中国科学院微生物研究所、中国微生物学会主办
文章信息
- 张宁, 阮亚男, 王姗姗, 刘洋, 赵宸, 王晶晶, 王凯玺, 王艳丽, 王红艳
- Zhang Ning, Ruan Yanan, Wang Shanshan, Liu Yang, Zhao Chen, Wang Jingjing, Wang Kaixi, Wang Yanli, Wang Hongyan
- MITE类转座子mPing在水稻不同亚种间的差异分析
- Comparison of MITE transposons mPing in different rice subspecies
- 生物工程学报, 2016, 32(9): 1264-1272
- Chin J Biotech, 2016, 32(9): 1264-1272
- 10.13345/j.cjb.150554
-
文章历史
- Received: December 25, 2015
- Accepted: April 12, 2016
2 西藏自治区农牧科学院农业研究所 品种资源研究室,西藏 拉萨 850000
3 辽宁省水土保持研究所,辽宁 朝阳 122000
2 Germplasm Resources Laboratory, Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa 850000, Tibet, China ;
3 Liaoning Institute of Soil and Water Conservation, Chaoyang 122000, Liaoning, China
转座子(Transpososable elements或transposons)是指基因组中可移动或复制自身DNA并整合到新位点的DNA片段[1]。转座子在基因组中常常具有很多拷贝,是真核生物基因组重要的组成部分[2]。人类基因组中转座子约占整个基因组的45%,植物中约为50%−90%[3]。由于转座子激活可造成基因的失活或缺失、基因组重排、甚至是着丝粒区序列的进化和分化[4-7],因此对于研究基因组结构、侧翼基因表达调控以及物种进化等都起到重要作用[3]。
根据转座方式可将转座子分为两类:Class Ⅰ称为反转录转座子(Retrotransposon),是利用RNA介导的转座[8]。根据其是否包含有长末端重复序列(LTR),分为LTR类反转座子(LTR retrotransposon)和非LTR类反转座子(Non-LTR retrotransposon),由于其利用复制/插入方式实现转座,因此可改变基因组的大小[3]。Class Ⅱ称为转座子(Transposons),是利用DNA介导转座。根据其转座的自主性,可分为自主性转座子(Autonomous element)、非自主性转座子(Non-autonomous element)及微型反向重复转座子(Miniature inverted-repeat transposable element, MITE),由于其利用切除/修复方式实现转座,因此基因组大小一般不会发生改变[9-10]。
MITE类转座子是一类在植物中分布广泛的非自主性转座子,自身不具编码转座酶的基因,序列只有100−500 bp[11],常常位于基因组的常染色质区[12-13]。水稻中第一个被鉴定的有活性的MITE类转座子是mPing (GenBank Accession No. BK000588),序列长430 bp[14],mPing的结构中包括15 bp的TIRs (Terminal inverted repeats),但不带有ORF (Open reading frame),它的转座依靠自主类转座子Ping或Pong的ORF2编码的转座酶进行转座[15-17]。mPing在水稻中的突变体有两种类型:mPing-1和mPing-2,其中mPing-1序列长430 bp,mPing-2序列长419 bp[18]。
水稻作为世界一半人口的食物来源,对人类的生存与发展有着重要的意义,在其9 000多年的驯化历史中[18-19],有20多个稻属物种,其主要分化为籼稻Indica和粳稻Japonica两种生态型[20]。籼稻和粳稻有着形态、生理、遗传等方面的差异。比如籼稻植株一般高于粳稻植株,抗倒伏能力较强,发芽较快,分蘖数较强,但籼稻比粳稻有更高的净光合速率[21];蔡星星等[20]利用籼稻93-11和粳稻日本晴基因组序列的差异片段进行研究,结果显示这两种生态型全基因序列具有很大的差异性。mPing的拷贝数差异就是其中的一个原因,为了更加完整地探寻mPing在水稻亚种分化以及基因组形成中的可能作用,本研究主要运用Southern杂交分析并结合生物信息学的方法,利用日本晴和93-11全基因组信息,对mPing在水稻两个亚种基因组中的结构特点、分布特征、以及mPing的插入对侧翼基因的影响进行研究,探寻mPing在水稻亚种形成过程中的可能作用。本研究为阐明粳稻和籼稻之间的遗传差异、探索粳稻和籼稻的进化历史提供初步理论基础。
1 材料与方法 1.1 材料来源本研究采用的实验材料为粳稻品种日本晴(Oryza sativa Japonica. cv. Nipponbare)和籼稻品种93-11 (Oryza sativa Indica. cv. 93-11)。
1.2 Southern杂交利用Hind Ⅲ (购自New England Biolabs公司)分别对日本晴和93-11的DNA样品进行酶切,选取这个酶的目的在于mPing中不存在Hind Ⅲ的酶切位点,因此我们可以估测mPing在这两个水稻亚种中的拷贝数。根据Shan等[5]提供的方法进行Southern杂交分析,以mPing-1 (AB087615)全长设计探针引物(由上海生工生物技术有限公司合成),具体序列为mPing-1 (positions 6-430):forward,5’-GTCACAATGGG GGTTTCACT-3’,reverse,5’-GGCCAGTCACA ATGGCTAGT-3’。
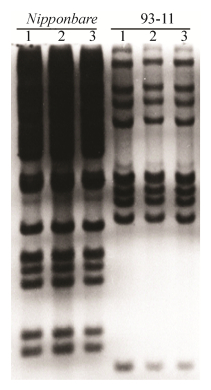
|
| 图 1 mPing在日本晴和93-11中的Southern杂交分析 Figure 1 Analysis of mPing in Nipponbare and 93-11 in the Southern blotting hybridization patterns. 1–3 means repeating three times. |
| |
mPing-1 (AB087615)的基因序列在NCBI网站(http://archive-dtd.ncbi.nlm.nih.gov/)中下载,根据mPing-1的序列,可得到mPing-2的序列[18]。根据GRAMENE (http://www.gramene.org/)网站,做Blast分析获得mPing在日本晴和93-11中的拷贝信息,根据RICE-MAP (http://www.ricemap.org/)数据库网站,可得到该mPing拷贝所在染色体的上下游5 kb的碱基序列信息,再根据KOME数据库(http://cdna01.dna.affrc.go.jp/cDNA/),E值设为0,即只有100%同源序列才被挑选出来,以此方法进行同源性探寻可得到侧翼序列的外显子信息及表达蛋白的功能信息。
2 结果与分析 2.1 日本晴和93-11的Southern杂交分析通过Hind
为了进一步确定mPing在两个亚种中的差别信息,通过同源性探寻方法,对mPing进行定位分析。结果表明,mPing在水稻亚种中分布数量和位置均不相同(表 1,表 2),mPing在93-11中的拷贝数是14 (图 2),在日本晴中的拷贝数是52 (图 3)。其中93-11中mPing-1的拷贝数是3,mPing-2的拷贝数是11,日本晴中的mPing均为mPing-1,mPing在日本晴中的拷贝数是93-11的3.7倍。此外,日本晴每条染色体均有mPing的分布,拷贝数最多的是10号染色体,最少的是1号染色体,93-11中7、8、10号染色体没有mPing的拷贝,1号染色体上的拷贝数最多,为3个拷贝。mPing在日本晴和93-11基因组中结构特点和分布特征的多样性,进一步说明它们是导致水稻不同亚种间基因组差异的原因。
| Copy | Ori | Chr | Start | End | %ID | Length | Type |
| 1 | + | 1 | 17 513 834 | 17 514 263 | 100.00 | 430 | mPing-1 |
| 2 | + | 1 | 23 332 547 | 23 332 976 | 100.00 | 430 | mPing-1 |
| 3 | + | 1 | 24 779 771 | 24 780 200 | 100.00 | 430 | mPing-1 |
| 4 | + | 1 | 25 261 112 | 25 261 541 | 100.00 | 430 | mPing-1 |
| 5 | + | 1 | 29 931 517 | 29 931 946 | 100.00 | 430 | mPing-1 |
| 6 | + | 2 | 214 437 | 214 866 | 100.00 | 430 | mPing-1 |
| 7 | + | 2 | 617 949 | 618 378 | 100.00 | 430 | mPing-1 |
| 8 | + | 2 | 13 161 938 | 13 162 367 | 100.00 | 430 | mPing-1 |
| 9 | + | 2 | 18 534 787 | 18 535 216 | 99.77 | 430 | mPing-1 |
| 10 | + | 2 | 22 549 115 | 22 549 544 | 100.00 | 430 | mPing-1 |
| 11 | + | 2 | 28 008 341 | 28 008 770 | 99.77 | 430 | mPing-1 |
| 12 | + | 2 | 29 244 327 | 29 244 756 | 100.00 | 430 | mPing-1 |
| 13 | + | 3 | 5 504 299 | 5 504 728 | 100.00 | 430 | mPing-1 |
| 14 | + | 3 | 6 513 589 | 6 514 018 | 100.00 | 430 | mPing-1 |
| 15 | + | 3 | 9 240 074 | 9 240 503 | 100.00 | 430 | mPing-1 |
| 16 | + | 3 | 9 427 120 | 9 427 549 | 100.00 | 430 | mPing-1 |
| 17 | + | 3 | 9 568 670 | 9 569 099 | 100.00 | 430 | mPing-1 |
| 18 | + | 3 | 12 735 756 | 12 736 185 | 99.77 | 430 | mPing-1 |
| 19 | + | 3 | 17 575 717 | 17 576 146 | 100.00 | 430 | mPing-1 |
| 20 | + | 3 | 21 360 120 | 21 360 549 | 100.00 | 430 | mPing-1 |
| 21 | + | 3 | 21 026 572 | 21 027 001 | 100.00 | 430 | mPing-1 |
| 22 | + | 3 | 34 592 545 | 34 592 974 | 99.77 | 430 | mPing-1 |
| 23 | + | 4 | 19 021 060 | 19 021 489 | 99.77 | 430 | mPing-1 |
| 24 | + | 4 | 33 021 868 | 33 022 297 | 100.00 | 430 | mPing-1 |
| 25 | + | 4 | 34 302 847 | 34 303 276 | 100.00 | 430 | mPing-1 |
| 26 | + | 4 | 34 688 306 | 34 688 735 | 100.00 | 430 | mPing-1 |
| 27 | + | 4 | 35 421 806 | 35 422 235 | 99.77 | 430 | mPing-1 |
| 28 | + | 5 | 18 747 498 | 18 747 927 | 100.00 | 430 | mPing-1 |
| 29 | + | 5 | 19 328 618 | 19 329 047 | 100.00 | 430 | mPing-1 |
| 30 | + | 5 | 22 235 594 | 22 236 023 | 100.00 | 430 | mPing-1 |
| 31 | + | 6 | 13 737 618 | 13 738 047 | 100.00 | 430 | mPing-1 |
| 32 | + | 6 | 18 136 422 | 18 136 851 | 100.00 | 430 | mPing-1 |
| 33 | + | 6 | 23 521 641 | 23 521 893 | 99.60 | 253 | mPing-1 |
| 33 | + | 6 | 23 526 804 | 23 526 981 | 100.00 | 178 | mPing-1 |
| 34 | + | 6 | 30 099 538 | 30 099 967 | 99.77 | 430 | mPing-1 |
| 35 | + | 7 | 4 560 975 | 4 561 404 | 100.00 | 430 | mPing-1 |
| 36 | + | 8 | 1 019 672 | 1 020 101 | 100.00 | 430 | mPing-1 |
| 37 | + | 8 | 4 712 970 | 4 713 399 | 100.00 | 430 | mPing-1 |
| 38 | + | 8 | 16 683 588 | 16 684 017 | 100.00 | 430 | mPing-1 |
| 39 | + | 8 | 20 442 416 | 20 442 845 | 100.00 | 430 | mPing-1 |
| 40 | + | 8 | 20 674 237 | 20 674 666 | 100.00 | 430 | mPing-1 |
| 41 | + | 8 | 28 186 241 | 28 186 670 | 100.00 | 430 | mPing-1 |
| 42 | + | 9 | 694 483 | 694 912 | 99.77 | 430 | mPing-1 |
| 43 | + | 9 | 16 698 141 | 16 698 570 | 99.77 | 430 | mPing-1 |
| 44 | + | 10 | 4 320 421 | 4 320 850 | 100.00 | 430 | mPing-1 |
| 45 | + | 10 | 21 716 391 | 21 716 819 | 99.77 | 430 | mPing-1 |
| 46 | + | 11 | 393 598 | 394 027 | 100.00 | 430 | mPing-1 |
| 47 | + | 11 | 23 200 105 | 23 200 534 | 100.00 | 430 | mPing-1 |
| 48 | + | 12 | 839 604 | 840 033 | 100.00 | 430 | mPing-1 |
| 49 | + | 12 | 1 045 463 | 1 045 892 | 100.00 | 430 | mPing-1 |
| 50 | + | 12 | 2 734 541 | 2 734 970 | 99.77 | 430 | mPing-1 |
| 51 | + | 12 | 3 285 787 | 3 286 216 | 100.00 | 430 | mPing-1 |
| 52 | + | 12 | 9 951 106 | 9 951 535 | 100.00 | 430 | mPing-1 |
| Copy | Ori | Chr | Start | End | %ID | Length | Type |
| 1 | + | 1 | 28 368 367 | 28 368 786 | 100.00 | 420 | mPing-2 |
| 2 | + | 1 | 12 119 746 | 12 119 983 | 100.00 | 238 | mPing-2 |
| 2 | + | 1 | 12 119 565 | 12 119 742 | 100.00 | 178 | mPing-2 |
| 3 | + | 1 | 13 010 583 | 13 010 823 | 99.59 | 241 | mPing-2 |
| 3 | + | 1 | 13 010 835 | 13 011 012 | 100.00 | 178 | mPing-2 |
| 4 | + | 2 | 30 104 358 | 30 104 595 | 100.00 | 238 | mPing-2 |
| 4 | + | 2 | 30 104 177 | 30 104 354 | 100.00 | 178 | mPing-2 |
| 5 | + | 3 | 31 477 707 | 3 477 884 | 99.44 | 178 | mPing-2 |
| 5 | + | 3 | 31 477 888 | 31 478 125 | 100.00 | 238 | mPing-2 |
| 6 | + | 4 | 31 613 909 | 31 614 146 | 100.00 | 238 | mPing-2 |
| 6 | + | 4 | 31 614 150 | 31 614 327 | 99.44 | 178 | mPing-2 |
| 7 | + | 4 | 34 250 349 | 34 250 586 | 100.00 | 238 | mPing-2 |
| 7 | + | 4 | 34 250 590 | 34 250 767 | 99.44 | 178 | mPing-2 |
| 8 | + | 5 | 23 911 981 | 23 912 158 | 100.00 | 178 | mPing-2 |
| 8 | + | 5 | 23 912 162 | 23 912 399 | 99.58 | 238 | mPing-2 |
| 9 | + | 5 | 3 467 412 | 3 467 589 | 100.00 | 178 | mPing-2 |
| 9 | + | 5 | 3 467 593 | 3 467 830 | 100.00 | 238 | mPing-2 |
| 10 | + | 6 | 5 018 844 | 5 019 273 | 100.00 | 430 | mPing-1 |
| 11 | + | 9 | 7 850 636 | 7 850 813 | 99.44 | 178 | mPing-2 |
| 11 | + | 9 | 7 850 817 | 7 851 054 | 100.00 | 238 | mPing-2 |
| 12 | + | 11 | 14 458 178 | 14 458 355 | 100.00 | 178 | mPing-2 |
| 12 | + | 11 | 14 458 359 | 14 458 596 | 100.00 | 238 | mPing-2 |
| 13 | + | 11 | 17 209 369 | 17 209 686 | 95.61 | 319 | mPing-1 |
| 13 | + | 11 | 17 209 678 | 17 209 814 | 93.43 | 137 | mPing-1 |
| 14 | + | 12 | 17 627 576 | 17 628 005 | 100.00 | 430 | mPing-1 |

|
| 图 2 mPing在93-11基因组染色体上的定位 Figure 2 Chromosomal location of mPing in 93-11 genome. |
| |

|
| 图 3 mPing在日本晴基因组染色体上的定位 Figure 3 Chromosomal location of mPing in Nipponbare genome. |
| |
转座子的转座会影响其侧翼基因的表达,在基因的结构和进化过程中具有重要的作用[8]。因此对mPing所有拷贝的侧翼序列上下游5 kb进行功能基因的同源性探寻。研究发现,mPing插入到了日本晴基因组中的某些基因的上游、下游以及内部,93-11基因组中的某些基因的上游和下游。日本晴中有23个mPing侧翼序列与已知功能基因有100%的同源性,这些已知基因中有与拟南芥相关的转座酶蛋白基因、与抗性相关的蛋白基因等(表 3)。同时,mPing插入到了1号染色体中的AK073838,11号染色体中的AK109523,12号染色体中的AK243131基因的内部。93-11中有3个mPing侧翼序列与已知的功能基因有100%的同源性,且mPing均插入到了基因的上游区域(表 4)。
| Chromsome | GenBank Accession No. | Flanking sequence | Putative protein function predicted by blast X |
| 1-1 | AK073838 | Up | Hordeum vulgare BRI1 mRNA for putative brassinosteroid-insensitive protein 1, complete cds. |
| 1-2 | AK100804 | Up | Arabidopsis thaliana clone 94974 mRNA, complete sequence. |
| AK073838 | Down | Hordeum vulgare BRI1 mRNA for putative brassinosteroid-insensitive protein 1, complete cds. | |
| 1-4 | AK071288 | Up | Arabidopsis thaliana mutator-like transposase-like protein (MQK4.25) mRNA, complete cds. |
| AK069334 | Down | Arabidopsis thaliana putative protein (At5g19590) mRNA, complete cds. | |
| 1-5 | AK122047 | Up | Triticum aestivum clone wlp1c.pk006.j5:fis, full insert mRNA sequence. |
| 3-1 | AK061148 | Up | Zea mays (clone wusl1032) mRNA sequence. |
| AK111899 | Down | Zea mays ZmRR8 mRNA for response regulator 8, complete cds. | |
| 4-2 | AK121689 | Up | Daucus carota pskr mRNA for phytosulfokine receptor, complete cds. |
| AK071468 | Down | Arabidopsis thaliana AT5g42940/MBD2_14 mRNA, complete cds. | |
| 4-4 | AK068257 | Up | Arabidopsis thaliana At1g71010 mRNA, complete cds. |
| 5-1 | AK288267 | Up | AY518220 Oryza sativa (Indica cultivar-group) NBS-LRR-like protein A (NL-A), NBS-LRR-like protein B (NL-B), NBS-LRR-like protein C (NL-C), and NBS-LRR-like protein D (NL-D) genes, complete cds. |
| 6-1 | AK242258 | Up | AY459336 Oryza sativa (Japonica cultivar-group) clone CL034244.2 R2R3-MYB gene region. |
| 6-5 | AK068363 | Up | Arabidopsis thaliana unknown protein (At3g15310) mRNA, complete cds. |
| 7-1 | AK063035 | Up | Triticum aestivum clone wr1.pk182.b10:fis, full insert mRNA sequence. |
| 8-1 | AK120080 | Up | Arabidopsis thaliana At1g71010 mRNA, complete cds. |
| 8-3 | AK100130 | Up | Arabidopsis thaliana putative DNA-binding protein (At1g50410) mRNA, complete cds. |
| 8-4 | AK288678 | Up | AF259976 Oryza sativa subsp. indica polyprotein mRNA, complete cds. |
| AK288293 | Down | AB110444 Oryza sativa (Japonica cultivar-group) rf-1gene for fertility restorer, hypothetical proteins, complete cds. | |
| 8-5 | AK105950 | Up | Lycopersicon esculentum putative anthocyanin permease mRNA, complete cds. |
| 11-1 | AK109523 | Up | Arabidopsis thaliana laccase (lac1) mRNA, complete cds. |
| AK109523 | Down | Arabidopsis thaliana laccase (lac1) mRNA, complete cds. | |
| 11-2 | AK070903 | Up | Arabidopsis thaliana unknown protein (At1g14330/F14L17_7) mRNA, complete cds. |
| 12-1 | AK120100 | Up | Prunus dulcis SLFd mRNA for S locus F-box protein d, complete cds. |
| 12-2 | AK243131 | Up | AB180961 Oryza sativa (Japonica cultivar-group) OsSCR mRNA for SCARECROW, complete cds. |
| AK243131 | Down | AB180961 Oryza sativa (Japonica cultivar-group) OsSCR mRNA for SCARECROW, complete cds. |
| Chromsome | GenBank Accession No. | Flanking sequence | Putative protein function predicted by blast X |
| 1-1 | AK100804 | Up | Arabidopsis thaliana clone 94974 mRNA, complete sequence. |
| 5-1 | AK102420 | Up | Arabidopsis thaliana MFP2 mRNA, complete cds. |
| 5-2 | AK120742 | Up | Physcomitrella patens mRNA for PPR986-12, complete cds. |
转座子的移动会影响宿主基因组的结构、功能和进化[2-5, 22-26]。在本研究中,我们以MITE类转座子mPing为研究对象,探讨了以其多样性为主要原因的水稻亚种间的基因组差异。结果表明,在日本晴和93-11基因组中,mPing的分布和类型均有较大差异。日本晴基因组中只存在mPing-1一种类型,共有52个拷贝;93-11基因组中有mPing 14个,包括3个mPing-1和11个mPing-2。这与Hu等的研究结果,粳稻品种中mPing-1所占比例高于mPing-2,而籼稻品种mPing-2所占比例高于mPing-1一致[18]。mPing在水稻亚种基因组中分布比例和位置的差异,暗示mPing在水稻驯化历史中有着不可忽视的作用;同时,mPing有望作为一种分子标签来快速鉴别水稻亚种。另一方面,mPing往往会插入到基因中或者基因附近[14],为了研究mPing与侧翼基因的关系,本研究对mPing所在位置的上下游延伸5 kb序列进行功能基因的同源性探寻,发现日本晴中有24个序列与已知基因序列100%同源,93-11中有3个序列与已知基因100%同源,并且日本晴中有3个基因内部插入了mPing,这3个基因与抗性基因、漆酶基因和细胞分裂基因有关,暗示着mPing在调控这3类基因表达的过程中存在一定的潜在影响。同时,日本晴和93-11中除了1个已知基因相同,剩余基因不相同,说明这些原因可能是导致日本晴和93-11基因组差异性的原因。然而mPing的插入机制、它在宿主基因组中对侧翼基因表达的影响、及其在水稻进化历史中的作用,有待进一步探索和研究。
| [1] | McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol,1951, 16 : 13 –47. DOI: 10.1101/SQB.1951.016.01.004 |
| [2] | Roffler S, Wicker T. Genome-wide comparison of Asian and African rice reveals high recent activity of DNA transposons. Mob DNA,2015, 6 : 8 . DOI: 10.1186/s13100-015-0040-x |
| [3] | Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet,2002, 3 (5) : 329 –341. DOI: 10.1038/nrg793 |
| [4] | McClintock B. The significance of responses of the genome to challenge. Science,1984, 226 (4676) : 792 –801. DOI: 10.1126/science.15739260 |
| [5] | Shan XH, Liu ZL, Dong ZY, et al. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol Biol Evol,2005, 22 (4) : 976 –990. DOI: 10.1093/molbev/msi082 |
| [6] | Michalak P. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity,2009, 102 (1) : 45 –50. DOI: 10.1038/hdy.2008.48 |
| [7] | Gao DY, Jiang N, Wing RA, et al. Transposons play an important role in the evolution and diversification of centromeres among closely related species. Front Plant Sci,2015, 6 : 216 . |
| [8] | Lisch D. How important are transposons for plant evolution?. Nat Rev Genet,2013, 14 (1) : 49 –61. |
| [9] | Shan XH, Lin XY, Long LK, et al. In planta mobilization of mPing and its putative autonomous element Pong in rice by hydrostatic pressurization. J Exp Bot,2006, 57 (10) : 2313 –2323. DOI: 10.1093/jxb/erj203 |
| [10] | Wang HY, Chai Y, Chu XC, et al. Molecular characterization of a rice mutator-phenotype derived from an incompatible cross-pollination reveals transgenerational mobilization of multiple transposable elements and extensive epigenetic instability. BMC Plant Biol,2009, 9 : 63 . DOI: 10.1186/1471-2229-9-63 |
| [11] | Jiang N, Feschotte C, Zhang XY, et al. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr Opin Plant Biol,2004, 7 (2) : 115 –119. DOI: 10.1016/j.pbi.2004.01.004 |
| [12] | Zhang Q, Arbuckle J, Wessler SR. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc Natl Acad Sci USA,2000, 97 (3) : 1160 –1165. DOI: 10.1073/pnas.97.3.1160 |
| [13] | Feng Q, Zhang YJ, Hao P, et al. Sequence and analysis of rice chromosome 4. Nature,2002, 420 (6913) : 316 –320. DOI: 10.1038/nature01183 |
| [14] | Yasuda K, Tsukiyama T, Karki S, et al. Mobilization of the active transposon mPing in interspecific hybrid rice between Oryza sativa and O. glaberrima. Euphytica,2013, 192 (1) : 17 –24. DOI: 10.1007/s10681-012-0810-1 |
| [15] | Jiang N, Bao ZR, Zhang XY, et al. An active DNA transposon family in rice. Nature,2003, 421 (6919) : 163 –167. DOI: 10.1038/nature01214 |
| [16] | Kikuchi K, Terauchi K, Wada M, et al. The plant MITE mPing is mobilized in anther culture. Nature,2003, 421 (6919) : 167 –170. DOI: 10.1038/nature01218 |
| [17] | Yang GJ, Zhang F, Hancock CN, et al. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA,2007, 104 (26) : 10962 –10967. DOI: 10.1073/pnas.0702080104 |
| [18] | Hu H, Mu J, Zhang HJ, et al. Differentiation of a miniature inverted transposable element (MITE) system in Asian rice cultivars and its inference for a diphyletic origin of two subspecies of Asian cultivated rice. J Integr Plant Biol,2006, 48 (3) : 260 –267. DOI: 10.1111/jipb.2006.48.issue-3 |
| [19] | Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol,1997, 35 (1/2) : 25 –34. DOI: 10.1023/A:1005810616885 |
| [20] |
Cai XX, Liu J, Chou YQ, et al. Differentiation of Indica-Japonica rice revealed by insertion/deletion fragments obtained from comparative genomic study of DNA sequences between 93-11 (Indica) and Nipponbare (Japonica).
J Fudan Univ: Nat Sci,2006, 45 (3) : 309 –315.
(in Chinese). 蔡星星, 刘晶, 仇吟秋, 等. 籼稻93-11和粳稻日本晴DNA插入缺失差异片段揭示的水稻籼-粳分化. 复旦学报:自然科学版, 2006 , 45 (3) : 309-315. |
| [21] |
Zhang Z, Mao T, Li X, et al. Research on difference and classification of Indica-japonica subspecies at Asian cultivated rice.
North Rice,2013, 43 (2) : 66 –69.
(in Chinese). 张战, 毛艇, 李鑫, 等. 亚洲栽培稻籼粳亚种差异及主要分类方法研究进展. 北方水稻, 2013 , 43 (2) : 66-69. |
| [22] | Marillonnet S, Wessler SR. Retrotransposon insertion into the maize waxy gene results in tissure-specific RNA processing. Plant Cell,1997, 9 (6) : 967 –978. DOI: 10.1105/tpc.9.6.967 |
| [23] | Wang HY, Tian Q, Ma YQ, et al. Transpositional reactivation of two LTR retrotransposons in rice-Zizania recombinant inbred lines (RILs). Hereditas,2010, 147 : 264 –277. DOI: 10.1111/more.2010.147.issue-6 |
| [24] | asacuberta E, Gonzalez J. The impact of transposable elements in environmental adaptation. Mol Ecol,2013, 22 : 1503 –1517. DOI: 10.1111/mec.12170 |
| [25] | Bennetzen JL, Wang H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol,2014, 65 : 505 –530. DOI: 10.1146/annurev-arplant-050213-035811 |
| [26] | Lisch D, Bennetzen JL. Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol,2011, 14 : 156 –161. DOI: 10.1016/j.pbi.2011.01.003 |
 2016, Vol. 32
2016, Vol. 32




