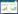中国科学院微生物研究所,中国微生物学会
文章信息
- 陈璐, 刘延琳, 秦义. 2024
- CHEN Lu, LIU Yanlin, QIN Yi.
- 酿酒酵母高级醇代谢的转录调控研究进展
- Transcriptional regulation of higher alcohol metabolism in Saccharomyces cerevisiae
- 微生物学报, 64(12): 4669-4680
- Acta Microbiologica Sinica, 64(12): 4669-4680
-
文章历史
- 收稿日期:2024-05-16
- 网络出版日期:2024-09-26
2. 国家林业和草原局葡萄与葡萄酒工程技术研究中心, 陕西 杨凌 712100;
3. 西北农林科技大学合阳葡萄试验示范站, 陕西 渭南 714000
2. National Forestry and Grassland Administration Engineering Research Center for Viti-viniculture, Yangling 712100, Shaanxi, China;
3. Heyang Viti-viniculture Station, Northwest A&F University, Weinan 714000, Shaanxi, China
全球气候变暖给葡萄酒产业带来了深刻影响,其中最为突出的是葡萄采收时的含糖量在过去20年里逐年攀升[1]。近年我国西北葡萄酒产区主栽红色酿酒葡萄的含糖量普遍超过260 g/L。葡萄的高含糖量不仅使发酵困难、酒度升高,还会导致葡萄酒高级醇含量显著升高。高级醇也称为“杂醇油”,指含有3个及以上碳原子的一元醇,是酿酒酵母的主要代谢副产物[2-5]。研究表明,葡萄酒中的高级醇含量小于300 mg/L时,能够赋予葡萄酒令人愉悦的风味,当超过400 mg/L则会对葡萄酒带来辛辣、腐臭等不愉悦的杂味,从而导致葡萄酒香气不纯净、感官品质劣变等问题[6]。更重要的是,高级醇含量过高易引发头痛、口渴、醒酒慢等不良症状,对饮用者构成安全隐患[7]。随着葡萄原料含糖量逐年增加,这一问题表现得越来越突出,已成为阻碍我国西北产区葡萄酒质量飞跃的重要问题。因此,亟须在葡萄酒酿造过程中合理控制高级醇含量。
葡萄酒中高级醇的产生与多种发酵环境和工艺条件相关,如发酵基质的同化氮含量和类型[7]、糖含量[8]和添加方法、发酵温度、发酵基质的初始pH值及溶氧量等[9-10]。面对复杂的发酵环境条件和多样的工艺措施,酿酒酵母可通过基因转录、转录后翻译和翻译后加工等多层次的调控体系,精细调节高级醇的合成代谢,其中转录水平的调控发生在基因表达的初期阶段,通常是代谢调控的主要方式之一。目前酿酒酵母的高级醇代谢途径已梳理清楚,代谢途径中的酶系及其编码基因已基本明确[11],针对酿酒酵母高级醇代谢的转录调控研究较少,在酿酒酵母高级醇合成代谢中发挥关键作用的转录因子及其转录调控机制尚不全面,这也导致对酿酒酵母的定向代谢工程育种存在代谢调控效果不理想等诸多问题。本文综述了酿酒酵母高级醇代谢途径和参与高级醇代谢的相关转录因子,旨在系统了解酿酒酵母高级醇代谢及转录调控机制,为改良和选育高级醇产量适中的优良酿酒酵母菌种提供理论支持。
1 高级醇代谢途径葡萄酒中高级醇的主体组分为正丙醇、异戊醇、异丁醇、苯乙醇和活性戊醇等,约占高级醇总量的70%。酿酒酵母的高级醇主要经由糖合成代谢途径(Harris途径)和氨基酸分解代谢途径(Ehrlich途径)合成(图 1)[12]。Harris途径主要指葡萄糖/果糖的糖酵解终产物α-酮酸,经2-酮基酸脱羧酶(KDCs)脱羧、脱氢酶(ADHs)脱氢还原成活性戊醇、异丁醇、异戊醇等高级醇;Ehrlich途径是氨基酸在氨基转移酶(BATs)的催化作用下生成α-酮酸,α-酮酸在脱羧酶的作用下生成相应的醛和CO2,醛经还原生成相应的醇。酿酒酵母所产生的高级醇约75%来自Harris途径,约25%来自Ehrlich途径[7]。
酵母的高级醇合成代谢是多基因控制的复杂性生物学过程,涉及多个调控因子。现有研究采用了过量表达/敲除高级醇类物质的代谢途径基因[13],删除/增强竞争途径[14],改变辅因子水平[15],或在线粒体或细胞质重构代谢途径[16]等策略,调控了酵母的高级醇代谢模式。其中,研究人员通过删除非特异性氨基酸转运蛋白编码基因GAP1、AGP1[10]以及支链氨基酸分解途径的BAT2[17],敲除α-酮酸合成代谢途径的LEU1、LEU2、GDH1[18],删除α-酮酸分解代谢途径的HOM2、PAD1、QCR2、SPE1、ALD6[19],过量表达乙酸酯代谢基因ATF1或同时敲除IAH1基因[20]等,均在一定程度上降低了酵母的高级醇产量。
对碳、氮等代谢通路上的单(多)基因敲除或过量表达,在削弱酿酒酵母高级醇合成途径代谢能力的同时,可能会导致乙酸等代谢副产物的增加,这些代谢副产物将严重影响微生物细胞生长和损伤葡萄酒感官品质,以及导致工程菌株发酵特性的改变。因此,通过对高级醇合成途径转录调控的精细优化,可能实现在基因组层面上定向进化或同时改变多个相关基因群,实现精准弱化高级醇合成能力的目的。
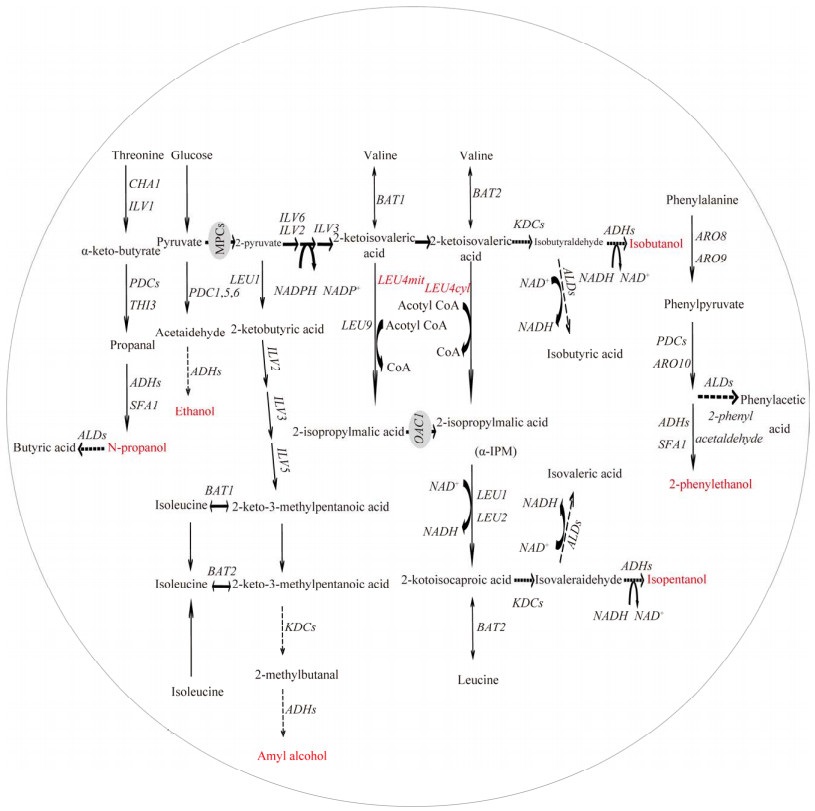
2 高级醇转录调控 2.1 转录因子结构转录因子(transcription factor, TF)通过识别并结合到DNA上的特定序列(顺式作用元件、增强子或沉默子),激活或抑制相关基因的转录,进而影响高级醇的合成。根据与DNA结合结构域类型[21],TF可分为3类,包括锌簇(Zn2+)稳定型、螺旋-旋转-螺旋型和拉链型。其中,Zn2+稳定型的DNA结合结构域在各种生物体中普遍存在。它的功能域模型如图 2A所示,整个DNA结合结构域(DNA-binding domain, DBD)分为3个区域:锌指区、连接体区和二聚区。锌簇蛋白有一个DBD,其中包含2个Zn2+,并以6个半胱氨酸(Cys)残基作为配体[22]。在多数锌簇TF的结构中存在一个显著特征,即DBD,位于TF的氨基端(N端)。这一结构域负责识别并结合特定的DNA序列,从而调控基因的表达。与此同时,与DNA序列直接相互作用并参与特异性结合的残基,则位于TF的羧基端(C端)[23] (图 2B)。这种结构使TF能够以高效且特异的方式与DNA相互作用,进而调控转录过程。
|
| 图 2 锌簇转录因子结构 Figure 2 The structure of Zinc finger transcription factors. A: Exploring the functional domain of Zinc cluster proteins. B: The TF-DNA binding spatial structure. MHR: Middle homology region. |
2.2 与高级醇代谢相关的转录因子
目前已知的与高级醇代谢相关的转录因子有Aro80p[24]、GATA[25]和Leu3p[26-29]。
2.2.1 Aro80pAro80p属于Zn2Cys6蛋白家族,由ARO80基因编码,其能够对芳香族氨基酸作出响应,并促进ARO9和ARO10基因的表达[30]。ARO8和ARO9编码的芳香族转氨酶I和II催化氨基酸形成相应的α-酮酸类似物,在ARO10编码的芳香族脱羧酶的作用下,进一步转化为2-苯乙醇、色醇和酪醇[31] (图 1)。
Lee等[32]研究发现,ARO9和ARO10基因的表达会被Aro80p激活,从而促进2-苯乙醇的合成;ARO80基因的缺失显著降低2-苯乙醇的合成。然而同时也发现,利用AgTEF1启动子过表达ARO80,会导致支链氨基酸分解代谢衍生的异戊醇和异丁醇水平增加2.5倍,而2-苯乙醇的水平基本不变[33]。
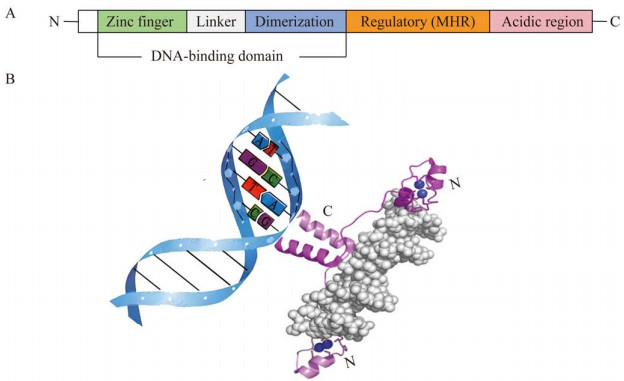
Aro80p的N末端具有1个DBD,能够与靶基因启动子序列上一段特定的36个碱基对(bp)长度的激活序列相结合。这一激活序列最初在ARO9和ARO10基因的启动子序列中被发现,它由4组CCG序列组成,每组CCG之间相隔7 bp (图 3);此外,在ARO80基因的启动子序列中也证实了这一激活序列的存在[30]。若要Aro80p与ARO9和ARO10启动子上的特定重复序列CCG相结合,还需芳香族氨基酸协助触发诱导信号。当培养基中氮源充足时,通过氮代谢物的阻遏抑制氨基酸通透酶编码基因GAP1表达,导致质膜上Gap1p失活。然而,当氮源只有l-苯丙氨酸存在时,Gap1p的转运活性将恢复,使得芳香族氨基酸在Gap1p的协助下进入细胞并参与代谢过程[34-35],进而促使Aro80p与靶基因启动子的特定重复序列相结合发挥作用(图 4)。这种特定的结合触发了ARO9与ARO10基因的表达活性,从而实现对2-苯乙醇的有效调控。因此,可以看出,Aro80p不仅直接参与了ARO9、ARO10和ARO80的调控,也间接影响了氨基酸转运酶。
|
| 图 4 转录因子Aro80p调控机制 Figure 4 Mechanism of Aro80p transcription factor regulation. When nitrogen sources are available in abundance, Gap1p becomes inactive, resulting in the inability of Aro80p to function. Conversely, when l-phenylalanine alone serves as the nitrogen source, the transport activity of Gap1p is restored, allowing it to enter the cell and thereby prompting Aro80p to function. |
2.2.2 GATA转录因子
GATA家族作为一类具有IV型锌指结构的转录调控因子,具有特异性识别并结合GATA基序(motif)的能力[36]。在酿酒酵母中,氮代谢相关基因的表达由4个GATA家族转录因子共同调控,它们是Gat1p、Gln3p、Gzf3p和Dal80p[37-38]。这4个转录因子具有一个共同的特征性锌指部分,使其能够结合到上游激活序列上,该区域位于受氮调控的基因上游约数百个碱基对处,并激活(Gat1p和Gln3p)或抑制(Gzf3p和Dal80p)转录的启动。
当敲除酿酒酵母GAT1和GLN3基因后,与高级醇代谢相关的基因表达水平显著降低[39]。Wang等[17]发现,GAT1缺失菌株中高级醇总含量增加28.36%,达到615.73 mg/L。GLN3的缺失可增加酵母对异丁醇的耐受性,异丁醇产量提高了4.9倍[40]。此外,尽管目前暂未发现Dal81p是否参与了酵母的高级醇代谢调控,但有研究显示Dal80p可能通过调节尿素代谢参与了氨基甲酸乙酯的调控[41]。Dal80p的同系物Dal81p与前者的作用明显不同,Dal81p是从尿囊素、尿素和亮氨酸获取氮的正调节因子[42]。
GATA家族Gat1p和Gln3p作为转录激活因子调控酿酒酵母中的氮分解代谢物抑制(nitrogen catabolism repression, NCR)。当微生物处于含有多种氮源的环境中时,它们会优先选择利用易于同化且能量效率高的氮源(如氨、谷氨酸等),而抑制利用成本较高或效率较低的氮源相关基因的表达[43]。GLN3基因的分离和测序结果表明,在306−330区域,Gln3p有1个锌指结构,与高等生物的GATA转录因子具有显著同源性[30]。免疫沉淀实验显示,Gln3p与GLN1基因启动子上游5′-GATAA/G-3′序列相结合[44]。在酵母中,除了已知的Gln3p调控因子外,还发现了氮代谢途径中的另一个重要激活因子Gat1p。该因子与Gln3p都具有GATA锌指基序,对于NCR敏感基因的表达至关重要,并呈现出轻微的转录激活潜力[45]。Gat1p能够与目标基因的启动子中位于310−334区域的特定序列“5′-GATAA/G-3′”进行结合[44]。在氮源充足的情况下,Tor1p及其他未知的磷酸激酶被激活,致使Gat1p和Gln3p发生磷酸化,并被核膜排斥至细胞质中,阻碍其参与靶基因的转录调控[46]。当Gat1p和Gln3p处于非活化状态时,多个基因的转录受到抑制,包括DUR1、DUR2 (编码脲基酰胺酶)、DUR3 (编码尿素渗透酶)、CAR1 (编码精氨酸酶);而在氮源匮乏时,Gat1p和Gln3p进入细胞核,并与NCR敏感基因启动子上的GATA序列结合,进而激活转录[47] (图 5)。
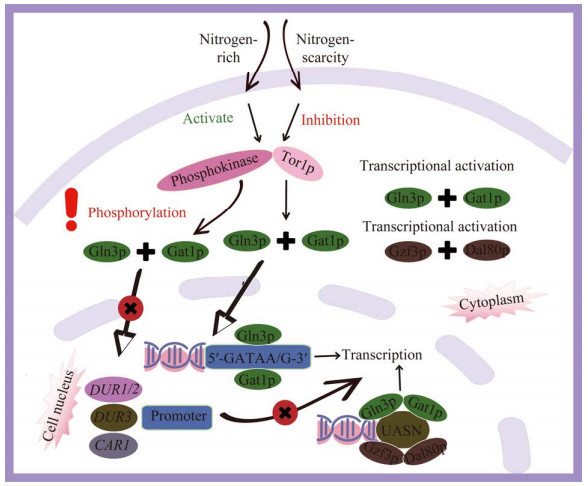
|
| 图 5 GATA家族转录因子调控机制 Figure 5 Mechanisms of regulation by GATA family transcription factors. When nitrogen sources are abundant, Gat1p and Gln3p undergo phosphorylation, preventing their entry into the nucleus and impeding their involvement in transcriptional regulation. Conversely, in a condition with limited nitrogen sources, Gat1p and Gln3p are allowed to enter the nucleus to participate in transcriptional regulation. |
2.2.3 Leu3p
Leu3p作为转录调控因子,可能参与了包括异戊醇和异丁醇等高级醇类物质的代谢调控[48-49]。Leu3p是一个Zn(II)2Cys6锌簇蛋白转录因子,结合DNA翻转重复序列CCG-N4-CGG,LEU3基因的表达受Gcn4介导的通用氨基酸控制(general amino acid control, GAAC);Leu3p自身的激活程度取决于细胞内Leu3p浓度,也受酵母细胞内亮氨酸/异戊醇生物合成途径的中间代谢物α-异丙基苹果酸(α-isopropylmalate, α-IPM)的调控;α-IPM是Leu3p的共激活因子,当α-IPM充足时,形成Leu3-α-IPM复合物,发挥激活因子功能,反之Leu3p则为转录阻遏物[50-51] (图 6)。
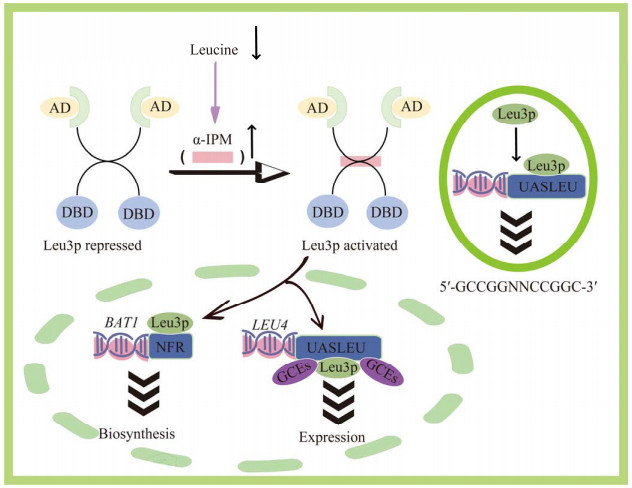
|
| 图 6 转录因子Leu3p调控机制 Figure 6 Mechanism of Leu3p transcription factor regulation. α-IPM serves as a coactivator for Leu3p. When α-IPM is abundant, it forms a Leu3-α-IPM complex, which then functions as an activator. Conversely, when α-IPM is in short supply, Leu3p acts as a transcriptional repressor. |
转录因子Leu3p对异戊醇合成途径编码基因BAT1和LEU4的调控机制相对清晰。BAT1编码线粒体支链氨基酸转氨酶,催化Ehrlich途径第1步反应(转氨作用);Bat1p主要参与支链氨基酸的生物合成[51],在对数生长期内表现为快速表达并大量积累,进入稳定期后其表达受到抑制。在生物合成阶段BAT1的表达主要受到Leu3-α-IPM的激活调控,其中,Leu3p结合于无核小体区域(nucleosome-free regions, NFR)[26]的BAT1侧翼区。LEU4编码的α-异丙基苹果酸合酶(Leu4p)是亮氨酸/异戊醇生物合成途径的关键调控酶,受亮氨酸反馈抑制[52]。LEU4启动子的主要调控元件是Leu3p结合元件(upstream active sequence, UAS)和2个一般控制响应Gcn4p结合元件(genetic code expansions, GCEs)[53]。亮氨酸对LEU4的调节是通过α-IPM进行的。当细胞内亮氨酸供应不足时,其对α-异丙基苹果酸合酶的反馈抑制作用减弱,引起α-IPM水平升高并与Leu3p相互作用,从而激活LEU4表达[52]。
尽管转录因子Leu3p对BAT1和LEU4的作用机制相对清晰,但通过YEASTRACT数据库[54]分析发现,Leu3p与异戊醇和异丁醇代谢途径的16个基因也有直接或间接的关系。其中,Leu3p激活OAC1、ALD5、BAP2、LEU1/2、ILV5、PDC5/6、THI3、ADH2/6、GAP1的表达(或者共表达),激活或抑制BAT2、ILV2/3表达,抑制ILV6的表达(图 7)。

|
| 图 7 酿酒酵母高级醇代谢途径编码基因与酿酒酵母转录因子(Leu3p)的关系 Figure 7 Relationship between coding genes involved in the higher alcohols metabolic pathway and transcription factors (Leu3p) in Saccharomyces cerevisiae. Solid lines indicate direct DNA-binding evidence derived from techniques such as ChIP/ChIP-seq, while dashed lines represent indirect associations inferred from transcriptome sequencing data. Green lines signify activation, red lines indicate inhibition, and brown lines imply a dual role of either activation or inhibition, depending on the context. |
3 展望
高级醇是包括葡萄酒在内的饮料酒重要风味物质和潜在健康危害因子,主要由酿酒酵母代谢产生。由于酿酒酵母高级醇代谢转录调控机制不明确,导致葡萄酒酿造中针对高级醇含量的控制缺乏有效策略。因此,需要进一步在转录、转录后、翻译和翻译后加工等水平探究高级醇代谢的精细调控。尽管目前已有实验证据表明转录因子Aro80p、GATA和Leu3p与酿酒酵母高级醇代谢有关,同时借助转录组学、基因组学等组学技术预测了更多与高级醇代谢具有潜在关系的转录因子,但是这些转录因子是在无约束条件下的分析结果,无法明确指出哪些转录因子是调控酿酒酵母高级醇合成代谢的关键转录因子。因此,在全基因组范围内挖掘和验证调控酿酒酵母高级醇的关键转录因子,是首要解决的关键科学问题;其次,通过关键转录因子靶向基因的解析,回答在不同发酵阶段关键转录因子通过何种方式调控酿酒酵母高级醇的合成代谢能力,从而揭示酿酒酵母高级醇合成代谢的调控机制,是需要解决的第2个关键科学问题。
对调控酿酒酵母关键转录因子的挖掘及其作用机制的解析,对于深刻系统理解酿酒酵母的高级醇代谢调控机制,为酿酒酵母高级醇代谢的精细调控提供新的思路,也为选育能够产出适量高级醇、适用于包括葡萄酒在内的各类饮料酒生产的优质酿酒酵母菌种,提供了重要的科学依据和实践指导,具有深远的意义。
作者贡献声明
陈璐:构思、论文撰写;刘延琳:修改;秦义:修改。
作者利益冲突公开声明
作者声明没有任何可能会影响本文所报告工作的已知经济利益或个人关系。
| [1] | GONEN LD, TAVOR T, SPIEGEL U. Adapting and thriving: global warming and the wine industry[J]. SAGE Open, 2024, 14(1): 21582440241227750 DOI:10.1177/21582440241227750. |
| [2] | QIN YN, XU HT, SUN JS, CHENG XY, LEI J, LIAN WJ, HAN C, HUANG WT, ZHANG MW, CHEN Y. Succession of microbiota and its influence on the dynamics of volatile compounds in the semi-artificial inoculation fermentation of mulberry wine[J]. Food Chemistry: X, 2024, 21: 101223 DOI:10.1016/j.fochx.2024.101223. |
| [3] | LIANG LH, MA YW, JIANG ZZ, SAM FE, PENG S, LI M, WANG J. Dynamic analysis of microbial communities and flavor properties in Merlot wines produced from inoculation and spontaneous fermentation[J]. Food Research International, 2023, 164: 112379 DOI:10.1016/j.foodres.2022.112379. |
| [4] |
孙细珍, 杜佳炜, 黄盼, 张帆, 刘源才. 现代工艺和传统工艺酿造小曲清香型白酒感官表征及风味成分分析[J]. 食品科学, 2021, 42(6): 282-290.
SUN XZ, DU JW, HUANG P, ZHANG F, LIU YC. Analysis of sensory characteristics and flavor components in light-flavor Chinese liquor (Baijiu) made with Xiaoqu starter by modern and traditional technologies[J]. Food Science, 2021, 42(6): 282-290 (in Chinese). |
| [5] |
王震, 叶宏, 朱婷婷, 黄明泉, 魏金旺, 吴继红, 张璟琳. 清香型白酒风味成分的研究进展[J]. 食品科学, 2022, 43(7): 232-244.
WANG Z, YE H, ZHU TT, HUANG MQ, WEI JW, WU JH, ZHANG JL. Progress in research on the flavor components of light-flavor Baijiu[J]. Food Science, 2022, 43(7): 232-244 (in Chinese). |
| [6] |
尹子迎, 关军锋, 赵江丽, 刘金龙, 赵国群. 水果发酵酒质量评价体系研究进展[J]. 食品与机械, 2023, 39(1): 234-240.
YIN ZY, GUAN JF, ZHAO JL, LIU JL, ZHAO GQ. Research progress on quality evaluation system of fruit fermented wine[J]. Food & Machinery, 2023, 39(1): 234-240 (in Chinese). |
| [7] | HUANG D, ZHONG Y, LIU YL, SONG YY, ZHAO XX, QIN Y. Reducing higher alcohols by integrating indigenous Saccharomyces cerevisiae, nitrogen compensation, and chaptalization methods during fermentation of kiwifruit wine[J]. LWT-Food Science and Technology, 2023, 184: 115059 DOI:10.1016/j.lwt.2023.115059. |
| [8] | JOSHI VK, KUMAR V. Influence of different sugar sources, nitrogen sources and inocula on the quality characteristics of apple tea wine[J]. Journal of the Institute of Brewing, 2017, 123(2): 268-276 DOI:10.1002/jib.417. |
| [9] |
廉苇佳, 雷静, 韩琛, 刘志刚, 苏含明, 阿依加马丽·加帕尔, 陈雅. 低产高级醇桑葚酒发酵工艺优化[J]. 江西农业学报, 2023, 35(5): 89-96.
LIAN WJ, LEI J, HAN C, LIU ZG, SU HM, AYIJIAMALI·J, CHEN Y. Optimization of fermentation process for low-yield and higher-alcohol mulberry wine[J]. Acta Agriculturae Jiangxi, 2023, 35(5): 89-96 (in Chinese). |
| [10] | WANG YP, SUN ZG, ZHANG CY, ZHANG QZ, GUO XW, XIAO DG. Comparative transcriptome analysis reveals the key regulatory genes for higher alcohol formation by yeast at different α-amino nitrogen concentrations[J]. Food Microbiology, 2021, 95: 103713 DOI:10.1016/j.fm.2020.103713. |
| [11] |
王倩, 冯文倩, 王雅楠, 宋育阳, 刘延琳, 秦义. 本土酿酒酵母发酵进程中异戊醇的合成代谢[J]. 中国食品学报, 2022, 22(6): 95-105.
WANG Q, FENG WQ, WANG YN, SONG YY, LIU YL, QIN Y. The synthetic metabolism of isoamyl alcohol in the fermentation process of native Saccharomyces cerevisiae[J]. Journal of Chinese Institute of Food Science and Technology, 2022, 22(6): 95-105 (in Chinese). |
| [12] |
孙中贯, 刘琳, 王亚平, 王雪山, 肖冬光. 酿酒酵母高级醇代谢研究进展[J]. 生物工程学报, 2021, 37(2): 429-447.
SUN ZG, LIU L, WANG YP, WANG XS, XIAO DG. Higher alcohols metabolism by Saccharomyces cerevisiae: a mini review[J]. Chinese Journal of Biotechnology, 2021, 37(2): 429-447 (in Chinese). |
| [13] | CUI DY, LIU L, ZHANG XY, LIN LC, LI X, CHENG T, WEI CH, ZHANG Y, ZHOU Z, LI W, ZHANG CY. Using transcriptomics to reveal the molecular mechanism of higher alcohol metabolism in Saccharomyces cerevisiae[J]. Food Bioscience, 2023, 51: 102227 DOI:10.1016/j.fbio.2022.102227. |
| [14] | MAURYA R, GOHIL N, NIXON S, KUMAR N, NORONHA SB, DHALI D, TRABELSI H, ALZAHRANI KJ, RESHAMWALA SMS, AWASTHI MK, RAMAKRISHNA S, SINGH V. Rewiring of metabolic pathways in yeasts for sustainable production of biofuels[J]. Bioresource Technology, 2023, 372: 128668 DOI:10.1016/j.biortech.2023.128668. |
| [15] | AVALOS JL, FINK GR, STEPHANOPOULOS G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols[J]. Nature Biotechnology, 2013, 31(4): 335-341 DOI:10.1038/nbt.2509. |
| [16] | YAN TS, WANG ZX, ZHOU HY, HE JJ, ZHOU SS. Effects of four critical gene deletions in Saccharomyces cerevisiae on fusel alcohols during red wine fermentation[J]. Fermentation, 2023, 9(4): 379 DOI:10.3390/fermentation9040379. |
| [17] | WANG YP, WEI XQ, GUO XW, XIAO DG. Effect of the deletion of genes related to amino acid metabolism on the production of higher alcohols by Saccharomyces cerevisiae[J]. BioMed Research International, 2020, 2020: 6802512. |
| [18] | ZHENG N, JIANG S, HE YH, CHEN YF, ZHANG CY, GUO XW, MA LJ, XIAO DG. Production of low-alcohol Huangjiu with improved acidity and reduced levels of higher alcohols by fermentation with scarless ALD6 overexpression yeast[J]. Food Chemistry, 2020, 321: 126691 DOI:10.1016/j.foodchem.2020.126691. |
| [19] | ZHANG JW, ZHANG CY, DAI LH, DONG J, LIU YL, GUO XW, XIAO DG. Effects of overexpression of the alcohol acetyltransferase-encoding gene ATF1 and disruption of the esterase-encoding gene IAH1 on the flavour profiles of Chinese yellow rice wine[J]. International Journal of Food Science & Technology, 2012, 47(12): 2590-2596. |
| [20] | MÁR M, NITSENKO K, HEIDARSSON PO. Multifunctional intrinsically disordered regions in transcription factors[J]. Chemistry, 2023, 29(21): e202203369 DOI:10.1002/chem.202203369. |
| [21] | HAHN S, YOUNG ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators[J]. Genetics, 2011, 189(3): 705-736 DOI:10.1534/genetics.111.127019. |
| [22] | TAPIA SM, PÉREZ-TORRADO R, ADAM AC, MACÍAS LG, BARRIO E, QUEROL A. Functional divergence in the proteins encoded by ARO80 from S. uvarum, S. kudriavzevii and S. cerevisiae explain differences in the aroma production during wine fermentation[J]. Microbial Biotechnology, 2022, 15(8): 2281-2291 DOI:10.1111/1751-7915.14071. |
| [23] | NISHIMURA A, ISOGAI S, MURAKAMI N, HOTTA N, KOTAKA A, MATSUMURA K, HATA Y, ISHIDA H, TAKAGI H. Isolation and analysis of a sake yeast mutant with phenylalanine accumulation[J]. Journal of Industrial Microbiology & Biotechnology, 2022, 49(3): kuab085. |
| [24] | MAGASANIK B, KAISER CA. Nitrogen regulation in Saccharomyces cerevisiae[J]. Gene, 2002, 290(1/2): 1-18. |
| [25] | GONZÁLEZ J, LÓPEZ G, ARGUETA S, ESCALERA-FANJUL X, EL HAFIDI M, CAMPERO-BASALDUA C, STRAUSS J, RIEGO-RUIZ L, GONZÁLEZ A. Diversification of transcriptional regulation determines subfunctionalization of paralogous branched chain aminotransferases in the yeast Saccharomyces cerevisiae[J]. Genetics, 2017, 207(3): 975-991 DOI:10.1534/genetics.117.300290. |
| [26] | YUAN JF, MISHRA P, CHING CB. Engineering the leucine biosynthetic pathway for isoamyl alcohol overproduction in Saccharomyces cerevisiae[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(1): 107-117. |
| [27] | FRIDEN P, SCHIMMEL P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence[J]. Molecular and Cellular Biology, 1988, 8(7): 2690-2697. |
| [28] | ZHANG YF, CORTEZ JD, HAMMER SK, CARRASCO-LÓPEZ C, GARCÍA ECHAURI SÁ, WIGGINS JB, WANG W, AVALOS JL. Biosensor for branched-chain amino acid metabolism in yeast and applications in isobutanol and isopentanol production[J]. Nature Communications, 2022, 13(1): 270 DOI:10.1038/s41467-021-27852-x. |
| [29] |
陈先锐, 王肇悦, 何秀萍. 酵母菌合成2-苯乙醇的研究进展[J]. 生物工程学报, 2016, 32(9): 1151-1163.
CHEN XR, WANG ZY, HE XP. Advances in biosynthesis of 2-phenylethanol by yeasts[J]. Chinese Journal of Biotechnology, 2016, 32(9): 1151-1163 (in Chinese). |
| [30] | DEED RC, HOU RY, KINZURIK MI, GARDNER RC, FEDRIZZI B. The role of yeast ARO8, ARO9 and ARO10 genes in the biosynthesis of 3-(methylthio)-1- propanol from l-methionine during fermentation in synthetic grape medium[J]. FEMS Yeast Research, 2019 DOI:10.1093/femsyr/foy109. |
| [31] | LEE K, HAHN JS. Interplay of Aro80 and GATA activators in regulation of genes for catabolism of aromatic amino acids in Saccharomyces cerevisiae[J]. Molecular Microbiology, 2013, 88(6): 1120-1134. |
| [32] | RAVASIO D, WENDLAND J, WALTHER A. Major contribution of the Ehrlich pathway for 2-phenylethanol/rose flavor production in Ashbya gossypii[J]. FEMS Yeast Research, 2014, 14(6): 833-844. |
| [33] | RISINGER AL, KAISER CA. Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking[J]. Molecular Biology of the Cell, 2008, 19(7): 2962-2972. |
| [34] | RUBIO-TEXEIRA M, KAISER CA. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway[J]. Molecular Biology of the Cell, 2006, 17(7): 3031-3050. |
| [35] |
史莹华, 许梓荣. GATA转录因子研究进展[J]. 生物学通报, 2005, 40(3): 1-2.
SHI YH, XU ZR. Recent advances in GATA transcription factor[J]. Bulletin of Biology, 2005, 40(3): 1-2 (in Chinese). |
| [36] | GEORIS I, FELLER A, VIERENDEELS F, DUBOIS E. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation[J]. Molecular and Cellular Biology, 2009, 29(13): 3803-3815. |
| [37] | CARDILLO SB, LEVI CE, BERMÚDEZ MORETTI M, CORREA GARCÍA S. Interplay between the transcription factors acting on the GATA- and GABA-responsive elements of Saccharomyces cerevisiae UGA promoters[J]. Microbiology, 2012, 158(Pt 4): 925-935. |
| [38] | WANG ZY, BAI XJ, GUO XN, HE XP. Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(1): 129-139. |
| [39] | HAMMER, SK. Metabolic engineering of Saccharomyces cerevisiae for enhanced production of branched-chain higher alcohols[D]. Princeton: Doctoral Dissertation of Princeton University, 2020. |
| [40] | WEI TY, JIAO ZH, HU JJ, LOU HH, CHEN QH. Chinese yellow rice wine processing with reduced ethyl carbamate formation by deleting transcriptional regulator Dal80p in Saccharomyces cerevisiae[J]. Molecules, 2020, 25(16): 3580. |
| [41] | TURNER SA, MA QX, OLA M, MARTINEZ de SAN VICENTE K, BUTLER G. Dal81 regulates expression of arginine metabolism genes in Candida parapsilosis[J]. mSphere, 2018, 3(2): e00028-18. |
| [42] | GEORIS I, RONSMANS A, VIERENDEELS F, DUBOIS E. Differing SAGA module requirements for NCR-sensitive gene transcription in yeast[J]. Yeast, 2024, 41(4): 207-221. |
| [43] | SCHERENS B, FELLER A, VIERENDEELS F, MESSENGUY F, DUBOIS E. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term[J]. FEMS Yeast Research, 2006, 6(5): 777-791. |
| [44] | GEORIS I, FAYYAD-KAZAN M, ZAREMBA E, VIERENDEELS F, ROOVERS M, DUBOIS E. Glutamine transport as a possible regulator of nitrogen catabolite repression in Saccharomyces cerevisiae[J]. Yeast, 2022, 39(9): 493-507. |
| [45] | AJIT K, THOMAS DB, RAJENDRA R, TERRANCE GC. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae[J]. FEMS Yeast Research, 6(2): 218-229. |
| [46] | COOPER TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots[J]. FEMS Microbiology Reviews, 2002, 26(3): 223-238. |
| [47] | STEYER JT, TODD RB. Branched-chain amino acid biosynthesis in fungi[J]. Essays in Biochemistry, 2023, 67(5): 865-876. |
| [48] | FITZGERALD MX, ROJAS JR, KIM JM, KOHLHAW GB, MARMORSTEIN R. Structure of a Leu3-DNA complex: recognition of everted CGG half-sites by a Zn2Cys6 binuclear cluster protein[J]. Structure, 2006, 14(4): 725-735. |
| [49] | LIU X, LEE CK, GRANEK JA, CLARKE ND, LIEB JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection[J]. Genome Research, 2006, 16(12): 1517-1528. |
| [50] | COLÓN M, HERNÁNDEZ F, LÓPEZ K, QUEZADA H, GONZÁLEZ J, LÓPEZ G, ARANDA C, GONZÁLEZ A. Saccharomyces cerevisiae Bat1 and Bat2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme[J]. PLoS One, 2011, 6(1): e16099. |
| [51] | LÓPEZ G, QUEZADA H, DUHNE M, GONZÁLEZ J, LEZAMA M, EL-HAFIDI M, COLÓN M, MARTÍNEZ deLa ESCALERA X, FLORES-VILLEGAS MC, SCAZZOCCHIO C, DeLUNA A, GONZÁLEZ A. Diversification of paralogous α-isopropylmalate synthases by modulation of feedback control and hetero-oligomerization in Saccharomyces cerevisiae[J]. Eukaryotic Cell, 2015, 14(6): 564-577. |
| [52] | ROLLERO S, MOURET JR, BLOEM A, SANCHEZ I, ORTIZ-JULIEN A, SABLAYROLLES JM, DEQUIN S, CAMARASA C. Quantitative 13C-isotope labelling-based analysis to elucidate the influence of environmental parameters on the production of fermentative aromas during wine fermentation[J]. Microbial Biotechnology, 2017, 10(6): 1649-1662. |
| [53] | MONTEIRO PT, OLIVEIRA J, PAIS P, ANTUNES M, PALMA M, CAVALHEIRO M, GALOCHA M, GODINHO CP, MARTINS LC, BOURBON N, MOTA MN, RIBEIRO RA, VIANA R, SÁ-CORREIA I, TEIXEIRA MC. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts[J]. Nucleic Acids Research, 2020, 48(D1): D642-D649. |
 2024, Vol. 64
2024, Vol. 64