, 2017, 89(3): 429–441. DOI: 10.1111/tpj.13410 |
| [33] | Yano S, 矢野伸一. Eucalyptus globulus monoterpene synthase gene: JP, WO/2007/018062A1. 2007-02-15. |
| [34] | Tang HB, Krishnakumar V, Bidwell S, et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics, 2014, 15: 312. DOI: 10.1186/1471-2164-15-312 |
| [35] | Vezzaro A, Krause ST, Nonis A, et al. Isolation and characterization of terpene synthases potentially involved in flavor development of ripening olive (Olea europaea) fruits. J Plant Physiol, 2012, 169(9): 908–914. DOI: 10.1016/j.jplph.2012.01.021 |
| [36] | Jia Q, Li G, Köllner TG, et al. Microbial-type terpene synthase genes occur widely in nonseed land plants, but not in seed plants. Proc Natl Acad Sci USA, 2016, 113(43): 12328–12333. DOI: 10.1073/pnas.1607973113 |
| [37] | Degenhardt J, Köllner TG, Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry, 2009, 70(15/16): 1621–1637. |
| [38] |
Tang L, Ma X, Zhou ZQ. An evolutionary study of plant terpene synthases.
J Southwest Univ: Nat Sci Ed, 2014, 36(4): 89–96.
(in Chinese). 唐亮, 马香, 周志钦. 植物萜类合成酶的进化研究. 西南大学学报:自然科学版, 2014, 36(4): 89-96. |
| [39] | Chen F, Tholl D, Bohlmann J, et al. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J, 2011, 66(1): 212–229. DOI: 10.1111/tpj.2011.66.issue-1 |
| [40] | Rife CL, Wells DH. Isoprene synthase variants with improved solubility for production of isoprene: US, 9273299B2. 2016-03-01. |
| [41] | Tarshis LC, Yan M, Poulter CD, et al. Crystalstructure of recombinant farnesyl diphosphate synthase at 2.6-A resolution. Biochemistry, 1994, 33(36): 10871–10877. DOI: 10.1021/bi00202a004 |
| [42] | Hyatt DC, Youn B, Zhao Y, et al. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci USA, 2007, 104(13): 5360–5365. DOI: 10.1073/pnas.0700915104 |
| [43] | Whittington DA, Wise ML, Urbansky M, et al. Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc Natl Acad Sci USA, 2002, 99(24): 15375–15380. DOI: 10.1073/pnas.232591099 |
| [44] | Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev, 2006, 106(8): 3412–3442. DOI: 10.1021/cr050286w |
| [45] | Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol, 2008, 12(2): 141–150. DOI: 10.1016/j.cbpa.2007.12.008 |
| [46] | Lesburg CA, Caruthers JM, Paschall CM, et al. Managing and manipulating carbocations in biology: terpenoid cyclase structure and mechanism. Curr Opin Struct Biol, 1998, 8(6): 695–703. DOI: 10.1016/S0959-440X(98)80088-2 |
| [47] | Faraldos JA, Gonzalez V, Li AM, et al. Probing the mechanism of 1, 4-conjugate elimination reactions catalyzed by terpene synthases. J Am Chem Soc, 2011, 134(51): 20844–20848. |
| [48] | Alonso WR, Rajaonarivony JI, Gershenzon J, et al. Purification of 4S-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha×piperita) and spearmint (Mentha spicata). J Biol Chem, 1992, 267(11): 7582–7587. |
| [49] | inslie Jr AG, Shill JP, Neet KE. Transients and cooperativity. A slow transition model for relating transients and cooperative kinetics of enzymes. J Biol Chem, 1972, 247(21): 7088–7096. |
| [50] | Wise ML, Savage TJ, Katahira E, et al. Monoterpene Synthases from Common Sage (Salvia officinalis): cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1, 8-cineole synthase, and (+)-bornyl diphosphate synthase. J Biol Chem, 1998, 273(24): 14891–14899. DOI: 10.1074/jbc.273.24.14891 |
| [51] | Guenther A, Karl T, Harley P, et al. Estimates of global terrestrial isoprene emissions using MEGAN (Model of Emissions of Gases and Aerosols from Nature). Atmos Chem Phys, 2006, 6(11): 3181–3210. DOI: 10.5194/acp-6-3181-2006 |
| [52] | Harley PC, Monson RK, Lerdau MT. Ecological and evolutionary aspects of isoprene emission from plants. Oecologia, 1999, 118(2): 109–123. DOI: 10.1007/s004420050709 |
| [53] | Hanson DT, Swanson S, Graham LE, et al. Evolutionary significance of isoprene emission from mosses. Am J Bot, 1999, 86(5): 634–639. DOI: 10.2307/2656571 |
| [54] | Zeinali N, Altarawneh M, Li D, et al. New mechanistic insights: why do plants produce isoprene?. ACS Omega, 2016, 1(2): 220–225. DOI: 10.1021/acsomega.6b00025 |
| [55] | Schnitzler JP, Louis S, Behnke K, et al. Poplar volatiles-biosynthesis, regulation and (eco)physiology of isoprene and stress-induced isoprenoids. Plant Biol, 2010, 12(2): 302–316. |
| [56] | Li ZR, Sharkey TD. Molecular and pathway controls on biogenic volatile organic compound emissions//Niinemets Ü, Monson RK. Biology, Controls and Models of Tree Volatile Organic Compound Emissions. Netherlands: Springer, 2013, 5: 119–151. |
| [57] | Monson RK, Grote R, Niinemets U, et al. Modeling the isoprene emission rate from leaves. New Phytol, 2012, 195(3): 541–559. DOI: 10.1111/j.1469-8137.2012.04204.x |
| [58] | Loreto F, Sharkey TD. A gas-exchange study of photosynthesis and isoprene emission in Quercus rubra L. Planta, 1990, 182(4): 523–531. DOI: 10.1007/BF02341027 |
| [59] | Monson RK, Fall R. Isoprene emission from aspen leaves: influence of environment and relation to photosynthesis and photorespiration. Plant Physiol, 1989, 90(1): 267–274. DOI: 10.1104/pp.90.1.267 |
| [60] | Monson RK, Trahan N, Rosenstiel TN, et al. Isoprene emission from terrestrial ecosystems in response to global change: minding the gap between models and observations. Philos Trans A Math Phys Eng Sci, 2007, 365(1856): 1677–1695. DOI: 10.1098/rsta.2007.2038 |
| [61] | Monson RK, Neice AA, Trahan NA, et al. Interactions between temperature and intercellular CO2 concentration in controlling leaf isoprene emission rates. Plant Cell Environ, 2016, 39(11): 2404–2413. DOI: 10.1111/pce.12787 |
| [62] | Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J Cell Mol Biol, 2008, 54(4): 656–669. DOI: 10.1111/j.1365-313X.2008.03449.x |
| [63] | Erickson B, Nelson JE, Winters P. Perspective on opportunities in industrial biotechnology in renewable chemicals. Biotechnol J, 2012, 7(2): 176–185. DOI: 10.1002/biot.201100069 |
| [64] | Rude MA, Schirmer A. New microbial fuels: a biotech perspective. Curr Opin Microbiol, 2009, 12(3): 274–281. DOI: 10.1016/j.mib.2009.04.004 |
| [65] |
Su SZ, Liu JZ, Yang JM, et al. Expression of isoprene synthase in Escherichia coli for isoprene production.
Chin J Bioprocess Eng, 2011, 9(3): 6–10.
(in Chinese). 苏思正, 刘建忠, 杨建明, 等. 异戊二烯合成酶(IspS)在大肠杆菌中的表达及其产异戊二烯的研究. 生物加工过程, 2011, 9(3): 6-10. |
| [66] | Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng, 2008, 10(6): 295–304. DOI: 10.1016/j.ymben.2008.06.009 |
| [67] | Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science, 2007, 315(5813): 801–804. DOI: 10.1126/science.1139612 |
| [68] | Yang JM, Xian M, Su SZ, et al. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PLoS ONE, 2012, 7(4): e33509. DOI: 10.1371/journal.pone.0033509 |
| [69] | Beck ZQ, Bott RR, Rife CL, et al. Three-dimensional structure of isoprene synthase and its use thereof for generating variants: US, 8518686. 2013-08-27. |
| [70] | Lü XM, Xie WP, Lu WQ, et al. Enhanced isoprene biosynthesis in Saccharomyces cerevisiae by engineering of the native acetyl-CoA and mevalonic acid pathways with a push-pull-restrain strategy. J Biotechnol, 2014, 186: 128–136. DOI: 10.1016/j.jbiotec.2014.06.024 |
| [71] | Wang F, Lv XM, Xie WP, et al. Combining Gal4p-mediated expression enhancement and directed evolution of isoprene synthase to improve isoprene production in Saccharomyces cerevisiae. Metab Eng, 2017, 39: 257–266. DOI: 10.1016/j.ymben.2016.12.011 |
| [72] | Bentley FK, Zurbriggen A, Melis A. Heterologous expression of the mevalonic acid pathway in cyanobacteria enhances endogenous carbon partitioning to isoprene. Mol Plant, 2014, 7(1): 71–86. DOI: 10.1093/mp/sst134 |
| [73] | Ilmén M, Oja M, Huuskonen A, et al. Identification of novel isoprene synthases through genome mining and expression in Escherichia coli. Metab Eng, 2015, 31: 153–162. DOI: 10.1016/j.ymben.2015.08.001 |
 2017, Vol. 33
2017, Vol. 33