服务
文章信息
- 刘仕平, 黄亚玺, 尹纪云, 吴小燕, 周兰庭, 王伟, 夏庆友
- Shiping Liu, Yaxi Huang, Jiyun Yin, Xiaoyan Wu, Lanting Zhou, Wei Wang, Qingyou Xia
- 家蚕Bmyan基因的克隆表达和作为microRNA 7靶基因的验证
- Cloning and expression profile of Bmyan in the silkworm (Bombyx mori) and experimental validation as one target of microRNA 7
- 生物工程学报, 2015, 31(11): 1612-1622
- Chin J Biotech, 2015, 31(11): 1612-1622
- 10.13345/j.cjb.140608
-
文章历史
- Received: December 8, 2014
- Accepted: March 13, 2015
复眼 (Compound eye) 出现于节肢动物门 (Arthropoda) 的昆虫纲 (Insecta)、甲壳纲 (Crustacea) 和部分双壳纲 (Bivalvia) 动物,是由一定数量的单眼 (Ocellus) 组成的视觉器官。果蝇 (Fruit fly) 复眼由约800个小眼 (Ommatidial unit) 组成,每个小眼由20个细胞构成,即8个光感受细胞 (R8) 和12个副细胞。Yan是在果蝇中发现的表皮生长因子受体 (Drosophila epidermal growth factor receptor,DER) 通路中的转录抑制因子[1]。果蝇的miR-7和yan基因形成一个负调控回路,使它们表达量处于动态平衡,保证了果蝇眼光感受器的正常分化和复眼的形成[2, 3, 4]。yan基因存 在于光感受器中,编码的核蛋白含有ETS (E twenty six) DNA结合基序、一个PNT (Pointed) 结构域、8个分裂素激活蛋白 (Mitogen-activated protein,MAP) 的激酶磷酸化位点和许多PEST信号区域[5, 6, 7]。miR-7在果蝇本体感受器官和嗅觉器官的分化中有重要作用[1, 2],在许多相互关联的前馈和反馈通路中起稳定器的作用,以缓冲环境变化对生物体的影响[8]。
家蚕Bombyx mori作为鳞翅目Lepidoptera昆虫的典型代表对于揭示一些生命活动的分子调控机理有着十分重要的作用。家蚕基因组测序的完成使家蚕成为研究一些重要生命机理的模式生物[9, 10]。家蚕眼睛也是由许多单眼构成的复眼,但关于其形成和发育的分子调控机制并没有研究报道。已经在家蚕体内鉴定了bmo-miR-7[11, 12, 13, 14],但还未见其靶基因的研究报道。本研究旨在通过克隆鉴定家蚕的yan基因,研究其表达模式,并验证它是否是bmo-miR-7的靶基因,以便为进一步揭示bmo-miR-7的生物学功能和研究家蚕光感受器的分化和复眼的发育调控机理奠定基础。
1 材料与方法 1.1 材料和宿主菌家蚕大造品种由西南大学家蚕基因库提供。将材料用液氮速冻后保存于-80 ℃备用。大肠杆菌DH5α菌株为本实验室所保存。家蚕胚胎细胞系BmE为本实验室经原代培养建成。
1.2 试剂提取试剂TRIzol购自Invitrogen公司;反转录试剂盒、PCR试剂、pMD19-T载体、DNA marker、限制性内切酶BamHⅠ、Hind Ⅲ、KpnⅠ、XbaⅠ和SYBR Premix Ex TaqTMⅡ均购于TaKaRa公司;胶回收试剂盒购自上海华舜生物技术有限公司;载体pGL3[luc- SV40]和大肠杆菌DH5α菌株由本实验室保存;定量PCR用的反转酶M-MLV、PCR产物回收试剂盒和双荧光素酶检测试剂盒购自Promega公司;质粒提取试剂盒购自上海生工生物工程技术服务有限公司 (生工生物);优质胎牛血清 (FBS) 购自PAA公司;bmo-miR-7 mimic购自广州锐博生物科技有限公司 (Ribobio)。
1.3 基因的克隆 1.3.1 所需的引物用Primer premier 5.0软件设计引物。所有引物均由生工生物合成,见表1。
| Primer name | Purpose | Primer sequence (5′−3′) |
| A3-F1 | RT-PCR for A3 | AACACCCCGTCCTGCTCACTG |
| A3-R1 | RT-PCR for A3 | GGGCGAGACGTGTGATTTCCT |
| yan-CDS-F2 | CDS | ATGAAAGTTGTAAGCTTACA |
| yan- CDS-R2 | CDS | TTACTGTTCACATTTTTCAC |
| yan-RT-F3 | RT-PCR | TCTTACAACAACTTCTGAATGAC |
| yan-RT-R3 | RT-PCR | TATCTCCGATAAGCGTACTG |
| yan-qPCR-F4 | qPCR | ATGAAAGAAGAAAGATGTGACGAAG |
| yan-qPCR-R4 | qPCR | ATAAGCGTACTGATCCGATGTTTG |
| sw22934-F5 | qPCR control | TTCGTACTGCTCTTCTCGT |
| sw22934-R5 | qPCR control | CAAAGTTGATAGCAATTCCCT |
| 3′-UTR-out-F6 | 3′-UTR | CTCTACTGCGACAACAAATGAGTCCCAC |
| 3′-UTR-in-R6 | 3′-UTR | GTTCCTTCCTCACCTGTTACTCCCACC |
| 3′-UTR-out-R7 | 3′-UTR | TACCGTCGTTCCACTAGTGATTT |
| 3′-UTR-in-F7 | 3′-UTR | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| A3-Prom-F8 | Promoter of A3 | CGGAATTCAGAGGTTACAAGCGACCG |
| A3- Prom-R8 | Promoter of A3 | CGGGATCCCTTGAATTAGTCTGCAAGAAAA |
| yan-3′UTR-F9 | Vector | TGCTCTAGACTTATTGAGATAGACTAAGGACA |
| yan-3′UTR-R9 | Vector | TGCTCTAGATAGTTGTGGCGGAAAAGTA |
参照TRIzol试剂盒说明书和已报道的方法提取总RNA[15, 16],溶解于无RNA酶水中,1.2%琼脂糖凝胶电泳检测RNA的完整性,紫外分光光度计 (Thermo Scientific NanoDrop Lite) 检测RNA纯度和浓度。
1.3.3 Bmyan基因克隆和序列分析用OligodT和五龄3 d总RNA合成cDNA第一链。分别用Actin3和Bmyan的引物进行PCR扩增。反应条件为:94 ℃预变性4 min;94 ℃变性30 s,56 ℃退火 30 s,72 ℃延伸 1 min,30个循环;72 ℃ 延伸 10 min。依次对目的片段进行连接、转化、筛选和验证。最后将阳性质粒送生工生物测序。
1.4 家蚕Bmyan基因的表达模式利用RT-PCR,以不同发育时期整蚕和五龄3 d组织的总RNA合成的cDNA第一链为模板,A3作内参,分析Bmyan的表达。参照试剂盒和仪器说明书,用7500 Fast Real-Time PCR System对Bmyan进行荧光定量PCR检测。从http://www.silkdb.org/microarray/download.html[17]下载Bmyan在家蚕不同组织中表达的芯片数据进行分析。
1.5 Bmyan基因3′UTR的克隆用3′-Full RACE Core Set Ver.2.0试剂盒克隆Bmyan基因3′-UTR全长。将3.0 µL (约3 µg)总RNA变性后进行逆转录合成cDNA第一链。用cDNA第一链为模板进行套式PCR扩增。Outer PCR反应条件为:94 ℃预变性4 min;94 ℃变性30 s,62 ℃退火30 s,72 ℃延伸1 min,20个循环;最后72 ℃延伸10 min。Inner PCR反应条件为:94 ℃预变性4 min;94 ℃变性30 s,65 ℃ 退火30 s,72 ℃延伸1 min,30个循环;72 ℃延伸10 min。然后依次进行目的条带的回收、连接、转化、阳性质粒的筛选和检测。
1.6 Bmyan基因和3′-UTR的序列分析用果蝇YAN的氨基酸序列检索家蚕基因组数据库,得到编号为BGIBMGA001167-TA的基因,将之命名为Bmyan。通过NCBI的Blast程序进行比对,下载其他物种的YAN氨基酸序列。用在线SMART程序对Bmyan基因的蛋白序列进行结构域分析。将Bmyan基因的氨基酸序列与其他物种YAN的氨基酸序列用MEGA 6.0软件进行多序列比对,构建分子进化树。bmo-miR-7的序列UGGAAGACUAGUGAUUUUGUUGU下载于MiRBase (http://www.mirbase.org/)[18]。用RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)[19]在线预测Bmyan 3′-UTR序列上bmo-miR-7的靶位点。
1.7 基因Bmyan 3′-UTR的载体构建以五龄3 d整蚕的DNA为模板从家蚕基因组DNA上扩增A3启动子序列。反应参数:94 ℃预变性4 min;94 ℃变性30 s、55 ℃退火30 s、72 ℃延伸1 min,30个循环;最后72 ℃ 延伸10 min。用限制性内切酶KpnⅠ和SacⅠ对A3启动子PCR产物进行双酶切。将回收的酶切产物连接到经KpnⅠ和SacⅠ双酶切处理的pGL3 [luc-SV40]载体上,构建成载体pGL3[A3-luc- SV40]。从Bmyan的3′-UTR质粒DNA上扩增得Bmyan 的3′-UTR片段。PCR条件:94℃预变性4 min;94 ℃变性30 s,58 ℃退火30 s,72 ℃延伸1 min,30个循环;72 ℃终延伸10 min。用XbaⅠ分别单酶切Bmyan 3′-UTR的PCR产物和载体pGL3[A3-luc-SV40],回收酶切产物,构建成载体pGL3[A3-luc-Bmyan 3′UTR-SV40]。
1.8 载体pGL3[A3-luc-Bmyan 3′UTR-SV40]与bmo-miR-7 mimics共转染用家蚕胚胎细胞系BmE进行转染实验。两个对照组分别转染pGL3[A3-luc-SV40]和pGL3[A3-luc-Bmyan 3′UTR-SV40]质粒;实验组用重组质粒pGL3[A3-luc-Bmyan 3′UTR-SV40]和bmo-miR-7 mimic进行共转染。都加入1/10 pGL3质粒量的海肾荧光素酶质粒作为内参,以消除转染时各孔之间的误差。每组设3个生物学重复。以萤火虫荧光素酶的酶活性值除以相应海肾荧光素酶的酶活性值作为最后的相对荧光值进行统计分析。
2 结果与分析 2.1 Bmyan基因的克隆和序列分析 2.1.1 Bmyan基因的CDS克隆以五龄3 d头部的cDNA为模板进行PCR扩增,得到一条约1 400 bp的片段 (图1A)。回收PCR扩增的目的片段,依次进行连接、转化、筛选和培养,然后抽提质粒DNA (图1B)。用Bam HⅠ和Hind Ⅲ双酶切以及质粒PCR鉴定滞后质粒,发现酶切片段与PCR扩增片段大小一致,初步说明该质粒是重组质粒,该克隆是阳性克隆 (图1C)。将此克隆进行测序验证。经克隆、测序和拼接得到Bmyan的CDS全长序列与SilkDB数据库里的编号为BGIBMGA001167-TA的CDS一致。

|
| 图1 Bmyan基因的克隆 Fig.1 Cloning of Bmyan. (A) PCR product. M: marker DL2 000; 2: PCR product. (B) Retarded plasmid. 1: retarded plasmid; 2: blue clone. (C) PCR of plasmid and plasmid digested by BamHⅠand Hind Ⅲ. 1: PCR of Bmyan plasmid; 2: Bmyan plasmid digested by BamH I and Hind Ⅲ. |
Bmyan基因定位于13号染色体上,CDS长1 431 bp,编码476个氨基酸。Bmyan基因有4个外显子和3个内含子。它有SAM-PNT蛋白结构域和ETs蛋白结构域。SAM-PNT蛋白结构域在信号传导中起作用,而ETS蛋白结构域是一种转录因子,结合在基因组上特异的DNA序列上,诱导或者抑制基因转录,它包含3个α-螺旋和4股β-片层,组成一个螺旋-转角-螺旋结构,第3个α-螺旋负责与DNA的大沟结合。
用Bmyan的氨基酸序列在NCBI上进行Blast在线比对,发现家蚕的Bmyan的氨基酸序列与黑麦金斑蝶Danaus plexippus的最相似,有87%的一致性,与赤拟谷盗Tribolium castaneu和毁侧沟茧蜂Microplitis demolitor有62%的一致性,与斑马鱼Danio rerio有35% 的一致性。Bmyan蛋白与其他真核生物特别是昆虫的YAN蛋白序列相似性较高。用MEGA 6对不同物种YAN的氨基酸序列进行多序列比对,构建分子进化树 (图2)。由进化树可以看出,不同目和科的昆虫聚在不同的分支上,家蚕和同为鳞翅目昆虫的黑麦金斑蝶聚在一起,进化距离最近,与赤拟谷盗Tribolium castaneum和山松甲虫Dendroctonus ponderosae等鞘翅目昆虫也有较近的距离,但与蛛形纲Arachnoidea中的寄螨目Parasitiformes和脊椎动物中的斑马鱼Danio rerio有较远的进化距离。
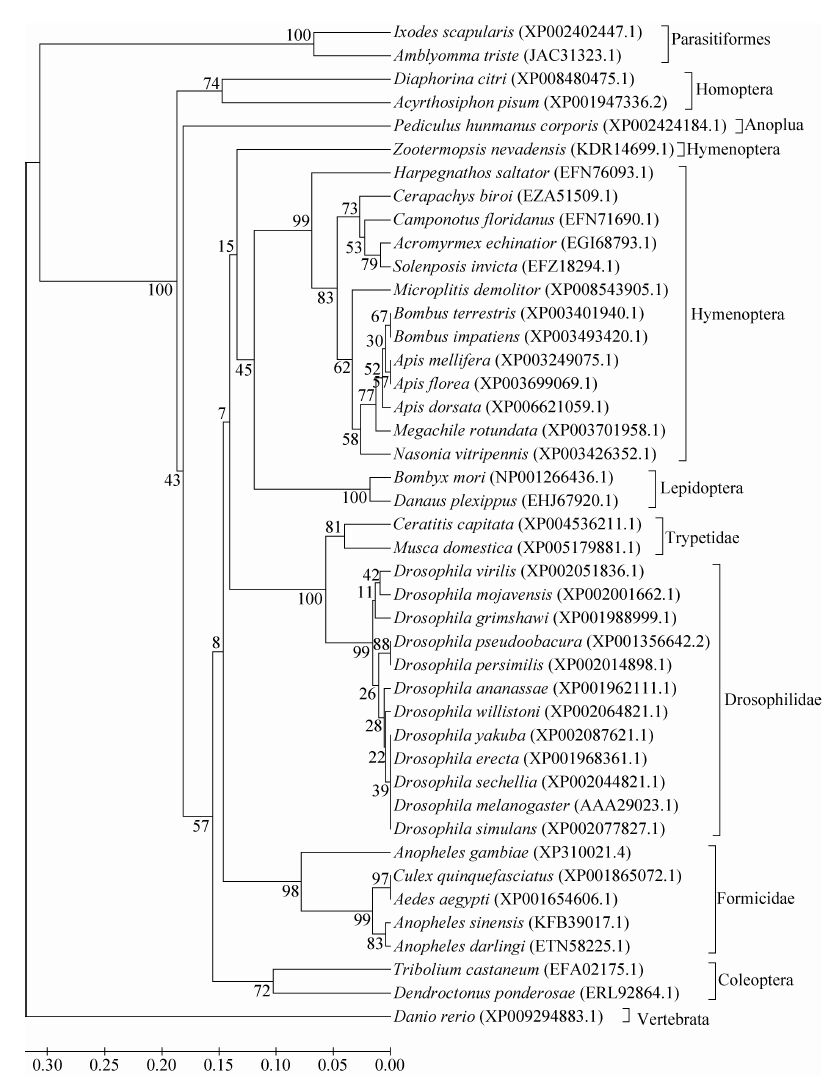
|
| 图2 家蚕Bmyan及其他生物同源体YAN蛋白氨基酸序列的分子系统进化树 Fig.2 Phylogenetic tree of Bmyan from B. mori and other species. |
基于RT-PCR检测,Bmyan在幼虫期,从一龄到五龄,Bmyan的表达量比较稳定,但在五
龄7 d和刚上蔟时无表达,从吐丝完毕到化蛹4 d时期高量表达,化蛹第5天和第6天低量表达,第7天又高量表达,蛹第8天低量表达 (图3A和3B),Bmyan在五龄3 d家蚕的头部、体壁和卵巢组织中高量表达,在中肠、精巢和血液中也有表达,而在其余组织中未检测到 (图3C)。下载家蚕全基因组芯片数据[17],分析Bmyan基因在家蚕五龄3 d不同组织中的表达模式,它在头部、体壁、血液和卵巢中表达量较高,在前中部丝腺、后部丝腺和脂肪体中低量表达或不表达 (图3D)。为了进一步验证Bmyan基因在家蚕不同组织中的表达,我们另外取了3组生物学重复材料,用定量PCR进行检测,结果与RT-PCR和芯片实验结果基本上是一致的,即在头部、体壁和卵巢中高量表达,在丝腺、脂肪体和精巢中表达量较低或不表达 (图3E)。

|
| 图3 家蚕Bmyan的表达模式 Fig.3 Expression profile of Bmyan in the silkworm. (A) RT-PCR based expression patterns of Bmyan in the larval stage of silkworm. eIL1: early 1st instar larva; eML1: early 1st molt larva; eIL2: early 2nd instar larva; eML2: early 2nd molt larva; eIL3: Early 3rd instar larva; eML3: early 3rd molt larva; eIL4: early 4th instar larva; eML4: early 4th molt larva; eIL5: early 5th instar larva. (B) RT-PCR based expression patterns of Bmyan in fifth instar and pupal stage. D3IL5: day-3 fifth instar larva; D4IL5: day-4 fifth instar larva; D5IL5: day-5 fifth instar larva; D6IL5: day-6 fifth instar larva; D7IL5: day-7 fifth instar larva; 0 h Sp: 0 h spinning; 12 h Sp: 12 h spinning; 24 h Sp: 24 h spinning; 36 h Sp: 36 h spinning; ePpu: early prepupa; ePu: early pupa; D1Pu: 1 d pupa; D2Pu: 2 d pupa; D3Pu: 3 d pupa; D4Pu: 4 d pupa; D5Pu: 5 d pupa; D6Pu: 6 d pupa; D7Pu: 7 d pupa; D8Pu: 8 d pupa; eAd: early adult moth. (C) RT-PCR based spatial expression profile of Bmyan in fifth instar larval silkworm. HD: head of silkworm; BW: body wall; MG: midgut; FB: fat body; SG: silk gland; MT: malpighian; HC: hemocyte; OV: ovary; TE: testis. (D) Microarray-based spatial expression of Bmyan in fifth instar larval silkworm. HD: head of female; BW: body wall; A/MSG: anterior/middle silk gland. PSG: posterior silk gland; MG: midgut; FB: fat body; HC: hemocyte; MT: malpighian; OV: ovary; TE: testis. The microarray data was downloaded from http://www.silkdb.org/microarray/download.html[17]. (E) qPCR-based spatial expression of Bmyan in fifth instar larval silkworm. HD: head of silkworm; BW: body wall; SG:anterior/middle silk gland; MG: midgut; FB: fat body; HC: hemocyte; MT: malpighian; OV: ovary; TE: testis. Error bars . |
按3′RACE试剂盒说明书对五龄3 d家蚕的头部总RNA进行反转录,然后对Bmyan的3′UTR进行套式PCR扩增,得到约1 000 nt的目的片段 (图4A)。将切胶回收的目的片段连接到pMD19-T载体,连接产物转化DH5α感受态细胞,再依次进行铺板、挑斑和培养。提取质粒DNA电泳检测滞后质粒 (图4B)。用质粒PCR对滞后质粒进行鉴定,质粒PCR片段与套式 PCR扩增片段大小一致 (图4C),说明该质粒是重组质粒,对应的克隆是阳性克隆。将此克隆测序得到Bmyan 3′UTR (长度为571 nt),序列提交至GenBank,序列号为KT934311。

|
| 图4 Bmyan的3′UTR克隆 Fig.4 Cloning of Bmyan 3′UTR. (A) 3′RACE PCR products. M: marker DL2 000; 1: 3′RACE PCR product of Bmyan. (B) retarded plasmid. M: marker DL2 000; 1: blue clone; 2: retarded plasmid of Bmyan 3′RACE. (C) PCR of plasmid. M: marker DL2000; 1: PCR product of Bmyan 3′RACE plasmid. |
用RNAhybrid在线软件预测bmo-miR-7的靶位点,发现在Bmyan 3′UTR的97-117和479-498碱基处分别有一个靶位点 (图5A)。97-117靶位点的mfe为-27.9 kcal/mol,479-498靶位点的mfe为-21.6 kcal/mol。基于预测,家蚕Bmyan可能是bmo-miR-7的靶基因。

|
| 图5 Bmyan 3′-UTR上bmo-miR-7靶位点的预测和验证 Fig.5 Target site prediction and experimental confirmation of bmo-miR-7 within the 3′-UTR of Bmyan. (A) Predicted target sites of bmo-miR-7 within Bmyan 3′-UTR. (a) target site at 97th nt of 3′-UTR. (b) target site at 479th nt of 3′-UTR. (B) Construction of luciferase reporter. (a) PCR of A3 promoter. M: marker DL2 000; 1: PCR product of A3 promoter. (b) plasmid digested with Kpn I and Sac I. M: marker DL2 000; 1: pGL3[A3-luc-SV40] plasmid; 2: pGL3[A3-luc-SV40] plasmid digested with Kpn I and Sac I. (c) PCR of Bmyan 3′-UTR. M: marker DL2 000; 1: PCR product of Bmyan 3′-UTR. (d) plasmid digested with Xba I. M: marker DL2 000; 1: pGL3[A3-luc-Bmyan 3′UTR-SV40]; 2: product of pGL3[A3-luc-Bmyan 3′UTR-SV40] digested with Xba I. (C) Target validation by luciferase reporter assay. RFV: relative fluorescence value; Control 1: pGL3[A3-luc-SV40]; Control2: pGL3[A3-luc-Bmyan 3′UTR-SV40]; 3′-UTR+miR-7: Co-transfection of pGL3[A3-luc-Bmyan 3′UTR-SV40] and bmo-miR-7 mimics. *P<0.05. Error bars . |
为了验证Bmyan是否是bmo-miR-7的靶基因,我们构建了含有Bmyan 3′UTR的荧光素酶报告基因载体。先以五龄3 d整蚕的cDNA为模板,扩增A3启动子序列,得到一条约750 bp的片段 (图5B-a),用限制性内切酶KpnⅠ和SacⅠ双酶切A3的PCR产物,将回收得到的A3启动子片段连接到经KpnⅠ和SacⅠ双酶切的pGL3 [luc- SV40]载体上,构建载体pGL3[A3-luc-SV40] (图5B-b)。以Bmyan 3′UTR质粒载体为模板,扩增得到Bmyan 3′-UTR片段 (图5B-c)。用限制性内切酶XbaⅠ单酶切Bmyan 3′-UTR的PCR产物,纯化后获得Bmyan 3′UTR片段,连接到经XbaⅠ酶切的pGL3[A3-luc-SV40]载体上,构建成转染载体pGL3[A3-luc-Bmyan 3′UTR-SV40] (图5B-d)。共转染48 h后测出每一组的荧光素酶活性值。与对照相比,实验组的荧光素酶活比值明显下降。bmo-miR-7 mimics对于pGL3[A3-luc-Bmyan 3′UTR-SV40]的表达是有抑制作用,所以Bmyan是bmo-miR-7的靶基因。
3 讨论Bmyan在家蚕头部表达量最高 (图3),暗示其在家蚕脑或神经系统以及眼睛中有重要的调控作用。在家蚕胚胎发生初期,控制形态发生的早期基因就开始表达,调控家蚕光感受器的分化、复眼的发育以及神经系统和其他器官的发生。在胚胎发生期间,Bmyan可能起着重要的调控作用。yan基因从果蝇胚胎发育早期开始表达,在果蝇三龄幼虫中,yan基因在眼成虫盘 (Imaginal disc) 形态发生沟的内部和后面以及脑叠状前体细胞中均有表达[20]。除了调控眼的发育[21, 22, 23],yan基因在果蝇形态发生中也有重要作用[24]。Bmyan在家蚕五龄3 d幼虫的体壁、中肠、卵巢、马氏管和血液中也有较高的表达量,但在丝腺和脂肪体中表达量很低或不表达,暗示除了神经系统和眼睛,Bmyan在家蚕幼虫期其他器官或非神经细胞中也有调控作用,或许与维持细胞的去分化状态有关。
YAN蛋白的组成和初级结构是高度保守的 (图2)。miR-7从无脊椎动到脊椎动物都具有高度的保守性。这暗示了它们在这些生物中都具有重要的调节作用。miR-7在脊椎动物脑的神经分泌细胞中特异表达,也存在于无脊椎动物神经系统中[8, 25, 26]。Bmo-miR-7存在于家蚕的头部、体壁、卵巢和精巢中[13],暗示它与家蚕这些组织和器官的分化和发育或细胞的代谢调节有关。我们找到了bmo-miR-7在Bmyan 3′-UTR的两个靶位点 (图5A),细胞水平实验证实了bmo-miR-7对Bmyan起到负调控作用 (图5C),后面有必要对这两个靶位点的效率进行比较,以更好地研究bmo-miR-7与Bmyan之间的调控关系和它们在家蚕体内的生物学功能。
4 结论本研究对家蚕Bmyan进行了克隆和表达分析,克隆了Bmyan的3′-UTR序列并预测了bmo-miR-7的靶位点;构建了含有家蚕特异启动子A3和Bmyan 3′-UTR序列的荧光素酶报告基因载体,在细胞水平上验证了bmo-miR-7和Bmyan基因之间的调控关系,为以后深入研究bmo-miR-7和Bmyan的功能奠定了基础。
| [1] | Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell, 1992, 70(4): 609-620. |
| [2] | Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell, 2005, 123(7): 1267-1277. |
| [3] | Boisclair Lachance JF, Peláez N, Cassidy JJ, et al. A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling. Dev Biol, 2014, 385(2): 263-278. |
| [4] | Vivekanand P, Tootle Tl, Rebay I. MAE, a dual regulator of the EGFR signaling pathway, is a target of the Ets transcription factors PNT and YAN. Mech Dev, 2004, 121(12): 1469-1479. |
| [5] | Lai ZC, Fetchko M, Li Y. Repression of Drosophila photoreceptor cell fate through cooperative action of two transcriptional repressors Yan and Tramtrack. Genetics, 1997, 147(3): 1131-1137. |
| [6] | Mavrothalassitis G, Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene, 2000, 19(55): 6524-6532. |
| [7] | Hsu T, Schulz RA. Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene, 2000, 19(55): 6409-6416. |
| [8] | Li X, Cassidy JJ, Reinke CA, et al. A microRNA imparts robustness against environmental fluctuation during development. Cell, 2009, 137(2): 273-282. |
| [9] | Xia QY, Zhou ZY, LU C, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science, 2004, 306(5703): 1937-1940. |
| [10] | Mita K, Kasahara M, Sasaki S, et al. The genome sequence of silkworm, Bombyx mori. DNA Res, 2004, 11(1): 27-35. |
| [11] | Liu SP, Zhang L, Li QB, et al. MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori). BMC Genomics, 2009, 10: 455. |
| [12] | Liu SP, Li D, Li QB, et al. MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genomics, 2010, 11: 148. |
| [13] | Liu SP, Gao S, Zhang DY, et al. MicroRNAs show diverse and dynamic expression patterns in multiple tissues of Bombyx mori. BMC Genomics, 2010, 11: 85. |
| [14] | Yu X, Zhou Q, Li SC, et al. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS ONE, 2008, 3(8): e2997. |
| [15] | Liu SP, Xia QY, Zhao P, et al. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev Biol, 2007, 7: 88. |
| [16] |
Liu SP, Xia QY. Protocol of Northern blotting hybridization for microRNA detection of silkworm (Bombyx mori). Sci Sericul, 2014, 40(4): 724-729 (in Chinese). 刘仕平, 夏庆友. Northern杂交检测家蚕microRNA的技术流程. 蚕业科学, 2014, 40(4): 724-729. |
| [17] | Xia QY, Cheng DJ, Duan J, et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol, 2007, 8(8): R162. |
| [18] | Kozomara A, Griffiths-jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res, 2014, 42 (Database issue): D68-D73. |
| [19] | Rehmsmeier M, steffen P, hochsmann M, et al. Fast and effective prediction of microRNA/target duplexes. RNA, 2004, 10(10): 1507-1517. |
| [20] | Gabay L, Scholz H, Golembo M, et al. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development, 1996, 122(11): 3355-3362. |
| [21] | Behan KJ, Nichols CD, Cheung TL, et al. Yan regulates Lozenge during Drosophila eye development. Dev Genes Evol, 2002, 212(6): 267-276. |
| [22] | Ramos E, Price M, Rohrbaugh M, et al. Identifying functional cis-acting regulatory modules of the yan gene in Drosophila melanogaster. Dev Genes Evol, 2003, 213(2): 83-89. |
| [23] | Rohrbaugh M, Ramos E, Nguyen D, et al. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr Biol, 2002, 12(7): 576-581. |
| [24] | Salzer CL, Elias y, Kumar JP, et al. The retinal determination gene eyes absent is regulated by the EGF receptor pathway throughout development in Drosophila. Genetics, 2010, 184(1): 185-197. |
| [25] | Lu ZJ, Liu SY, Yao YQ, et al. The effect of miR-7 on behavior and global protein expression in glioma cell lines. Electrophoresis, 2011, 32(24): 3612-3620. |
| [26] | Chen H, Shalom-Feuerstein R, Riley J, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun, 2010, 394(4): 921-927. |
 2015, Vol. 31
2015, Vol. 31




