中国科学院微生物研究所、中国微生物学会主办
文章信息
- 杨震, 彭选明, 张逸妍, 张源海, 张勇
- Yang Zhen, Peng Xuanming, Zhang Yiyan, Zhang Yuanhai, Zhang Yong
- 植物DNA甲基化及胁迫诱导的变异
- Research progress and stress-induced variation of DNA methylation in plants
- 生物工程学报, 2016, 32(12): 1642-1653
- Chin J Biotech, 2016, 32(12): 1642-1653
- 10.13345/j.cjb.160226
-
文章历史
- Received: June 11, 2016
- Accepted: September 14, 2016
2 湖南省农业生物辐照工程技术研究中心,湖南 长沙 410125
2 Hunan Province Engineering Technology Research Center for Agricultural Bio-irradiation, Changsha 410125, Hunan, China
DNA甲基化是一种常见的表观遗传修饰方式,在大多数真核生物和原核生物中,主要有3种类型的甲基化形式,即N6-甲基腺嘌呤、N4-甲基胞嘧啶和5-甲基胞嘧啶[1]。在真核生物体中,5-甲基胞嘧啶(5-mC)是DNA甲基化的主要存在形式,即特定的甲基转移酶从供体S-腺苷-L-甲硫氨酸上转移一个甲基基团到嘧啶环的第5位C位置上[2-3],有时也被称作第4种碱基[4]。在真核生物植物、真菌和动物中DNA甲基化主要发生在基因组中的重复序列和转座子,部分出现在主要引起转录抑制和基因表达沉默的5'端调控区[5-7]。DNA甲基化相对稳定,且在DNA复制过程中甲基化状态能被维持[8],在植物体中这种相对稳定的甲基化状态对转座子自身移动引起的突变和染色质畸变能起到防御作用[9],因而在动植物发育过程中起重要的调控作用。此外,胞嘧啶甲基化在原核和真核生物中扮演不同的生物角色,当细菌进入宿主细胞内,胞嘧啶甲基化的方式能起到防止被宿主细胞内限制性内切酶酶解的作用,而在大多数真核生物体中,胞嘧啶甲基化方式通过改变基因组结构和组蛋白修饰的形式影响基因的表达调控[10]。
1 植物DNA甲基转移酶哺乳动物中,DNA甲基化模式在世代之间需要消除和重建,即在完成受精后不久发生全基因组范围的DNA去甲基化,随后在生长发育过程中通过特定的甲基化转移酶又重新建立甲基化状态[11]。由于植物体没有单独的生殖细胞系,因此在DNA甲基化突变体中即使DNA甲基化机能被恢复,引起的表观遗传的改变也能在植物发育和后几代中稳定遗传[12]。
哺乳动物中,目前主要有4种DNA甲基转移酶(DNA methyltransferases;Dnmts),根据其自身结构和在甲基化过程中功能的差异分为3大类:分别以Dnmt1和Dnmt3为代表,在细胞分化的DNA复制期间,Dnmt1主要参与DNA甲基化状态的维持,也是非CpG位点从头甲基化所必需,并与甲基化状态的延伸有关;Dnmt3包括Dnmt3a、Dnmt3b、Dnmt3L等,Dnmt3a和Dnmt3b是负责从头合成的DNA甲基化的酶[13],Dnmt3L自身无催化活性,主要对Dnmt3a和Dnmt3b起调控作用[14-15]。而Dnmt2的归属和功能起初尚不十分明确,后来研究者发现Dnmt2是一种tRNA甲基转移酶,而非DNA甲基转移酶,被重新命名为tRNA天门冬氨酸甲基转移酶[16]。在植物中,DNA甲基转移酶介导的DNA甲基化主要发生在对称的CG、CHG和非对称的CHH位点(H代表任意的A、T或C)[10-17]。植物体中主要发现的有2种维持甲基转移酶和1种从头重新甲基转移酶,包括MET1、CMT3和DRM2[18-20]。MET1与哺乳动物中的Dnmt1同源,负责CG双核苷酸位点甲基化的维持[21]。CMT3是植物中特有的一种DNA甲基转移酶,主要维持植物体中CHG位点的甲基化状态[19]。在DNA姊妹链复制期间,由于CG和CHG位点的对称性,两者位点的甲基化模式能被有活性的MET1、CMT3两种甲基转移酶转移和维持,而CMT3和DRM2参与催化非对称CHH位点的重新从头甲基化的过程,其中DRM2与哺乳动物中的重新从头甲基转移酶Dnmt3a/3b系同源物[22-23]。近年来也有研究者在植物体中发现一种具有催化活性的突变DNA甲基转移酶DRM3,在RNA介导的甲基化(RNA-directed DNA methylation;RdDM)途径中参与刺激DRM2甲基转移酶的活性而发挥重要作用[24]。
2 植物体基因组的DNA甲基化哺乳动物中,DNA甲基化主要出现在对称的CG双核苷酸位点,整个基因组被甲基化的CG位点大约占70%-80%[25],在胚胎干细胞中出现少量的非CG位点的甲基化[8, 26],而其余未甲基化的CG双核甘酸主要聚簇在基因的启动子附近,即CpG岛[27-28]。在植物中,整个基因组序列内的胞嘧啶碱基通常都可以发生DNA甲基化:对称的CG和CHG位点(H代表除G以外的任意3种碱基),非对称的CHH位点[16]。CG位点的胞嘧啶甲基化状态相对最为稳定[29]。在模式植物拟南芥基因组中,CG、CHG和CHH位点胞嘧啶甲基化含量大约分别为24%、6.7%和1.7%[30]。与动物基因组不同,植物基因组中的DNA甲基化主要出现在转录子和DNA重复序列[31]。
DNA甲基化序列或区域在动植物基因组上的位置是特定而非随机的,在异染色质区域含有大量的甲基化胞嘧啶,且常常富集在重复DNAs上,如在重复DNAs中,着丝粒的180 bp重复序列和rDNAs处的CpG位点通常被甲基化,甲基化胞嘧啶在染色质中心大量存在,而在染色体臂上稀疏分布[32]。Doerfler等[31]、Zilberman等[33]使用Tilling array技术对高等模式真核生物体拟南芥绘制出第一张DNA甲基化和小RNAs的表观基因组图,表明在拟南芥基因组的异染色质的转座元件和重复序列区碱基上存在高密度的DNA甲基化(超过90%被甲基化),研究也显示约30%的转录基因序列被甲基化,进一步通过拟南芥表观基因组的研究,使用重亚硫酸序列转化的方法在单碱基分辨率的水平下发现甲基化主要发生在基因编码区的CG位点,然而,这些甲基化的功能还是未知的[30, 34]。通过比较出现在转录编码区CG位点5mC的数据,发现转录编码区内以适当量水平表达的基因往往更容易被甲基化,然而以极端量(过高水平和过低水平)表达的基因通常是较少出现甲基化现象[30, 34]。启动子区的5mC含量相当低(拟南芥中小于5%),且常具有基因表达的组织特异性,拟南芥和水稻胚乳的甲基化测定表明,转录起始位点位置CG位点的甲基化水平的降低导致胚乳特定基因的表达[35-37]。虽然基因编码转录区5mC的生物学功能目前尚不清楚,但很有可能抑制基因组中启动子的异常转录[33, 38]。异常转录有时出现在基因表达区是由于RNA聚合酶Ⅱ (RNAPⅡ)的延伸传输期间使染色体断裂,并允许前起始复合物聚集在非正确启动子旁,引起异常转录能通过sRNA途径导致同源染色体DNA甲基化[29, 33, 38],它们的产物能激起5mC在转录编码区的聚集,进一步损害畸变转录物结构的循环。转录基因区过强或过弱的转录不需要转录编码区5mC,由于RNAPII的过快或过慢的延伸速率,降低了前起始复合物的结合机率。转录编码区5mC也可能与外显子和内含子的边界区有一定关系。Matzke等[39]通过对植物拟南芥全基因组中核小体的定位和单碱基分辨率的DNA甲基化分布图分析发现,由于DNA甲基转移酶优先以与核小体结合的DNA为目标,因而核小体位置能影响基因组中的DNA甲基化。
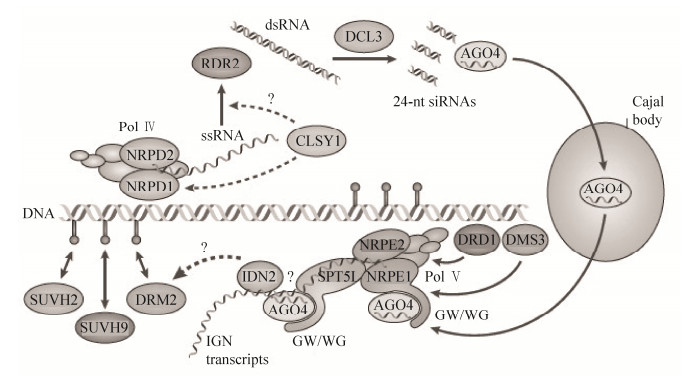
3 植物中RNA介导的DNA甲基化RNA介导的DNA甲基化(RNA-directed DNA methylation,RdDM)是植物中特有的一种重新从头甲基化途径,小分子RNAs靶定从头甲基转移酶DRM2到同源染色体的基因组位点建立DNA甲基化[40],它涉及转基因沉默、转座子抑制、基因组印记和副突变等表观遗传现象[29, 41-43]。RdDM是植物体一种保守的基因序列从头甲基化机制,这种现象首次在含类病毒的转基因烟草中被发现,Wassenegger等[44]利用农杆菌介导的叶圆盘转化法将类病毒cDNA序列导入烟草基因组中,当类病毒再次感染烟草并进行复制的时候,烟草基因组中的cDNA序列(与病毒RNA同源)产生了甲基化,而cDNA序列两侧的T-DNA序列和烟草基因组DNA并没有被甲基化,在转基因植物整个生长发育时期,重组类病毒基因序列的甲基化过程伴随着RNA类病毒的复制,即小分子RNAs靶标同源基因组序列的DNA胞嘧啶甲基化。在模式植物拟南芥中,DNA甲基化高度富集在着丝粒区域和基因组的重复序列中,大约30%的DNA甲基化位点富含大量的小干扰RNA (Small interfering RNA;siRNAs),也表明siRNAs在DNA甲基化过程中的重要性[33-34]。Wierzbicki等[45]对拟南芥利用全基因组扫描的叠瓦芯片技术研究发现siRNAs和长的非编码的RNAs均参与基因组的DNA从头甲基化过程。RdDM主要涉及RNA干扰机制(Dicer和Argonaute蛋白家族成员)和DRM2的从头甲基化,2种在植物体中特有的RNA聚合酶Pol Ⅳ、Pol Ⅴ、2个染色质重塑因子和一些近期研究发现的未知蛋白参与RdDM过程[46-47]。
RdDM也被认为是一种转录水平的基因组沉默机制,已经通过正向遗传学筛查和亲和提纯法进行了研究,多亚基蛋白复合体Pol Ⅳ、Pol Ⅴ与依赖DNA介导的RNA聚合酶Ⅱ (RNA PolⅡ)相似,两者彼此有各自特定的亚基并分享共同的亚基,且与RNA PolⅡ分享其他亚基[48-49],两者在从头甲基化和转录水平基因沉默过程中发挥不同的功能[50-53]。Pol Ⅳ (NRPD1和NRPD2亚基)利用DNA上的RdDM靶向位点转座子和重复序列为模板合成单链核糖核酸(Single-stranded ribonucleic acid;ssRNA)转录物,染色质重塑因子CLASSY1 (CLSY1)可能参与招募Pol Ⅳ结合到染色质上和辅助ssRNA转录物的产生。ssRNA转录物通过依赖于RNA的RNA聚合酶RDR2合成双链RNA (dsRNA)。dsRNA被DICER-LIKE 3蛋白DCL3剪辑成24-nt的初始siRNAs (24-nt siRNAs)。成熟的siRNAs需要一个保守的依赖S-腺苷-L-甲硫氨酸RNA甲基化转移酶HUAENHANCER1 (HEN1)使初始siRNAs产物核苷酸3′-端的2′-羟基基团甲基化[45, 54-55]。Argonaute蛋白家族的AGO4通过碱基配对结合siRNAs将其定位到卡哈尔体(Cajal bodies)中,这个过程的功能目前还是未知。AGO4同Pol Ⅴ的2个亚基(NRPE1和NRPE2)及结构域重排甲基转移酶DRM2处在一个特定的核心重点区,AGO4-NRPE1复合体可能涉及活化RdDM途径位点的作用,Pol Ⅴ参与整个基因组基因间非编码区的转录,NRPE1结合到染色质上需要另一种染色质重塑因子DRD1和一种染色体结构维持域蛋白DMS3,基因间非编码(Intergenic noncoding;IGN)转录物作为一种支架结构招募结合NRPE1和SPT5L的GW/WG基序列的AGO4结合到成熟siRNAs转录产物上[52, 56]。识别AGO4-siRNAs复合体的RNA结合蛋白IDN2,协助重新从头甲基转移酶DRM2靶定到产生24-nt siRNAs和IGN转录物的基因组位点,DNA甲基化结合蛋白SUVH9、SUVH2参与招募和固定DRM2到基因组该位点,DRM2催化胞嘧啶的从头甲基化,从而调控和影响基因的表达[45, 52, 57-60]。
4 植物中DNA甲基化的维持 4.1 CG位点甲基化的维持动物细胞中,CG位点胞嘧啶甲基化主要通过DNA甲基化转移酶Dnmt1进行维持[10]。有丝分裂间期S阶段,DNA甲基化转移酶Dnmt1首先以半甲基化DNA链为酶作用底物,同DNA复制因子中的增殖细胞核抗原(PCNA)结合在一起,恢复新合成DNA链从半甲基化到全甲基化状态[61-62],从而通过维持型甲基化酶使先前存在母细胞中的甲基化方式能够在DNA复制过程中连续多代保留[63-64]。在植物中,通过转反义MET1基因或者MET1基因突变导致CG位点甲基化的减少[17, 65-66],且MET1或DDM1突变体引起的CG位点甲基化的缺失或减少不能回复,并且这种低甲基化水平的状态能保持多代遗传[65-67]。虽然无明确的生物化学数据表明植物MET1酶有一个维持DNA甲基化活性功能,但遗传证据表明MET1是Dnmt1的正向同源物,MET1在植物生长发育过程中对于维持CG位点甲基化是必需的[66, 68-69],且同Dnmt1在维持DNA甲基化过程中的方式具有相似性[70]。
拟南芥基因组中大约1/3基因的编码区中包含CG甲基化位点,这种甲基化状态依靠MET1来维持[30]。与转座子中的甲基化能引起基因沉默、抑制基因的表达不同,转录区域中一些甲基化基因(Body-methylated genes)并不受甲基化影响,但是某些甲基化位点的改变能影响组织中基因的表达,随着环境和种群的不同表现出特异性差异基因[31],如met1突变体中body-methylated genes表达水平上升[8],且body-methylated genes表达水平的高低趋向于缺乏body甲基化,表明转录和body甲基化两者之间互相影响。这种转录区域基因中CG位点甲基化的这种作用方式在其他无脊椎动物中也有报道,说明它可能是真核生物中的一种普遍特性[28]。最初研究认为拟南芥body-methylated genes中CG位点甲基化可能抑制来自于隐藏在启动子中的反义转录子产物,而对met1突变体的研究发现,其反义转录子的增加很少,且和body-methylated genes无关[31]。
4.2 CHG/CHH位点甲基化的维持植物基因组中存在对称的CHG和非对称的CHH位点的甲基化,拟南芥中cmt3的突变和负责H3K9双甲基化的组蛋白甲基转移酶SUVH4缺失能引起植物基因组中CHG位点,尤其是重复序列和转座子序列中CHG位点甲基化的明显减少[71-73],表明CMT3主要负责CHG位点的甲基化[19],也说明组蛋白H3K9的甲基化修饰能影响CHG位点的甲基化,同时另2种H3K9双甲基化的组蛋白甲基转移酶SUVH5、SUVH6也同样影响全基因组CHG位点的甲基化水平[74-75]。SUVH4具有SRA域结构,能特定地包裹CHG甲基化,与此同时CMT3具有绑定组蛋白H3尾部的染色质域结构[76],DNA和组蛋白甲基化这种结构上的相互衔接说明甲基化修饰可能涉及组蛋白和DNA甲基转移酶两个蛋白之间的相互作用[27],然而目前对CMT3和SUVH4两种蛋白直接相互作用如何维持CHG位点胞嘧啶甲基化机理还不是很清楚。非对称的CHH位点甲基化主要依赖于DRM2和RdDM的不断从头甲基化过程来维持。也有研究显示CMT3和DRM2参与某些CHH位点胞嘧啶甲基化[22]。与维持CG、CHG位点甲基化类似,RdDM需要SRA域蛋白,SUVH9和SUVH2具有SRA域蛋白更容易结合到CHH、CG甲基化位点,SUVH9和SUVH2在RdDM途经后开始作用,被认为可能发挥一种保留DRM2靶结合此位点进行甲基化的功能[77]。
5 DNA甲基化检测方法根据检测样本不同,分为DNA和mRNA,现有方法大部分都是针对基因组DNA进行检测。根据研究水平,主要分为3大类,即:基因组甲基化水平的分析,候选基因甲基化的分析,以及基因组层次的DNA甲基化模式与甲基化谱的分析。
甲基化DNA免疫沉淀法(MeDIP)是基于特异抗体对全基因组5meC DNA片段的富集,这种方法相对便宜并广泛用于全基因组甲基化的检测,但分辨率有限,往往不能反映真实的甲基化水平。基于HpaⅡ和MspⅡ甲基化敏感限制性酶切法通常被使用来检测甲基化的方式(即:MSAP法),这种方法的主要缺点是分辨率的数据依赖于限制性酶切位点在全基因组的分布。当前最好的对于检测全基因组甲基化信息的手段是基于高通量测序的重亚硫酸盐转化DNA,然而这种方法对于大基因组来说费用非常昂贵。
6 胁迫诱导植物DNA甲基化的变异 6.1 诱变胁迫诱导的DNA甲基化变异胞嘧啶甲基化能增加基因组重排,各种DNA损伤因子,包括离子辐射能影响基因组DNA甲基化方式,从而可能导致基因组失稳效应[78-79]。在动物中,许多证据表明直接辐射和辐射旁效应均能导致DNA甲基化改变的表观遗传现象,且体内实验表明辐射诱导的DNA甲基化水平的变化具有组织和性别特异性[80-83]。本课题组采用高能7Li离子束注入小麦品种京411干种子,在M1代幼苗时期出现叶脉失绿(MS)和叶脉开裂(MD) 2种诱发畸变体。通过MSAP技术对2种畸变体的基因组进行DNA甲基化变异分析。发现2种畸变体的甲基化水平(49.01%、49.18%)与野生型比较(43.74%)均有所升高,野生型及2种畸变类型的基因组DNA胞嘧啶甲基化的全甲基化率均高于半甲基化率。甲基化模式变异方面,与野生型相比,2种畸变体中单态性的甲基化条带比率分别为58.94% (MD)和56.43% (MS),MD中甲基化增加带型与去甲基化带型分别为25.55%和15.49%,MS中甲基化增加带型与去甲基化带型分别为20.61%和22.94%。利用MSAP技术对60CO-γ射线诱变小麦产生的M3代株高(高秆和矮秆)稳定突变体进行检测DNA甲基化模式的改变。结果表明,与对照植株相比,γ辐射诱导小麦的M3代矮秆突变体中发生在CG、CHG位点的甲基化率(CG/CHG hyper-)均高于相应位点的去甲基化率(CG/CHG hypo-),从而导致整体矮秆植株的基因组的甲基化率(hyper-)高于去甲基化率(hypo-),高秆突变体中则呈现相反,且变异的幅度也不相同。矮秆基因组中CG位点发生甲基化变异的幅度高于CHG位点,但差异不明显,而高秆突变体中DNA甲基化变异发生在CG位点的相对变异率要远高于发生在CHG位点的变异,通过对甲基化位点的甲基化状态和模式的变异分析,有助于更好地理解诱发畸变体、稳定突变体表型和DNA甲基化之间的关联,为DNA甲基化可能参与涉及植物的表型可塑性提供一定的依据。本课题组下一步研究思路是对诱变胁迫后畸变体、稳定突变体中产生的大量差异甲基化修饰的CCGG/CCGG片段序列进行分离、测序,和已知功能基因比对才能了解基因组中这些发生了DNA甲基化变化的位点具体涉及到哪些基因,有何生物功能,以及更深入地理解诱变胁迫基因组DNA的甲基化及逆境适应的机制。
6.2 其他胁迫与DNA甲基化DNA甲基化对环境胁迫的影响非常敏感,生物体基因组甲基化的改变能形成一种可遗传的胁迫适应性,即使这种隔代遗传效应在后代中是非必需的[84]。这种能被暂时遗传到下一代中的DNA甲基化形式也能引起很大的表观遗传多样性,特别是表观遗传差异很高的植物群体,如果生活在截然不同栖息地,它们中的一些物种能利用表观遗传的改变来适应不同的环境[85]。耐盐碱方面:Catarina等[86]利用MSAP检测生活在河边(RS)和盐沼泽附近(SM)两种不同环境下L. racemosa植物群体发现:尽管不同环境下的L. racemosa植物群体表现出明显的形态差异(株高、叶型),基因组结构未发生变化,但出现了丰富的DNA甲基化差异,表明表观遗传变异有利于自然界植物群体的个体适应不同的栖息环境。植物抗性方面:水稻品种Wase Aikoku 3在幼苗期时不抗枯萎病Xanthomonasoryzaepv.oryzae而在成株期则表现为抗性,人工接种幼苗期和成株期的水稻,利用MSAP方法检测到在成株期的水稻苗比幼苗存在更多高度甲基化的多态性位点[87],这些多态性的甲基化位点很可能影响了水稻的抗性。Akimoto等[88]利用去甲基化试剂5-氮脱氧胞嘧啶诱导水稻line-2株系产生抗枯萎病能力,并利用MSAP技术筛选line-2和野生型水稻的多态性片段,得到一个Xa21G克隆体,发现与水稻的抗枯萎病基因Xa21很相似。line-2中Xa21G的启动子没有被甲基化,而野生型中则被完全甲基化,这引起了line-2中Xa21G的高度表达。因此,可以通过降低Xa21G启动子的甲基化水平提高该基因的表达从而达到抗枯萎病的效果。
7 展望植物界中存在大量的遗传变异,这些变异是自然突变和自然选择的结果,也是植物种群赖以生存和进化的动力。大量实验表明,自然环境中的各种生物与非生物因子对植物产生的胁迫效应是产生表观遗传变异的一个重要来源。辐射诱变能促进植物体细胞内同源染色体重组,创制农作物新材料,因此也是产生新的遗传变异的重要途径。DNA甲基化是不基于DNA序列差异的变异,被称为表观遗传变异(Epigenetic variation),植物诱发突变过程可以产生大量的表观遗传变异,这种表观遗传变异主要是编码基因和转座子DNA甲基化水平和模式的改变,其与之相关的组蛋白修饰和染色质结构也随之发生变化。这些表观遗传变异能产生基因表达的改变并由此产生新表型,从而丰富和扩大植物遗传资源的生物多样性,这些不能用经典遗传学理论解释的表观遗传变异在植物育种上可能有重要的利用价值。
| [1] | Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem, 2002, 3 (4) : 274–293. DOI: 10.1002/(ISSN)1439-7633 |
| [2] | Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci, 2004, 61 (19/20) : 2571–2587. |
| [3] | Wada Y. Physiological functions of plant DNA methyltransferases. Plant Biotech, 2005, 22 (2) : 71–80. DOI: 10.5511/plantbiotechnology.22.71 |
| [4] | Johnson EL. Effects of X-rays upon growth, development, and oxidizing enzymes of Helianthus annuus. Bot Gaz, 1926, 82 (4) : 373–402. DOI: 10.1086/333674 |
| [5] | Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet, 1997, 13 (8) : 335–340. DOI: 10.1016/S0168-9525(97)01181-5 |
| [6] | Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci, 2006, 31 (2) : 89–97. DOI: 10.1016/j.tibs.2005.12.008 |
| [7] | Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science, 2001, 293 (5532) : 1070–1074. DOI: 10.1126/science.293.5532.1070 |
| [8] | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev, 2002, 16 (1) : 6–21. DOI: 10.1101/gad.947102 |
| [9] | Robertson KD. DNA methylation and human disease. Nat Rev Genet, 2005, 6 (8) : 597–610. |
| [10] | Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem, 2005, 74 (1) : 481–514. DOI: 10.1146/annurev.biochem.74.010904.153721 |
| [11] | Guo M, Rupe MA, Yang XF, et al. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor Appl Genet, 2006, 113 (5) : 831–845. DOI: 10.1007/s00122-006-0335-x |
| [12] | Reinders J, Wulff BBH, Mirouze M, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev, 2009, 23 (8) : 939–950. DOI: 10.1101/gad.524609 |
| [13] | Chen TP, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Topics Dev Biol, 2004, 60 : 55–89. DOI: 10.1016/S0070-2153(04)60003-2 |
| [14] | Bourc'his D, Xu GL, Lin CS, et al. Dnmt3L and the establishment of maternal genomic imprints. Science, 2001, 294 (5551) : 2536–2539. DOI: 10.1126/science.1065848 |
| [15] | Hata K, Okano M, Lei H, et al. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development, 2002, 129 (8) : 1983–1993. |
| [16] | Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science, 2006, 311 (5759) : 395–398. DOI: 10.1126/science.1120976 |
| [17] | Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature, 2007, 447 (7143) : 418–424. DOI: 10.1038/nature05917 |
| [18] | Ronemus MJ, Galbiati M, Ticknor C, et al. Demethylation-induced developmental pleiotropy in Arabidopsis. Science, 1996, 273 (5275) : 654–657. DOI: 10.1126/science.273.5275.654 |
| [19] | Lindroth AM, Cao XF, Jackson JP, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science, 2001, 292 (5524) : 2077–2080. DOI: 10.1126/science.1059745 |
| [20] | Cao XF, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA, 2002, 99 (4) : 16491–16498. |
| [21] | Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol, 2001, 11 (10) : 747–757. DOI: 10.1016/S0960-9822(01)00226-3 |
| [22] | Cao XF, Aufsatz W, Zilberman D, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol, 2003, 13 (24) : 2212–2217. DOI: 10.1016/j.cub.2003.11.052 |
| [23] | Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet, 2005, 6 (5) : 351–360. DOI: 10.1038/nrg1601 |
| [24] | Henderson IR, Deleris A, Wong W, et al. The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet, 2010, 6 (10) : e1001182. DOI: 10.1371/journal.pgen.1001182 |
| [25] | Wu CT, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science, 2001, 293 (5532) : 1103–1105. DOI: 10.1126/science.293.5532.1103 |
| [26] | Ramsahoye BH, Biniszkiewicz D, Lyko F, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA, 2000, 97 (10) : 5237–5242. DOI: 10.1073/pnas.97.10.5237 |
| [27] | Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet, 2009, 10 (5) : 295–304. DOI: 10.1038/nrg2540 |
| [28] | Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet, 2008, 9 (6) : 465–476. DOI: 10.1038/nrg2341 |
| [29] | Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet, 2010, 11 (3) : 204–220. DOI: 10.1038/nrg2719 |
| [30] | Cokus SJ, Feng SH, Zhang XY, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature, 2008, 452 (7184) : 215–219. DOI: 10.1038/nature06745 |
| [31] | Zhang XY, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell, 2006, 126 (6) : 1189–1201. DOI: 10.1016/j.cell.2006.08.003 |
| [32] | Doerfler W. DNA methylation and gene activity. Annu Rev Biochem, 1983, 52 (1) : 93–124. DOI: 10.1146/annurev.bi.52.070183.000521 |
| [33] | Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet, 2007, 39 (1) : 61–69. DOI: 10.1038/ng1929 |
| [34] | Lister R, O'Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell, 2008, 133 (3) : 523–536. DOI: 10.1016/j.cell.2008.03.029 |
| [35] | Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science, 2009, 324 (5933) : 1447–1451. DOI: 10.1126/science.1171609 |
| [36] | Hsieh TF, Ibarra CA, Silva P, et al. Genome-wide demethylation of Arabidopsis endosperm. Science, 2009, 324 (5933) : 1451–1454. DOI: 10.1126/science.1172417 |
| [37] | Zemach A, Kim MY, Silva P, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA, 2010, 107 (43) : 18729–18734. DOI: 10.1073/pnas.1009695107 |
| [38] | Tran RK, Henikoff JG, Zilberman D, et al. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol, 2005, 15 (2) : 154–159. DOI: 10.1016/j.cub.2005.01.008 |
| [39] | Chodavarapu RK, Feng SH, Bernatavichute YV, et al. Relationship between nucleosome positioning and DNA methylation. Nature, 2010, 466 (7304) : 388–392. DOI: 10.1038/nature09147 |
| [40] | Matzke M, Kanno T, Daxinger L, et al. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol, 2009, 21 (3) : 367–376. DOI: 10.1016/j.ceb.2009.01.025 |
| [41] | Xie ZX, Johansen LK, Gustafson AM, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol, 2004, 2 (5) : e104. DOI: 10.1371/journal.pbio.0020104 |
| [42] | Herr AJ, Jensen MB, Dalmay T, et al. RNA polymerase IV directs silencing of endogenous DNA. Science, 2005, 308 (5718) : 118–120. DOI: 10.1126/science.1106910 |
| [43] | Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell, 2007, 128 (4) : 763–776. DOI: 10.1016/j.cell.2007.02.016 |
| [44] | Wassenegger M, Heimes S, Riedel L, et al. RNA-directed de novo methylation of genomic sequences in plants. Cell, 1994, 76 (3) : 567–576. DOI: 10.1016/0092-8674(94)90119-8 |
| [45] | Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell, 2008, 135 (4) : 635–648. DOI: 10.1016/j.cell.2008.09.035 |
| [46] | Huettel B, Kanno T, Daxinger L, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta, 2007, 1769 (5/6) : 358–374. |
| [47] | Pikaard CS, Haag JR, Ream T, et al. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci, 2008, 13 (7) : 390–397. DOI: 10.1016/j.tplants.2008.04.008 |
| [48] | Ream TS, Haag JR, Wierzbicki AT, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell, 2008, 33 (2) : 192–203. |
| [49] | Huang LF, Jones AME, Searle I, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol, 2009, 16 (1) : 91–93. DOI: 10.1038/nsmb.1539 |
| [50] | Kanno T, Huettel B, Mette MF, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet, 2005, 37 (7) : 761–765. DOI: 10.1038/ng1580 |
| [51] | Onodera Y, Haag JR, Ream T, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell, 2005, 120 (5) : 613–622. DOI: 10.1016/j.cell.2005.02.007 |
| [52] | He XJ, Su YF, Pontes O, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev, 2009, 23 (3) : 318–330. DOI: 10.1101/gad.1765209 |
| [53] | Lahmy S, Pontier D, Cavel E, et al. PolV (PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci USA, 2009, 106 (3) : 941–946. DOI: 10.1073/pnas.0810310106 |
| [54] | Yu HC, Zhao J, Xu J, et al. Detection of changes in DNA methylation induced by low-energy ion implantation in Arabidopsis thaliana. Radiat Res, 2011, 175 (5) : 599–609. DOI: 10.1667/RR2209.1 |
| [55] | Yang ZY, Ebright YW, Yu B, et al. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2' OH of the 3' terminal nucleotide. Nucleic Acids Res, 2006, 34 (2) : 667–675. DOI: 10.1093/nar/gkj474 |
| [56] | Wierzbicki AT, Ream TS, Haag JR, et al. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet, 2009, 41 (5) : 630–634. DOI: 10.1038/ng.365 |
| [57] | Zheng XW, Zhu JH, Kapoor A, et al. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J, 2007, 26 (6) : 1691–1701. DOI: 10.1038/sj.emboj.7601603 |
| [58] | Zilberman M, Tavares H, el-Guebaly N. Gender similarities and differences: the prevalence and course of alcohol-and other substance-related disorders. J Addict Dis, 2003, 22 (4) : 61–74. |
| [59] | Ausin I, Mockler TC, Chory J, et al. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol, 2009, 16 (12) : 1325–1327. DOI: 10.1038/nsmb.1690 |
| [60] | Zheng ZM, Xing Y, He XJ, et al. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J, 2010, 62 (1) : 92–99. DOI: 10.1111/j.1365-313X.2010.04130.x |
| [61] | Chuang LSH, Ian HI, Koh TW, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science, 1997, 277 (5334) : 1996–2000. DOI: 10.1126/science.277.5334.1996 |
| [62] | Iida T, Ishihara H. Study of the mechanical interaction between an electromagnetic field and a nanoscopic thin film near electronic resonance. Opt Lett, 2002, 27 (9) : 754–756. DOI: 10.1364/OL.27.000754 |
| [63] | Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science, 1975, 187 (4173) : 226–232. DOI: 10.1126/science.1111098 |
| [64] | Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Genome Res, 1975, 14 (1) : 9–25. DOI: 10.1159/000130315 |
| [65] | Kankel MW, Ramsey DE, Stokes TL, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics, 2003, 163 (3) : 1109–1122. |
| [66] | Saze H, Mittelsten ScheidO, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet, 2003, 34 (1) : 65–69. DOI: 10.1038/ng1138 |
| [67] | Kakutani T, Munakata K, Richards EJ, et al. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics, 1999, 151 (2) : 831–838. |
| [68] | Takeda SI, Funaki S, Yumiba T, et al. Gastropleural fistula due to gastric perforation after lobectomy for lung cancer. Interact Cardiovasc Thorac Surg, 2005, 4 (5) : 420–422. DOI: 10.1510/icvts.2005.108779 |
| [69] | Jullien PE, Kinoshita T, Ohad N, et al. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell, 2006, 18 (6) : 1360–1372. DOI: 10.1105/tpc.106.041178 |
| [70] | Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell, 2000, 12 (3) : 357–368. DOI: 10.1105/tpc.12.3.357 |
| [71] | Jackson JP, Lindroth AM, Cao XF, et al. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature, 2002, 416 (6880) : 556–560. DOI: 10.1038/nature731 |
| [72] | Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J, 2002, 21 (24) : 6842–6852. DOI: 10.1093/emboj/cdf687 |
| [73] | Jackson JP, Johnson L, Jasencakova Z, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma, 2004, 112 (6) : 308–315. DOI: 10.1007/s00412-004-0275-7 |
| [74] | Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol, 2005, 25 (23) : 10507–10515. DOI: 10.1128/MCB.25.23.10507-10515.2005 |
| [75] | Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell, 2006, 18 (5) : 1166–1176. DOI: 10.1105/tpc.106.041400 |
| [76] | Lindroth AM, Shultis D, Jasencakova Z, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J, 2004, 23 (21) : 4286–4296. DOI: 10.1038/sj.emboj.7600428 |
| [77] | Johnson LM, Law JA, Khattar A, et al. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet, 2008, 4 (11) : e1000280. DOI: 10.1371/journal.pgen.1000280 |
| [78] | Pogribny I, Raiche J, Slovack M, et al. Dose-dependence, sex-and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun, 2004, 320 (4) : 1253–1261. DOI: 10.1016/j.bbrc.2004.06.081 |
| [79] | Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue-a possible mechanism contributing to radiation carcinogenesis?. Biochem Biophys Res Commun, 2005, 337 (2) : 526–533. DOI: 10.1016/j.bbrc.2005.09.084 |
| [80] | Kalinich JF, Catravas GN, Snyder SL. The effect of γ-radiation on DNA methylation. Radiat Res, 1989, 117 (2) : 185–197. DOI: 10.2307/3577319 |
| [81] | Tawa R, Kimura Y, Komura JI, et al. Effects of X-ray irradiation on genomic DNA methylation levels in mouse tissues. J Radiat Res, 1998, 39 (4) : 271–278. DOI: 10.1269/jrr.39.271 |
| [82] | Kovalchuk O, Burke P, Arkhipov A, et al. Genome hypermethylation in Pinus silvestris of Chernobyl-a mechanism for radiation adaptation?. Mutat Res, 2003, 529 (1/2) : 13–20. |
| [83] | Raiche J, Rodriguez-Juarez R, Pogribny I, et al. Sex-and tissue-specific expression of maintenance andde novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem Biophys Res Commun, 2004, 325 (1) : 39–47. DOI: 10.1016/j.bbrc.2004.10.002 |
| [84] | Boyko A, Blevins T, Yao YL, et al. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLoS ONE, 2010, 5 (3) : e9514. DOI: 10.1371/journal.pone.0009514 |
| [85] | Medeiros AM, Barbosa JE, Medeiros PR, et al. Salinity and freshwater discharge determine rotifer distribution at the Mossoró River Estuary (Semiarid Region of Brazil). Braz J Biol, 2010, 70 (3) : 551–557. |
| [86] | Lira-Medeiros CF, Parisod C, Fernandes RA, et al. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE, 2010, 5 (4) : e10326. DOI: 10.1371/journal.pone.0010326 |
| [87] | Sha AH, Lin XH, Huang JB, et al. Analysis of DNA methylation related to rice adult plant resistance to bacterial blight based on methylation-sensitive AFLP (MSAP) analysis. Mol Genet Genomics, 2005, 273 (6) : 484–490. DOI: 10.1007/s00438-005-1148-3 |
| [88] | Akimoto K, Katakami H, Kim HJ, et al. Epigenetic inheritance in rice plants. Ann Bot (Lond), 2007, 100 (2) : 205–217. DOI: 10.1093/aob/mcm110 |
 2016, Vol. 32
2016, Vol. 32