中国科学院微生物研究所,中国微生物学会
文章信息
- 张博, 杨玉凤, 贺周霖, 章誉琼, 柳志强, 郑裕国. 2023
- ZHANG Bo, YANG Yufeng, HE Zhoulin, ZHANG Yuqiong, LIU Zhiqiang, ZHENG Yuguo.
- 利用CRISPRi挖掘大肠杆菌生物合成D-泛酸的关键基因
- Mining key genes for biosynthesis of D-pantothenic acid in Escherichia coli by CRISPRi
- 微生物学报, 63(12): 4625-4643
- Acta Microbiologica Sinica, 63(12): 4625-4643
-
文章历史
- 收稿日期:2023-04-17
- 网络出版日期:2023-08-29
2. 浙江工业大学生物工程学院 浙江省生物有机合成技术研究重点实验室, 浙江 杭州 310014;
3. 华东合成生物学产业技术研究院, 浙江 杭州 310014
2. Key Laboratory of Bioorganic Synthesis of Zhejiang Province, College of Biotechnology and Bioengineering, Zhejiang University of Technology, Hangzhou 310014, Zhejiang, China;
3. Huadong Industrial Technology Research Institute of Synthetic Biology, Hangzhou 310014, Zhejiang, China
D-泛酸(D-pantothenic acid, DPA)又称维生素B5,作为一种水溶性维生素,自然界中泛酸主要存在两种构型,D型和L型,其中,只有D型具有生物活性。在生物体内,DPA主要通过参与辅酶A (coenzyme A, CoA)以及酰基载体蛋白(acyl carrier protein, ACP)的形成,从而在蛋白质代谢、脂质代谢以及许多次生代谢物的合成过程中发挥着重要作用[1]。因此DPA被广泛应用于制药行业、食品工业以及饲料添加剂。目前,工业上DPA主要以泛酸内酯和β-丙氨酸的钙盐为原料通过化学法结合酶法拆分生产,但过程中存在底物毒性、操作繁琐、催化效率低等缺点,在一定程度上限制了其发展。相较于运用化学法和酶法合成DPA,微生物发酵在生产DPA在过程中则更具安全性,且无需水解和拆分等繁琐过程。当前,用于微生物发酵生产DPA得到宿主菌株主要有大肠杆菌、谷氨酸棒杆菌、枯草芽孢杆菌,且人们围绕上述菌株已进行了相关研究,例如Zou等[2]利用代谢工程结合发酵工程技术构建了一株大肠杆菌DPA高产菌株,发酵产量达到了68.3 g/L;Chassagnole等[3]在谷氨酸棒状杆菌中通过对ilvE启动子突变降低其活性,提高酮异戊酸的积累,并增加关键基因panB、panC的拷贝数,在pH调节的分批培养过程中,积累了不到2 g/L的DPA;Yocum等[4]通过应用代谢工程策略,构建了以枯草芽孢杆菌为底盘的DPA生产菌株PA824,并在此菌株的基础上,通过强化glyA提高亚甲基四氢叶酸(methylenetetrahydrofolate, CH2-THF)供应得到的工程菌株PA1014-3,在外源添加β-丙氨酸的条件下,能生产86 g/L的DPA。
途径设计和优化对于构建目标产品的高产菌株至关重要。随着基因工程技术的成熟,研究人员可以迅速对底盘细胞进行一系列改造,以生产新的生物化学物质。近年来本课题组Zhang等[1, 5]主要通过敲除、下调和过表达多个基因靶点的代谢工程手段来构建DPA高产菌株,但随着对菌株的不断代谢改造,由于缺少对DPA生物合成与其他细胞过程之间联系的了解,DPA产量的进一步提高受到阻碍,因此需要挖掘更多基因靶点来提高DPA产量。CRISPR干扰(clustered regularly interspaced palindromic repeats interference, CRISPRi)技术可用于高通量的功能基因筛查,CRISPRi由成簇的规律间隔短回文重复序列及CRISPR相关蛋白(clustered regularly interspaced palindromic repeats and CRISPR-associated proteins system, CRISPR-Cas)衍生而来,通过在核酸酶的催化残基中引入点突变(D10A和H840A),设计出一种催化活性部分失活的Cas9蛋白(dCas9),这些突变使Cas9内切酶不能切断双链DNA,但允许使用RNA进行引导[6-8],在sgRNA的作用下,dCas9蛋白可精确定位目标靶点,空间上阻碍RNA聚合酶(RNA polymerase, RNAP)的转录过程,实现特定基因的表达下调。CRISPRi已经广泛应用于微生物代谢工程,例如Fang等[9]利用该技术在大肠杆菌中鉴定得到提高游离脂肪酸的有益基因,Li等[10]在谷氨酸棒状杆菌通过构建的CRISPR-dCas9系统得到高效生产O-乙酰高丝氨酸的关键基因。本研究以实验室前期构建的DPA工程菌DPAP10为出发菌株进行CRISPRi筛选,目前报道的大肠杆菌DPA的最高产菌株DPA02/pT-ppnk在摇瓶中积累DPA为6.89 g/L,在5 L罐中积累DPA产量为68.3 g/L[2],但由于该菌株携带质粒限制了其工业化生产。而DPAP10菌株则为不带质粒的无抗菌株,在摇瓶中积累DPA为3.57 g/L,在5 L罐中积累DPA为63.1 g/L,相比菌株DPA02/pT-ppnk更具潜力且更适合于工业生产。
1 材料与方法 1.1 菌种、质粒和引物本研究以实验室前期构建的大肠杆菌(Escherichia coli) DPAP10 (DPAL6[11], Trc-ilvD/ Trc-panB/Trc-panC/Trc-alsS)作为出发菌株,大肠杆菌DH5α为克隆宿主,用于pTarget质粒构建。pTarget质粒(Addgene Plasmid #62226)由中国科学院上海生命科学研究院植物生理生态研究所杨晟研究员馈赠;pdCas9质粒由实验室菌种库保存,dCas9基因表达由trc启动子控制,需异丙基-β-D-硫代半乳糖苷(isopropyl-β-D-galactothioglycoside, IPTG)诱导。PCR引物由北京擎科生物科技有限公司合成。本文所用pTarget突变引物详见表 1。
| Primers | Sequences (5′→3′) | |
| pT-avtA-F | TAATACTAGTGATGCACTGTGTAACTACGAGTTTTAGAGCTAGAAATAGC | |
| pT-avtA-R | GCTCTAAAACTCGTAGTTACACAGTGCATCACTAGTATTATACCTAGGAC | |
| pT-fadR-F | TAATACTAGTCATTAAGGCGCAAAGCCCGGGTTTTAGAGCTAGAAATAGC | |
| pT-fadR-R | GCTCTAAAACCCGGGCTTTGCGCCTTAATGACTAGTATTATACCTAGGAC | |
| pT-fabR-F | TAATACTAGTTATAGTGCCAGCAGTACTTTGTTTTAGAGCTAGAAATAGC | |
| pT-fabR-R | GCTCTAAAACAAAGTACTGCTGGCACTATAACTAGTATTATACCTAGGAC | |
| pT-mdh-F | TAATACTAGTGATATCGCTCCAGTGACTCCGTTTTAGAGCTAGAAATAGC | |
| pT-mdh-R | GCTCTAAAACGGAGTCACTGGAGCGATATCACTAGTATTATACCTAGGAC | |
| pT-ihfA-F | TAATACTAGTATAAGCTTGGGCTTAGCAAGGTTTTAGAGCTAGAAATAGC | |
| pT-ihfA-R | GCTCTAAAACCTTGCTAAGCCCAAGCTTATACTAGTATTATACCTAGGAC | |
| pT-ihfB-F | TAATACTAGTATCGCACATTCCCGCCAAGAGTTTTAGAGCTAGAAATAGC | |
| pT-ihfB-R | GCTCTAAAACTCTTGGCGGGAATGTGCGATACTAGTATTATACCTAGGAC | |
| pT-gapC-F | TAATACTAGTCTGAAACATGATTCAAACTAGTTTTAGAGCTAGAAATAGC | |
| pT-gapC-R | GCTCTAAAACTAGTTTGAATCATGTTTCAGACTAGTATTATACCTAGGAC | |
| pT-adiY-F | TAATACTAGTATGTCTGGATAAGGGTGAATGTTTTAGAGCTAGAAATAGC | |
| pT-adiY-R | GCTCTAAAACATTCACCCTTATCCAGACATACTAGTATTATACCTAGGAC | |
| pT-deoR-F | TAATACTAGTACACGTCGCGAAGAGCGTATGTTTTAGAGCTAGAAATAGC | |
| pT-deoR-R | GCTCTAAAACATACGCTCTTCGCGACGTGTACTAGTATTATACCTAGGAC | |
| pT-asnC-F | TAATACTAGTCTGATCGACAATCTGGACCGGTTTTAGAGCTAGAAATAGC | |
| pT-asnC-R | GCTCTAAAACCGGTCCAGATTGTCGATCAGACTAGTATTATACCTAGGAC | |
| pT-ilvY-F | TAATACTAGTGAAAACCTTCCTGCATCTGGGTTTTAGAGCTAGAAATAGC | |
| pT-ilvY-R | GCTCTAAAACCCAGATGCAGGAAGGTTTTCACTAGTATTATACCTAGGAC | |
| pT-crp-F | TAATACTAGTAAACAGACCCGACTCTCGAAGTTTTAGAGCTAGA AATAGC | |
| pT-crp-R | GCTCTAAAACTTCGAGAGTCGGGTCTGTTTACTAGTATTATACCTAGGAC | |
| pT-narL-F | TAATACTAGTGATCACCCGATGCTGCGAACGTTTTAGAGCTAGAAATAGC | |
| pT-narL-R | GCTCTAAAACGTTCGCAGCATCGGGTGATCACTAGTATTATACCTAGGAC | |
| pT-metJ-F | TAATACTAGTATCAGCCCATACGCTGAGCAGTTTTAGAGCTAGAAATAGC | |
| pT-metJ-R | GCTCTAAAACTGCTCAGCGTATGGGCTGATACTAGTATTATACCTAGGAC | |
| pT-lrp-F | TAATACTAGTGTAGATAGCAAGAAGCGCCCGTTTTAGAGCTAGAAATAGC | |
| pT-lrp-R | GCTCTAAAACGGGCGCTTCTTGCTATCTACACTAGTATTATACCTAGGAC | |
| pT-nac-F | TAATACTAGTAGATATTGGTAGCCTGACCCGTTTTAGAGCTAGAAATAGC | |
| pT-nac-R | GCTCTAAAACGGGTCAGGCTACCAATATCTACTAGTATTATACCTAGGAC | |
| pT-nsrR-F | TAATACTAGTTTAACGAGTTTCACTGATTAGTTTTAGAGCTAGAAATAGC | |
| pT-nsrR-R | GCTCTAAAACTAATCAGTGAAACTCGTTAAACTAGTATTATACCTAGGAC | |
| pT-rbsB-F | TAATACTAGTCACCGTCAGTGCGAATGCGAGTTTTAGAGCTAGAAATAGC | |
| pT-rbsB-R | GCTCTAAAACTCGCATTCGCACTGACGGTGACTAGTATTATACCTAGGAC | |
| pT-rob-F | TAATACTAGTTCGCGACCTTTTAATCTGGCGTTTTAGAGCTAGAAATAGC | |
| pT-rob-R | GCTCTAAAACGCCAGATTAAAAGGTCGCGAACTAGTATTATACCTAGGAC | |
| pT-lexA-F | TAATACTAGTGTTAACGGCCAGGCAACAAGGTTTTAGAGCTAGAAATAGC | |
| pT-lexA-R | GCTCTAAAACCTTGTTGCCTGGCCGTTAACACTAGTATTATACCTAGGAC | |
| pT-soxS-F | TAATACTAGTGTCCCATCAGAAAATTATTCGTTTTAGAGCTAGAAATAGC | |
| pT-soxS-R | GCTCTAAAACGAATAATTTTCTGATGGGACACTAGTATTATACCTAGGAC | |
| pT-phoP-F | TAATACTAGTCCACCTTAAAGTTCAGATTCGTTTTAGAGCTAGAAATAGC | |
| pT-phoP-R | GCTCTAAAACGAATCTGAACTTTAAGGTGGACTAGTATTATACCTAGGAC | |
| pT-lysC-F | TAATACTAGTGCGCGCTTTGAGAAGGTGATGTTTTAGAGCTAGAAATAGC | |
| pT-lysC-R | GCTCTAAAACATCACCTTCTCAAAGCGCGCACTAGTATTATACCTAGGAC | |
| pT-tktA-F | TAATACTAGTGAAAGCCAAATCCGGTCACCGTTTTAGAGCTAGAAATAGC | |
| pT-tktA-R | GCTCTAAAACGGTGACCGGATTTGGCTTTCACTAGTATTATACCTAGGAC | |
| pT-tktB-F | TAATACTAGTAAAGCCAACTCTGGTCATCCGTTTTAGAGCTAGAAATAGC | |
| pT-tktB-R | GCTCTAAAACGGATGACCAGAGTTGGCTTTACTAGTATTATACCTAGGAC | |
| pT-talA-F | TAATACTAGTTCGCTGTTACTCAAAGCTGCGTTTTAGAGCTAGAAATAGC | |
| pT-talA-R | GCTCTAAAACGCAGCTTTGAGTAACAGCGAACTAGTATTATACCTAGGAC | |
| pT-talB-F | TAATACTAGTTCTTAACGCAGCGCAGATTCGTTTTAGAGCTAGAAATAGC | |
| pT-talB-R | GCTCTAAAACGAATCTGCGCTGCGTTAAGAACTAGTATTATACCTAGGAC | |
| pT-ptsI-F | TAATACTAGTTTCAGGCATTTTAGCATCCCGTTTTAGAGCTAGAAATAGC | |
| pT-ptsI-R | GCTCTAAAACGGGATGCTAAAATGCCTGAAACTAGTATTATACCTAGGAC | |
| pT-ptsH-F | TAATACTAGTGTTACCATTACCGCTCCGAAGTTTTAGAGCTAGAAATAGC | |
| pT-ptsH-R | GCTCTAAAACGGGATGCTAAAATGCCTGAAACTAGTATTATACCTAGGAC | |
| pT-rpe-F | TAATACTAGTGATTGCCCCCTCAATTCTGTGTTTTAGAGCTAGAAATAGC | |
| pT-rpe-R | GCTCTAAAACACAGAATTGAGGGGGCAATCACTAGTATTATACCTAGGAC | |
| pT-pgk-F | TAATACTAGTCTGAACGTACCAGTAAAAGAGTTTTAGAGCTAGAAATAGC | |
| pT-pgk-R | GCTCTAAAACTCTTTTACTGGTACGTTCAGACTAGTATTATACCTAGGAC | |
| pT-gnd-F | TAATACTAGTCAACCGTTCCCGTGAGAAGAGTTTTAGAGCTAGAAATAGC | |
| pT-gnd-R | GCTCTAAAACTCTTCTCACGGGAACGGTTGACTAGTATTATACCTAGGAC | |
| pT-zwf-F | TAATACTAGTGCCTGTGACCTGGTCATTTTGTTTTAGAGCTAGAAATAGC | |
| pT-zwf-R | GCTCTAAAACAAAATGACCAGGTCACAGGCACTAGTATTATACCTAGGAC | |
| pT-gapA-F | TAATACTAGTCTGAAATATGACTCCACTCAGTTTTAGAGCTAGAAATAGC | |
| pT-gapA-R | GCTCTAAAACTGAGTGGAGTCATATTTCAGACTAGTATTATACCTAGGAC | |
| pT-rpiA-F | TAATACTAGTGAAAAAAGCAGTAGGATGGGGTTTTAGAGCTAGAAATAGC | |
| pT-rpiA-R | GCTCTAAAACCCCATCCTACTGCTTTTTTCACTAGTATTATACCTAGGAC | |
| pT-rpiB-F | TAATACTAGTGCATTTGGCTGTGATCATGTGTTTTAGAGCTAGAAATAGC | |
| pT-rpiB-R | GCTCTAAAACACATGATCACAGCCAAAGTCACTAGTATTATACCTAGGAC | |
| pT-crr-F | TAATACTAGTTCCGACGACAAGAAGGATACGTTTTAGAGCTAGAAATAGC | |
| pT-crr-R | GCTCTAAAACGTATCCTTCTTGTCGTCGGAACTAGTATTATACCTAGGAC | |
| pT-serA-F | TAATACTAGTAGACAAGATTAAGTTTCTGCGTTTTAGAGCTAGAAATAGC | |
| pT-serA-R | GCTCTAAAACGCAGAAACTTAATCTTGTCTACTAGTATTATACCTAGGAC | |
| pT-serB-F | GCTCTAAAACGCAGAAACTTAATCTTGTCTACTAGTATTATACCTAGGAC | |
| pT-serB-R | GCTCTAAAACTAAAGAGACATCTTCAGGCAACTAGTATTATACCTAGGAC | |
| pT-serC-F | TAATACTAGTCTTCAATTTTAGTTCTGGTCGTTTTAGAGCTAGAAATAGC | |
| pT-serC-R | GCTCTAAAACGACCAGAACTAAAATTGAAGACTAGTATTATACCTAGGAC | |
| pT-leuA-F | TAATACTAGTTTCGATACCACATTGCGCGAGTTTTAGAGCTAGAAATAGC | |
| pT-leuA-R | GCTCTAAAACTCGCGCAATGTGGTATCGAAACTAGTATTATACCTAGGAC | |
| pT-leuB-F | TAATACTAGTGCCGGGGGACGGTATTGGTCGTTTTAGAGCTAGAAATAGC | |
| pT-leuB-R | GCTCTAAAACGACCAATACCGTCCCCCGGCACTAGTATTATACCTAGGAC | |
| pT-leuC-F | TAATACTAGTTTCGATGGTCTGCGCGCCCAGTTTTAGAGCTAGAAATAGC | |
| pT-leuC-R | GCTCTAAAACTGGGCGCGCAGACCATCGAAACTAGTATTATACCTAGGAC | |
| pT-leuD-F | TAATACTAGTTTTGCAGAAAGTGACCCGTAGTTTTAGAGCTAGAAATAGC | |
| pT-leuD-R | GCTCTAAAACTACGGGTCACTTTCTGCAAAACTAGTATTATACCTAGGAC | |
| pT-ilvI-F | TAATACTAGTCAGGGCGTTAAACAAGTATTGTTTTAGAGCTAGAAATAGC | |
| pT-ilvI-R | GCTCTAAAACAATACTTGTTTAACGCCCTGACTAGTATTATACCTAGGAC | |
| pT-ilvH-F | TAATACTAGTGGCGCGTTATCCCGCGTGATGTTTTAGAGCTAGAAATAGC | |
| pT-ilvH-R | GCTCTAAAACATCACGCGGGATAACGCGCCACTAGTATTATACCTAGGAC | |
| pT-ilvB-F | TAATACTAGTTCGACGCGTAAGCGCTTTACGTTTTAGAGCTAGAAATAGC | |
| pT-ilvB-R | GCTCTAAAACGTAAAGCGCTTACGCGTCGAACTAGTATTATACCTAGGAC | |
| pT-ilvN-F | TAATACTAGTGCTCACCGTTCGCAACCATCGTTTTAGAGCTAGAAATAGC | |
| pT-ilvN-R | GCTCTAAAACGATGGTTGCGAACGGTGAGCACTAGTATTATACCTAGGAC | |
| pT-ilvG-F | TAATACTAGTAGTGGGTGGTACATGCGTTGGTTTTAGAGCTAGAAATAGC | |
| pT-ilvG-R | GCTCTAAAACCAACGCATGTACCACCCACTACTAGTATTATACCTAGGAC | |
| pT-ilvM-F | TAATACTAGTGCAACATCAGGTCAATGTATGTTTTAGAGCTAGAAATAGC | |
| pT-ilvM-R | GCTCTAAAACATACATTGACCTGATGTTGCACTAGTATTATACCTAGGAC | |
| pT-ilvE-F | TAATACTAGTCGCGCTGCACTATGGCACTTGTTTTAGAGCTAGAAATAGC | |
| pT-ilvE-R | GCTCTAAAACAAGTGCCATAGTGCAGCGCGACTAGTATTATACCTAGGAC | |
| pT-ilvC-F | TAATACTAGTGGTAAAAAAGTAGTCATCGTGTTTTAGAGCTAGAAATAGC | |
| pT-ilvC-R | GCTCTAAAACACGATGACTACTTTTTTACCACTAGTATTATACCTAGGAC | |
| pT-ilvD-F | TAATACTAGTCACTCATGGTCGTAATATGGGTTTTAGAGCTAGAAATAGC | |
| pT-ilvD-R | GCTCTAAAACCCATATTACGACCATGAGTGACTAGTATTATACCTAGGAC | |
| pT-yfbQ-F | TAATACTAGTCTGTTATGACATCCGTGGTCGTTTTAGAGCTAGAAATAGC | |
| pT-yfbQ-R | GCTCTAAAACGACCACGGATGTCATAACAGACTAGTATTATACCTAGGAC | |
| pT-lpd-F | TAATACTAGTGAACGTTACAACACCCTTGGGTTTTAGAGCTAGAAATAGC | |
| pT-lpd-R | GCTCTAAAACCCAAGGGTGTTGTAACGTTCACTAGTATTATACCTAGGAC | |
| pT-gcvP-F | TAATACTAGTTTAAGCCAGCTTGAAAACAGGTTTTAGAGCTAGAAATAGC | |
| pT-gcvP-R | GCTCTAAAACCTGTTTTCAAGCTGGCTTAAACTAGTATTATACCTAGGAC | |
| pT-gcvT-F | TAATACTAGTTACGAACAACACACGCTTTGGTTTTAGAGCTAGAAATAGC | |
| pT-gcvT-R | GCTCTAAAACCAAAGCGTGTGTTGTTCGTAACTAGTATTATACCTAGGAC | |
| pT-puuE-F | TAATACTAGTCGTCTTTCTGCCACTCCGCGGTTTTAGAGCTAGAAATAGC | |
| pT-puuE-R | GCTCTAAAACCGCGGAGTGGCAGAAAGACGACTAGTATTATACCTAGGAC | |
| pT-metL-F | TAATACTAGTGAAGTGTTATTTGCGTGTCGGTTTTAGAGCTAGAAATAGC | |
| pT-metL-R | GCTCTAAAACCGACACGCAAATAACACTTCACTAGTATTATACCTAGGAC | |
| pT-gadA-F | TAATACTAGTAAAGGCCATTTCTACTATCGGTTTTAGAGCTAGAAATAGC | |
| pT-gadA-R | GCTCTAAAACCGATAGTAGAAATGGCCTTTACTAGTATTATACCTAGGAC | |
| pT-gadB-F | TAATACTAGTGCAAGTAACGGATTTAAGGTGTTTTAGAGCTAGAAATAGC | |
| pT-gadB-R | GCTCTAAAACACCTTAAATCCGTTACTTGCACTAGTATTATACCTAGGAC | |
| pT-gabT-F | TAATACTAGTCCCGATTTTCGCTGACCGCGGTTTTAGAGCTAGAAATAGC | |
| pT-gabT-R | GCTCTAAAACCGCGGTCAGCGAAAATCGGGACTAGTATTATACCTAGGAC | |
| pT-sad-F | TAATACTAGTAATTTCGATAAATCCTGCCAGTTTTAGAGCTAGAAATAGC | |
| pT-sad-R | GCTCTAAAACTGGCAGGATTTATCGAAATTACTAGTATTATACCTAGGAC | |
| pT-thrA-F | TAATACTAGTATGCGAGTGTTGAAGTTCGGGTTTTAGAGCTAGAAATAGC | |
| pT-thrA-R | GCTCTAAAACCCGAACTTCAACACTCGCATACTAGTATTATACCTAGGAC | |
| pT-thrB-F | TAATACTAGTTCCAGTGCCAATATGAGCGTGTTTTAGAGCTAGAAATAGC | |
| pT-thrB-R | GCTCTAAAACACGCTCATATTGGCACTGGAACTAGTATTATACCTAGGAC | |
| pT-thrC-F | TAATACTAGTGTTTTTTCCGCACGACCTGCGTTTTAGAGCTAGAAATAGC | |
| pT-thrC-R | GCTCTAAAACGCAGGTCGTGCGGAAAAAACACTAGTATTATACCTAGGAC | |
| pT-aceE-F | TAATACTAGTCTGGCTCCAGGCGATCGAATGTTTTAGAGCTAGAAATAGC | |
| pT-aceE-R | GCTCTAAAACATTCGATCGCCTGGAGCCAGACTAGTATTATACCTAGGAC | |
| pT-sucC-F | TAATACTAGTCCGCTATGGCTTACCAGCACGTTTTAGAGCTAGAAATAGC | |
| pT-sucC-R | GCTCTAAAACGTGCTGGTAAGCCATAGCGGACTAGTATTATACCTAGGAC | |
| pT-asd-F | TAATACTAGTGTTGGTTTTATCGGCTGGCGGTTTTAGAGCTAGAAATAGC | |
| pT-asd-R | GCTCTAAAACCGCCAGCCGATAAAACCAACACTAGTATTATACCTAGGAC | |
| pT-fumA-F | TAATACTAGTTGCGTCGTTCATGCTGCGTCGTTTTAGAGCTAGAAATAGC | |
| pT-fumA-R | GCTCTAAAACGACGCAGCATGAACGACGCAACTAGTATTATACCTAGGAC | |
| pT-fumC-F | TAATACTAGTGATGGGGGCGATTGATGTCCGTTTTAGAGCTAGAAATAGC | |
| pT-fumC-R | GCTCTAAAACGGACATCAATCGCCCCCATCACTAGTATTATACCTAGGAC | |
| pT-argG-F | TAATACTAGTGACGATTCTCAAGCATCTCCGTTTTAGAGCTAGAAATAGC | |
| pT-argG-R | GCTCTAAAACGGAGATGCTTGAGAATCGTCACTAGTATTATACCTAGGAC | |
| pT-argH-F | TAATACTAGTTTACCCAGGCAGCAGATCAAGTTTTAGAGCTAGAAATAGC | |
| pT-argH-R | GCTCTAAAACTTGATCTGCTGCCTGGGTAAACTAGTATTATACCTAGGAC | |
| pT-ftsZ-F | TAATACTAGTCAGACGATTCAAATCGGTAGGTTTTAGAGCTAGAAATAGC | |
| pT-ftsZ-R | GCTCTAAAACCTACCGATTTGAATCGTCTGACTAGTATTATACCTAGGAC | |
| pT-sulA-F | TAATACTAGTTAAAATTGCGCGTGTCTCTAGTTTTAGAGCTAGAAATAGC | |
| pT-sulA-R | GCTCTAAAACTAGAGACACGCGCAATTTTAACTAGTATTATACCTAGGAC | |
| pT-rssB-F | TAATACTAGTATCGCGATGCCACGAATGAAGTTTTAGAGCTAGAAATAGC | |
| pT-rssB-R | GCTCTAAAACTTCATTCGTGGCATCGCGATACTAGTATTATACCTAGGAC | |
| pT-rodZ-F | TAATACTAGTCTTGCTTCAACATTCCTGCGGTTTTAGAGCTAGAAATAGC | |
| pT-rodZ-R | GCTCTAAAACCGCAGGAATGTTGAAGCAAGACTAGTATTATACCTAGGAC | |
| pT-rpoS-F | TAATACTAGTTGGTTATTCACCACTGTTAAGTTTTAGAGCTAGAAATAGC | |
| pT-rpoS-R | GCTCTAAAACTTAACAGTGGTGAATAACCAACTAGTATTATACCTAGGAC | |
| pT-minC-F | TAATACTAGTCTGGTCAGCGATGCATAAGGGTTTTAGAGCTAGAAATAGC | |
| pT-minC-R | GCTCTAAAACCCTTATGCATCGCTGACCAGACTAGTATTATACCTAGGAC | |
| pT-fnr-F | TAATACTAGTATACGGCGCATTCAGTCTGGGTTTTAGAGCTAGAAATAGC | |
| pT-fnr-R | GCTCTAAAACCCAGACTGAATGCGCCGTATACTAGTATTATACCTAGGAC | |
| pT-nadR-F | TAATACTAGTCAGCAGGTAGCTGATGCCAGGTTTTAGAGCTAGAAATAGC | |
| pT-nadR-R | GCTCTAAAACCTGGCATCAGCTACCTGCTGACTAGTATTATACCTAGGAC | |
| pT-recQ-F | TAATACTAGTATTATCGACACTGTGCTTTCGTTTTAGAGCTAGAAATAGC | |
| pT-recQ-R | GCTCTAAAACGAAAGCACAGTGTCGATAATACTAGTATTATACCTAGGAC | |
| pT-groC-F | TAATACTAGTGTAGTTCTGGATAAATCTTTGTTTTAGAGCTAGAAATAGC | |
| pT-groC-R | GCTCTAAAACAAAGATTTATCCAGAACTACACTAGTATTATACCTAGGAC | |
| pT-cysK-F | TAATACTAGTCGCCTGAATCGCATCGGTAAGTTTTAGAGCTAGAAATAGC | |
| pT-cysK-R | GCTCTAAAACTTACCGATGCGATTCAGGCGACTAGTATTATACCTAGGAC | |
| pT-glnA-F | TAATACTAGTGAAGAAGGCAAAATGTTTGAGTTTTAGAGCTAGAAATAGC | |
| pT-glnA-R | GCTCTAAAACTCAAACATTTTGCCTTCTTCACTAGTATTATACCTAGGAC | |
| pT-proB-F | TAATACTAGTTGCGCGCAGTTACATGCCGCGTTTTAGAGCTAGAAATAGC | |
| pT-proB-R | GCTCTAAAACGCGGCATGTAACTGCGCGCAACTAGTATTATACCTAGGAC | |
| pT-gltB-F | TAATACTAGTATTCTCGCCGATGGTAAAACGTTTTAGAGCTAGAAATAGC | |
| pT-gltB-R | GCTCTAAAACGTTTTACCATCGGCGAGAATACTAGTATTATACCTAGGAC | |
| pT-pfkA-F | TAATACTAGTCGTATCAAATGGAAAATGGAGTTTTAGAGCTAGAAATAGC | |
| pT-pfkA-R | GCTCTAAAACCGAATTGCGGCGTTCATGCCACTAGTATTATACCTAGGAC | |
| pT-pfkB-F | TAATACTAGTAAAACTGCGCTGTACCGCACGTTTTAGAGCTAGAAATAGC | |
| pT-pfkB-R | GCTCTAAAACGTGCGGTACAGCGCAGTTTTACTAGTATTATACCTAGGAC | |
| pT-pykA-F | TAATACTAGTTAATCTTGAAAAAGTTATCGGTTTTAGAGCTAGAAATAGC | |
| pT-pykA-R | GCTCTAAAACCGATAACTTTTTCAAGATTAACTAGTATTATACCTAGGAC | |
| pT-acnA-F | TAATACTAGTGCGCTGGCAGGATGGTAACTGTTTTAGAGCTAGAAATAGC | |
| pT-acnA-R | GCTCTAAAACAGTTACCATCCTGCCAGCGCACTAGTATTATACCTAGGAC | |
| pT-acnB-F | TAATACTAGTGCTGCTGAAAAACCCGCCCGGTTTTAGAGCTAGAAATAGC | |
| pT-acnB-R | GCTCTAAAACCGGGCGGGTTTTTCAGCAGCACTAGTATTATACCTAGGAC | |
| Pt-rpoE-F | TAATACTAGTGGTTTCCCGCTATGTGCCGTGTTTTAGAGCTAGAAATAGC | |
| pT-rpoE-R | GCTCTAAAACACGGCACATAGCGGGAAACCACTAGTATTATACCTAGGAC | |
| pT-rpoH-F | TAATACTAGTTAACGCGTGGCCGATGTTGTGTTTTAGAGCTAGAAATAGC | |
| pT-rpoH-R | GCTCTAAAACACAACATCGGCCACGCGTTAACTAGTATTATACCTAGGAC | |
| pT-rpoD-F | TAATACTAGTCGAGGTCAATGACCATCTGCGTTTTAGAGCTAGAAATAGC | |
| pT-rpoD-R | GCTCTAAAACGCAGATGGTCATTGACCTCGACTAGTATTATACCTAGGAC | |
1.2 培养基
低盐LB液体培养基(low salt Luria Bertani, LLB, g/L):酵母粉5,氯化钠5,蛋白胨10。固体培养基另添加20 g/L琼脂,高压蒸汽灭菌121 ℃、20 min。
MS1发酵培养基:葡萄糖20 g/L,硫酸铵16 g/L,酵母粉2 g/L,磷酸二氢钾2 g/L,硫酸镁0.5 g/L,盐溶液1 mL/L,(盐溶液母液:CuCl2 10 g/L、FeSO4·7H2O 10 g/L、ZnSO4·7H2O 10 g/L、CuSO4 0.2 g/L、NiCl2·7H2O 0.02 g/L),500 mL摇瓶装液量50 mL,高压蒸汽灭菌115 ℃、30 min。
5 L发酵培养基:葡萄糖20 g/L,硫酸铵16 g/L,酵母粉2 g/L,磷酸二氢钾2 g/L,硫酸镁0.5 g/L,甜菜碱2 g/L,β-丙氨酸1.5 g/L,盐溶液1 mL/L (盐溶液母液:CuCl2 10 g/L、FeSO4·7H2O 10 g/L、ZnSO4·7H2O 10 g/L、CuSO4 0.2 g/L、NiCl2·7H2O 0.02 g/L),消泡剂2.5 mL/L,加蒸馏水定容至1.8 L (发酵总体积为2 L),高压蒸汽灭菌115 ℃、30 min。
5 L发酵补料培养基:葡萄糖500 g/L、磷酸二氢钾14 g/L、甜菜碱4 g/L、酵母粉2 g/L、硫酸铵10 g/L、硫酸镁8 g/L、盐溶液2 mL/L、β-丙氨酸40 g/L,高压蒸汽灭菌115 ℃、30 min。
抗生素浓度为:盐酸壮观霉素(spectinomycin hydrochloride, SD) 50 mg/L;氯霉素(chloramphenicol, CM) 30 mg/L。
1.3 质粒和菌株构建pTarget突变质粒pT-gene构建:以构建pT-avtA为例,在基因avtA上确定紧邻前间区序列邻近基序(protospacer adjacent motif, PAM)位点(NGG)位置,以PAM位点前20 bp作为突变序列,设计突变引物pT-avtA-F/pT-avtA-R,以实验室保藏原始pTarget为模板,通过Phanta Max (P505)试剂盒PCR获得线性化突变质粒pT-avtA。
pdCas9质粒为本实验室保藏。
抑制菌株构建:首先制备底盘菌株DPAP10的感受态,其次转化pdCas9质粒得到重组菌株DPAP10/pdCas9,最后制备DPAP10/pdCas9的电转感受态,电转突变pTarget质粒得到最终抑制菌株DPAP10/pdCas9+pT-gene。
1.4 发酵培养从固体平板上挑取单菌落接种于LLB试管中,37 ℃、180 r/min培养12 h。取1.5 mL种子液接种于装液量50 mL发酵培养基的250 mL摇瓶中,加入0.5 g灭菌过的CaCO3,同时额外添加(每50 mL):VB1 (2 g/L) 50 μL,VB12 (5 g/L) 50 μL,IPTG (1 mol/L) 10 μL,SD (50 g/L) 50 μL,CM (30 g/L) 50 μL,β-丙氨酸(250 g/L) 450 μL,异亮氨酸(isoleucine, Ile) (40 g/L) 50 μL,30 ℃摇床180 r/min振荡培养48 h。
1.5 发酵液处理摇瓶发酵结束后,取发酵液1 mL置于2 mL EP管中,12 000 r/min离心2 min,分离沉淀与上清,上清用于后续产物检测。沉淀用于检测生物量,吸取1 mL去离子水对其吹打混匀,12 000 r/min离心2 min,去除上清;再次吸取800 μL去离子水对EP管中的沉淀物进行吹打混匀,并加入200 μL乙酸溶液用以中和沉淀中残余碳酸钙,过程中需不定时上下翻转EP管,待EP管中无气泡产生后,吸取部分溶液稀释10−20倍,控制OD600在0.2−0.8间,使用紫外分光光度计对其进行检测,根据最终OD600检测值乘以相应稀释倍数即为发酵液中所含菌体生物量。
1.6 高效液相色谱分析发酵液中DPA含量使用高效液相色谱(high performance liquid chromatography, HPLC)进行检测。发酵液上清液处理,使用去离子水将1.5中所得上清液稀释一定倍数后,使用0.22 μm无机滤膜过膜除杂或置于高速离心机12 000 r/min离心15 min,吸取200 μL上清置于液相瓶中。
流动相准备,C泵:93% (1 000 mL超纯水+ 1 mL磷酸)使用0.20 μm微孔水系滤膜过膜;A泵:7% (纯乙腈),使用0.20 μm微孔有机系滤膜过膜,并超声除去气泡。
色谱柱型号:ACQUITYUPLC BEHC18 column (100 mm×2.1 mm, 1.7 μm, Waters)。
检测仪:赛默飞UltiMate 3 000。
参数设置:样品进样量10 μL,液相柱柱温度30 ℃,流速0.75 mL/min,检测波长200 nm。
1.7 实时荧光定量聚合酶链式反应(real-time fluorescence quantitative polymerase chain reaction, RT-qPCR)用核酸提取试剂盒提取所有样本的RNA,检测提取的总RNA的浓度,然后用逆转录试剂盒按照说明书的要求将RNA逆转录成cDNA,产物立即用于RT-qPCR,最后以16S RNA为内参基因,根据采集的数据进行相对定量,计算出2−ΔΔCt值。关于RT-qPCR的引物如表 2所示。
| Primers | Sequences (5′→3′) |
| 16S-F | CCTTACGACCAGGGCTACAC |
| 16S-R | CAATCCGGACTACGACGC |
| RT-gltA-F | CGGTGAACATGGAAGACGGA |
| RT-gltA-R | AAAACATCGCGCTGAACGAC |
| RT-pgk-F | TAGCCACCTTCGTCACCAAC |
| RT-pgk-R | TGGCGCGAAAACTGTGAAAG |
| RT-ptsH-F | ACTGTGACTTCCAACGGCAA |
| RT-ptsH-R | TTTCTGCTCGTCTTCGCCTT |
| RT-ptsI-F | GAAGAGAACCCGTTCCTCGG |
| RT-ptsI-R | TGCGCAATTTACCGAAAGCC |
| RT-crp-F | CAGACCCGACTCTCGAATGG |
| RT-crp-R | AGCGTTTCCGCTTTTTCACC |
所需试剂盒均购置于南京诺唯赞生物科技股份有限公司,实时荧光定量PCR仪型号为赛默飞StepOne Plus,荧光染料ChamQ Universal SYBR qPCR Master Mix。
2 结果与分析 2.1 大肠杆菌CRISPRi系统的构建与功能验证利用本实验室保藏的pTarget质粒和pdCas9质粒构建CRISPRi双质粒系统(图 1A)。pTarget质粒为sgRNA的表达载体,pdCas9质粒在trc启动子控制下转录dCas9基因,其表达产物dCas9为Cas9核酸酶的催化失活突变体。首先将dCas9的表达载体整合到出发菌株DPAP10中,得到重组菌株DPAP10/pdCas9;随后将sgRNA的表达载体整合到重组菌株DPAP10/pdCas9中得到抑制菌株DPAP10/pdCas9+ pT-gene。CRISPRi系统的使用需注意对照组的选择及dCas9蛋白对产量的影响,由于DPAP10基因组上已经敲除了avtA基因,pT-avtA表达的sgRNA无法引导dCas9与基因组结合,因此选择菌株DPAP10/ pdCas+pT-avtA为对照菌株。分别对DPAP10、DPAP10/pdCas9、DPAP10/pdCas9+pT-avtA菌株进行摇瓶发酵,结果表明pdCas9导入使得菌株的产量下降1.55%,pdCas9和pT-avtA双质粒的导入使得菌株的产量下降4.19% (图 1B),说明CRISPRi系统的引入会对宿主细胞造成一定的代谢压力,但对DPA的合成影响是有限的,适用于大肠杆菌宿主合成途径系统优化的筛选实验。
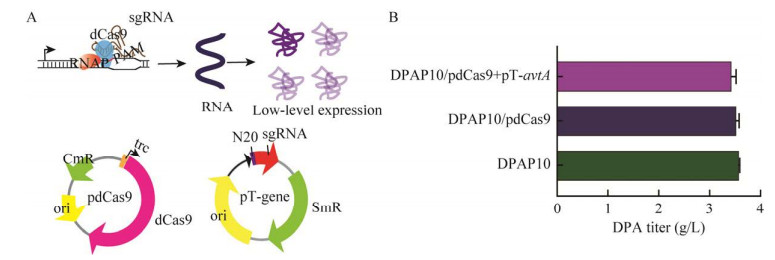
|
| 图 1 CRISPRi系统构建(A)、CRISPRi系统功能验证(B) Figure 1 Construction of CRISPRi system (A), functional verification of the CRISPRi system (B). |
2.2 sgRNA表达质粒文库的构建
在大肠杆菌中,DPA的合成途径可分为β-丙氨酸模块和D-泛解酸模块,以葡萄糖作为碳源,经磷酸烯醇式丙酮酸糖磷酸转移酶系统(phosphotransferase system, PTS)或非磷酸烯醇丙酮酸糖磷酸转移酶系统(no-PTS)转运系统进入细胞,在糖酵解途径(Embden-Meyerhof-Parnas, EMP)作用下生成磷酸烯醇式丙酮酸(phosphoenolpyruvate, PEP)以及丙酮酸(pyruvic acid, PYR),其中PEP通过磷酸烯醇式丙酮酸羧化酶(phosphoenolpyruvate carboxylase, PEPC,由ppc编码)直接转化为草酰乙酸,在天冬氨酸转氨酶(由aspC编码)作用下形成天冬氨酸,然后在天冬氨酸脱羧酶(由panD编码)作用下发生脱羧反应生成β-丙氨酸;PYR是D-泛解酸合成的重要前体,在乙酰乳酸合酶(由ilvGMIHBN编码)催化下生成乙酰乳酸,再经酮醇酸还原异构酶(ilvC基因编码)、二羟基酸脱水酶(ilvD基因编码)生成α-酮异戊酸(α-ketoisovaleric acid, 2-KIV),2-KIV在3-甲基-2-氧代丁酸羟甲基转移酶(panB基因编码)和2-脱氢泛酸2-还原酶(由panE和ilvC基因编码)作用下生成D-泛解酸,最后D-泛解酸和β-丙氨酸通过泛酸合成酶(panC编码)缩合反应形成DPA。
DPA合成过程中的中间体同时也参与了其他代谢途径,为了进一步强化DPA合成途径,寻找影响D-泛酸合成的潜在基因靶点,选择了与DPA合成直接或间接相关的126个基因靶点,构建sgRNA质粒表达文库,基因靶点包括糖分解代谢(糖酵解、磷酸戊糖途径、三羧酸循环)中的38个基因、氨基酸代谢中的39个基因、一碳单位合成中的9个基因、辅因子代谢中的26个基因、σ因子中的4个基因,以及与细胞生长形态控制相关的10个基因,选点分布如图 2所示。
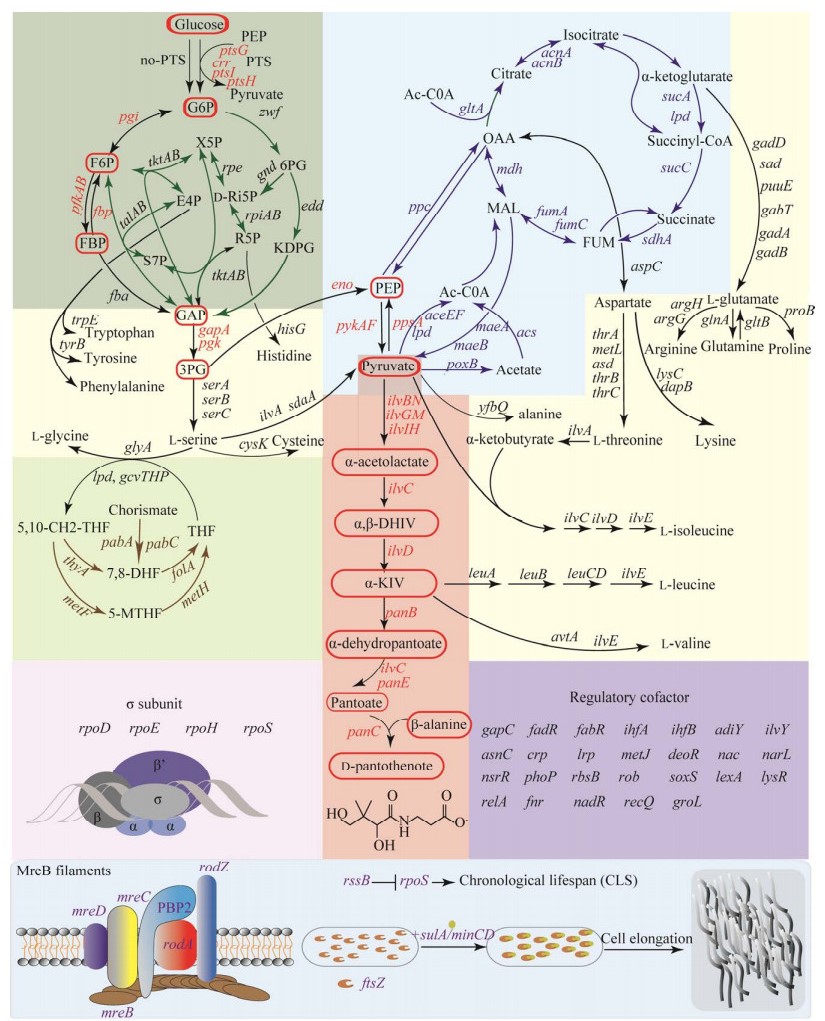
|
| 图 2 基于CRISPRi筛选DPA代谢相关靶基因位点图 Figure 2 Screening of DPA metabolism related target gene loci based on CRISPRi. |
2.3 CRISPRi抑制单基因表达对DPA生产的影响
将上述126个选点构建相应的sgRNA表达质粒pT-gene,分别导入DPAP10/pdCas9菌株得到相应基因的抑制菌株,摇瓶发酵48 h,测定DPA产量,发酵结果显示(图 3A),抑制氨基酸合成途径、干扰一碳单位循环和阻断σ因子、抑制细胞生长形态相关基因的表达不利于DPA的合成。在氨基酸代谢模块中,干扰后导致DPA产量大幅降低40%以上的基因(serA、serC、yfbQ、ilvHGM、leuD、ilvE)主要集中在以EMP途径中间产物3-磷酸甘油酸(3PG)、PYR为前体的氨基酸合成路径上,3PG同时也会转化为PYR进入TCA循环,为菌体生长提供必需的物质和能量[12],当这些基因被抑制后,代谢流从3PG和PYR大量进入TCA循环可能是DPA产量降低的原因之一,其次氨基酸是细胞存活所必需的,受到CRISPRi抑制后,减少的氨基酸数量不足以维持正常的细胞生理也会造成DPA合成减弱。研究表明[13],通向DPA生产通量受到途径中间体5, 10-亚甲基四氢叶酸(5, 10-CH2-THF)再生速度的限制,L-丝氨酸转化为甘氨酸是L-丝氨酸的主要降解途径,该反应为细胞生长提供了大部分的C1单位,对于大肠杆菌,甘氨酸裂解(glycine cleavage, GCV)系统将甘氨酸转化为5, 10-CH2-THF,该系统在维持细胞内甘氨酸和C1单位浓度之间的动态平衡方面起着重要作用,因此干扰一碳循环后,5, 10-CH2-THF再生速度降低,同时也削弱了DPA合成能力。σ因子是原核生物RNA聚合酶的一个亚基,rpoD编码σ70、rpoS编码σ38、rpoE编码σ24和rpoH编码σ32,都是转录所必需的因子,阻断σ因子影响RNA聚合酶对转录起始位点的正确识别,阻碍细胞全基因组转录,从而影响了DPA的生物合成。工业菌株的细胞大小对微生物细胞工厂的效率发挥重要作用,已有文章报道为了提高目标产物的效价,已经开发了一系列的形态工程策略,包括过表达参与细胞分裂的基因,如ftsZ,参与二进制分裂的基因的过度表达,sulA和minCD,调节肽聚糖细胞壁和类肌动蛋白mreB细胞骨架结构,以加速生长,可以增加细胞密度[14],DPA生产与细胞生长成正相关,细胞生长受抑制导致DPA产量降低。而对DPA合成具有促进作用基因主要分布在糖代谢途径(gapA、pgk、crr、ptsI、ptsH、aceE和gltA),调控辅因子(ilvY、crp、nac)模块中,说明减少其他物质对碳源葡萄糖的竞争有助于提高DPA的产量。受益于基因表达的全局调控,减少一些转录因子的表达也在一定程度上加强了DPA的合成。综上,最终确定了抑制后对DPA产量提高10%以上的基因(图 3B):pgk (29.5%)、gltA (22.1%)、ptsH (24.6%)、ptsI (10.9%)和crp (15.4%),上述基因的RT-qPCR结果显示(图 3C),相比于对照菌株DPAP10,所有干扰基因的转录水平都显著降低,证明了CRISPRi能够正常工作并用于后续研究。
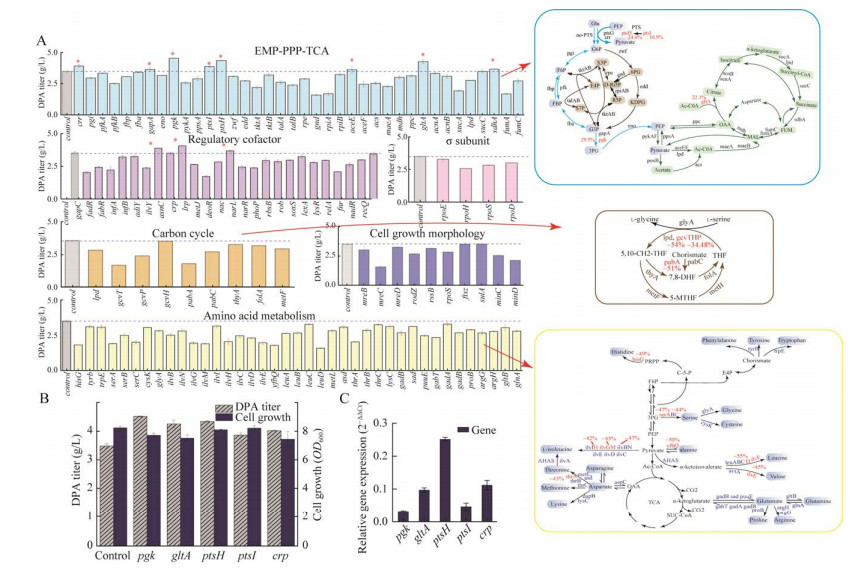
|
| 图 3 基于CRISPRi的DPA生产有益基因的筛选(A)、产量提高10%以上的抑制菌株(B)、RT-qPCR验证产量提高10%以上的抑制菌株中pgk、gltA、ptsH、ptsI和crp的转录水平强度变化(C) Figure 3 CRISPRi-based identification of beneficial genes for DPA production (A), inhibition strains that increased yield by more than 10% (B), transcriptional intensity changes of pgk, gltA, ptsH, ptsI and crp in inhibited strains whose yield was increased by more than 10% were verified by RT-qPCR (C). Error bars represent the deviation of the standard from the triplicate experiment. |
为了进一步探究pgk、gltA、ptsH、ptsI和crp基因被抑制后造成DPA合成增强的原因,对菌株的关键中间体PEP和PYR含量进行测定,发现在发酵液中几乎没有检测到PYR和PEP的残留,PYR是各种化合物和有机酸合成的重要节点,PEP可通过磷酸烯醇式丙酮酸羧化酶直接转化为草酰乙酸进入TCA循环,于是继续检测了PYR分支途径中的有机酸(甲酸、乙酸和乳酸)以及TCA循环中的琥珀酸、草酰乙酸和α-酮戊二酸的含量。结果显示(图 4①②③,相较于对照组,抑制菌株中PYR流向3种有机酸的通量分布发生了变化,抑制菌株中乳酸含量增加且多于甲酸含量(图 4①③),已有报道证明,PYR节点的甲酸裂解酶(PYR formate lyase, PFL)活力比乳酸脱氢酶(lactate dehydrogenase, LDH)高[15],上述基因抑制后,可能由于PYR节点附近的代谢通量发生变化,从而促进了乳酸的合成。乙酸含量除了在ptsH抑制菌株中降低,在其他基因抑制菌株中含量均有不同程度增加,尤其在gltA抑制菌株中乙酸增加最为显著(图 4②)。研究表明[16],当大肠杆菌在以葡萄糖为碳源的培养基中生长时,PTS能消耗50%的有效PEP生成丙酮酸。此时,PTS对葡萄糖的高摄取率结合EMP途径的葡萄糖分解代谢使乙酰辅酶A (Ac-CoA)的合成速度超过TCA循环的消耗能力,最终导致过量的Ac-CoA生成乙酸,因此抑制ptsH会导致乙酸合成速率下降。草酰乙酸和Ac-CoA是柠檬酸合成酶gltA的底物,抑制gltA表达后,进入TCA循环的Ac-CoA减少,于是更多Ac-CoA转移到乙酸生产途径。不同于PYR节点通量或增或减的变化,TCA循环中的琥珀酸、草酰乙酸和α-酮戊二酸的含量都有不同程度的降低(图 4④⑤⑥),有研究证明,3-磷酸甘油酸激酶pgk的过度表达将磷酸戊糖途径(pentose phosphate pathway, PPP)的通量导向EMP途径[17],而EMP的产物PYR是TCA循环的基础,抑制pgk后,进入TCA循环的碳流减少,因此pgk抑制菌株中,草酰乙酸和琥珀酸含量大幅降低(图 4④⑤⑥);结果显示gltA抑制菌株中草酰乙酸含量最低(图 4⑤),原因是gltA的表达水平决定了TCA循环速率[10],gltA转录水平降低,减缓TCA代谢速率,进入TCA循环的草酰乙酸减少;PTS组分的修饰或消除将对中心代谢中的碳通量分布产生重大影响[16],考虑到ptsHI的抑制导致葡萄糖消耗速率降低,在大多数情况下葡萄糖是TCA循环的主要营养来源[18],并且图中结果表明,ptsH的抑制对TCA循环的影响大于ptsI抑制,3种TCA循环中间体含量都显著降低;全局调控因子crp在整个碳源的选择性运输中起着关键的调节作用[19],在稳定期,crp-cAMP浓度随游离crp浓度的升高而升高,crp-cAMP的降低抑制了TCA循环的代谢基因[20]。综上,抑制pgk、gltA、ptsH、ptsI和crp基因后,改变了PYR的通量同时降低了TCA循环速率,进而促进了DPA的生物合成。
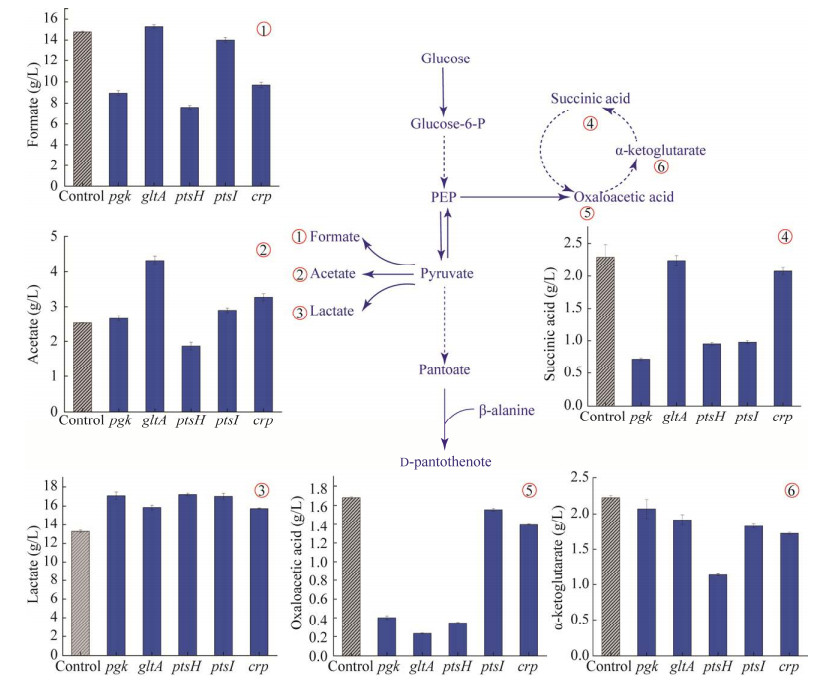
|
| 图 4 DPA合成中间代谢物变化 Figure 4 Accumulation of intermediate metabolites in DPA biosynthesis. Error bars represent the deviation of the standard from the triplicate experiment. ①: Formate. ②: Acetate. ③: Lactate. ④: Succinic acid. ⑤: Oxaloacetic acid. ⑥: α-ketoglutarate. |
2.4 CRISPRi抑制双基因表达对DPA生产的影响
在对单基因的干扰实验中,成功筛选得到了5个对DPA合成具有促进作用的基因:pgk、gltA、ptsH、ptsI和crp,为了进一步加强DPA合成能力,利用CRISPRi系统同时对双基因进行干扰。分别构建了多种基因组合pgk-gltA、pgk-ptsH、gltA-ptsH、pgk-ptsI、gltA-ptsI、pgk-crp和gltA-crp,将构建好的pTarget-sg-1-2质粒导入菌株DPAP10/pdCas9得到双基因抑制菌株DPAP10/pdCas9+pT-sg-1-2。以DPAP10/pdCas9+pT-avtA菌株为对照,对DPAP10/pdCas9+pT-sg-1-2菌株中被抑制的双基因进行了RT-qPCR检测,以验证相关基因的转录水平,结果表明,基因组合抑制的转录水平(图 5B)与基因单独抑制的转录水平(图 3C)有较大差异,2.3中结果表明这5个基因的单独抑制会改变DPA合成代谢网络,由于生物体内复杂的代谢网络,导致了双基因抑制后的转录水平与单基因抑制差别较大。图 5B显示,相比于对照菌株,抑制菌株中的两个基因转录水平均降低,并且同一个菌株中的两个基因被抑制水平相近,证明质粒pTarget-sg-1-2在CRISPRi系统中可以同时发挥两个sgRNA作用,从而指导dCas9蛋白实现对双基因的抑制。
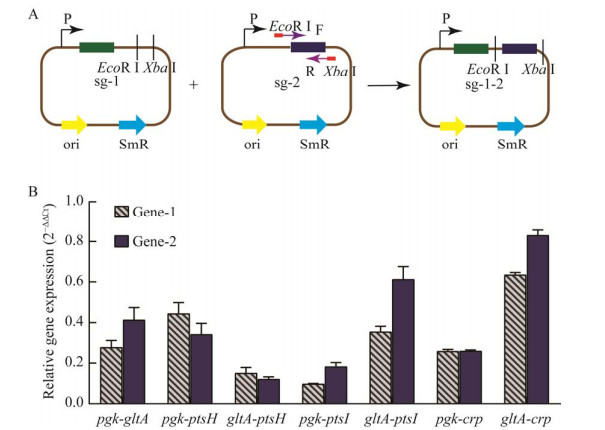
|
| 图 5 双基因抑制质粒构建(A)、RT-qPCR验证pgk-gltA、pgk-ptsH、gltA-ptsH、pgk-ptsI、gltA-ptsI、pgk-crp和gltA-crp的转录水平强度变化(B) Figure 5 Construction of double gene inhibitory plasmid (A), RT-qPCR verified the transcription levels of pgk-gltA, pgk-ptsH, gltA-ptsH, pgk-ptsI, gltA-ptsI, pgk-crp, gltA-crp (B). Error bars represent the deviation of the standard from the triplicate experiment. |
在接下来的研究中,利用构建的双基因抑制CRISPRi系统,验证其对DPA合成的影响。结果显示(图 6A),相较于对照菌株(3.50 g/L),DPAP10/pdCas9+pT-pgk-gltA产量提高了25.5%, ,DPAP10/pdCas9+pT-pgk-ptsH产量提高了40.5%,DPAP10/pdCas9+pT-gltA-ptsH产量提高了49.5%,DPAP10/pdCas9+pT-pgk-ptsI产量提高了35.3%,DPAP10/pdCas9+pT-gltA-ptsI产量提高了24.2%,DPAP10/pdCas9+pT-pgk-crp产量提高了11.9%,DPAP10/pdCas9+pT-gltA-crp产量提高了17.6%。对比单基因的抑制,双基因组合抑制对DPA生物合成更为有益。对双基因抑制菌株同单基因抑制菌株测定相同的中间代谢物,结果显示(图 6B),双基因抑制同样改变了PYR流向3种有机酸的通量分布,降低了TCA循环速率。与单基因抑制不同的是,双基因抑制菌株中有机酸含量变化更加统一,都表现为甲酸含量减少,乙酸和乳酸含量增加;TCA循环中的琥珀酸、草酰乙酸和α-酮戊二酸的含量的降低程度要低于单基因抑制,当进行双基因抑制后,TCA循环速率发生变化,PYR节点通量更多的流向泛解酸路径,促进了DPA的生物合成。本研究中得到的最优双基因抑制组合是gltA-ptsH,该组合抑制有效提升了DPA生产能力,摇瓶总产量提高49.5%达到了5.3 g/L。
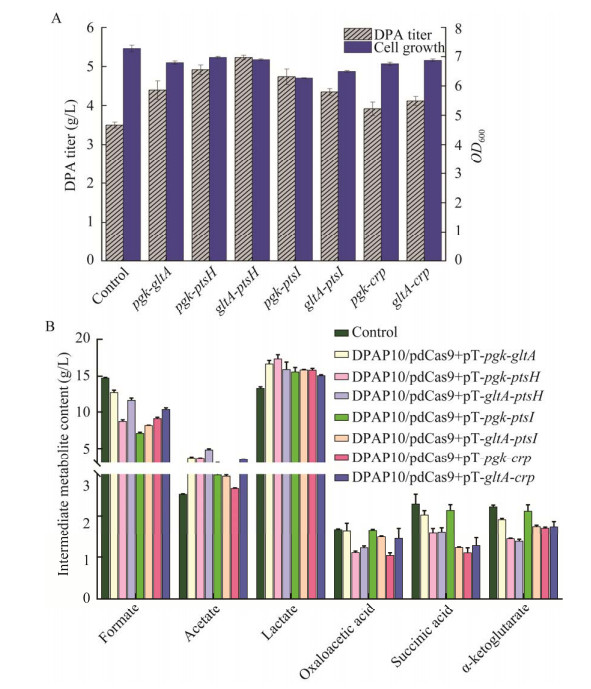
|
| 图 6 双位点阻断对DPA产量的影响(A)、双位点抑制菌株中间代谢物变化(B) Figure 6 Effect of two-site blockade on the yield of DPA (A), inhibition of intermediate metabolites by double blocking (B). Error bars represent the deviation of the standard from the triplicate experiment. |
2.5 5 L罐发酵验证
为了进一步提高DPA的产量,利用5 L发酵罐进行分批补料发酵实验。本研究通过CRISPRi确定了当gltA和ptsH基因受到组合抑制后,能够有效提高DPA的合成。但抑制菌株DPAP10/pdCas9+pT-gltA-ptsH由于含有双质粒会造成代谢压力[21],不适用于罐上放大发酵生产,于是在基因组上通过起始密码子替换实现基因的下调表达。gltA和ptsH基因的起始密码子都是ATG,起始密码子强度由大到小依次为ATG、GTG、TTG[22]。CRISPRi系统中,基因表达的抑制水平与转录起始点的靶点距离一般呈负相关,设计sgRNA分别在前端、中端、末端与非模板DNA链结合,分别对应基因抑制的高、中、低效率[9]。本研究中设计的sgRNA都在靠近转录起始位点处对基因进行高效抑制,为了在基因组上下调基因的表达水平更接近CRISPRi抑制效率,选择最低强度的TTG起始密码子替换ATG,从而下调基因ptsH和gltA的表达,得到无质粒菌株DPAP10-gltATTG-ptsHTTG。
以出发菌株DPAP10作为对照,DPAP10是异亮氨酸缺陷型菌株,在发酵过程中需要回补异亮氨酸(Ile)维持细胞生长,发酵结果显示(图 7),DPA产量随菌株的生长同步提高,DPAP10菌株在87 h产量最高,达到63.1 g/L,DPAP10-gltATTG-ptsHTTG在83 h产量最高,达到75.4 g/L,提高了19.5%。可以看出,在5 L发酵罐的放大过程中,对照菌株前期生长速率快,70 h细胞生长进入衰退期,DPA产量增长速率减慢(图 7A),改造菌株前期缓慢生长,70 h后反而出现一个快速生长和生产的趋势(图 7B),改造菌株中下调了gltA和ptsH基因的表达,造成TCA循环速率降低,草酰乙酸减少,Ile的供应更加不足,于是在发酵后期添加了40 mg/L Ile后,改造菌株刚好得到Ile补给,于是出现了一个快速生长和生产的趋势,尽管对照菌株后期也添加了同样的异亮氨酸,但并没有出现快速生长和生产的趋势,说明Ile的补给需要适时适量,当菌株并不缺乏Ile时,增加Ile并不能提高菌株产能。
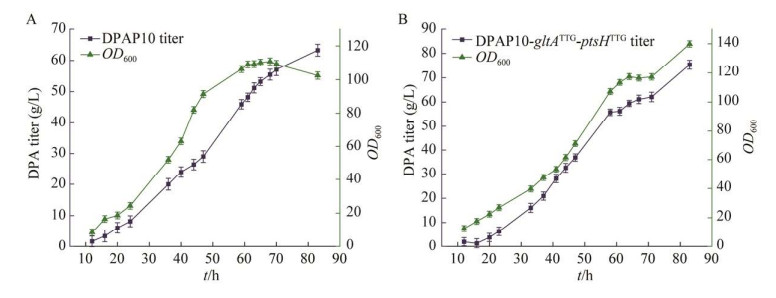
|
| 图 7 DPAP10 (A)与DPAP10-gltATTG-ptsHTTG (B)的分批补料发酵过程的比较 Figure 7 Comparison of DPAP10 (A) and DPAP10-gltATTG-ptsHTTG (B) in a batch replenishment fermentation process. Error bars represent the deviation of the standard from the triplicate experiment. |
3 讨论与结论
本研究通过CRISPRi技术,根据与DPA合成直接或间接相关代谢途径,在糖分解代谢(糖酵解、磷酸戊糖途径、三羧酸循环)模块、氨基酸代谢模块、调控辅因子模块、σ因子以及细胞生长形态模块共126个基因靶点中,最终筛选得到DPA产量提高10%以上的基因:pgk (29.5%)、gltA (22.1%)、ptsH (24.6%)、ptsI (10.9%)和crp (15.4%);在此基础上,构建了CRISPRi双基因抑制系统,同时干扰双基因pgk-gltA、pgk-ptsH、gltA-ptsH、pgk-ptsI、gltA-ptsI、pgk-crp和gltA-crp,组合抑制后得到菌株DPAP10/pdCas9+pT-gltA-ptsH的DPA产量最高,达到了5.3 g/L,是对照菌株DPAP10/pdCas9+pT-avtA产量的1.5倍;最后依据筛选结果,在基因组上下调基因ptsH和gltA的表达,得到工程菌株DPAP10-gltATTG-ptsHTTG,在5 L发酵罐中放大培养,最终产量达到75.4 g/L,相比于出发菌株DPAP10 (63.1 g/L)提高了19.5%。
根据上文单基因抑制的结果与分析,发现PYR和TCA循环是影响DPA合成的关键节点,于是对以PYR为节点的DPA合成竞争支路的有机酸和TCA循环的关键中间体进行检测,根据这些中间代谢物发生的变化来反映代谢通路的变化,本研究中间代谢物检测结果表明,无论是单基因抑制菌株还是双基因抑制菌株,在改变了PYR通量分布的同时降低了TCA循环速率,从而导致了更加高效的DPA合成,这一结果也说明通过重新分配代谢通量可以实现目标产物的高效合成,利用体外模块化工程与体内CRISPRi技术相结合可以调控多基因表达从而协调路径通量分布[23],受此启发,在未来的研究中,使用CRISPRi干扰,为DPA合成的通路模块合理分配基因表达来消除约束,最大限度提高产量。
在大规模筛选DPA生长关键基因方面,CRISPRi抑制方法显然比基因敲除方法更有优势。基因敲除更适合操作少量基因和对生长不关键的基因,CRISPRi抑制不能完全去除基因功能,残留功能可能会导致对结果的误解。此外,多拷贝质粒的存在给细胞带来代谢负担,这是由于质粒复制和质粒基因翻译对核苷酸的需求增加。本研究描述的DPA生产实验中,使用了基因抑制,但对少数不是生长关键的基因,没有进行敲除。然而,根据实验目的,结合CRISPRi抑制和基因敲除是必要的。在之后的研究中,应继续扩大DPA的筛选基因文库,对从中确定的非关键基因但对产量有提高的基因进行敲除,对确定的关键基因进行单基因的精细调控以及多基因的组合优化以实现代谢通量的合理分配。另外,利用筛选得到的DPA合成有益基因在基因组上进一步进行代谢改造,构建无质粒的工程菌,并对发酵条件进行优化,以实现产物合成的最优策略。
| [1] | ZHANG B, ZHANG XM, WANG W, LIU ZQ, ZHENG YG. Metabolic engineering of Escherichia coli for D-pantothenic acid production[J]. Food Chemistry, 2019, 294: 267-275 DOI:10.1016/j.foodchem.2019.05.044. |
| [2] | ZOU SP, ZHAO K, WANG ZJ, ZHANG B, LIU ZQ, ZHENG YG. Overproduction of D-pantothenic acid via fermentation conditions optimization and isoleucine feeding from recombinant Escherichia coli W3110[J]. 3 Biotech, 2021, 11(6): 295 DOI:10.1007/s13205-021-02773-0. |
| [3] | CHASSAGNOLE C, DIANO A, LÉTISSE F, LINDLEY, ND. Metabolic network analysis during fed-batch cultivation of Corynebacterium glutamicum for pantothenic acid production: first quantitative data and analysis of by-product formation[J]. Journal of Biotechnology, 2003, 104(1/2/3): 261-272. |
| [4] | YOCUM RR, PATTERSON TA, PERO JG, HERMANN T. Microorganisms and processes for enhanced production of pantothenate[P]. US: 7989187B2. 2011-08-02. |
| [5] | ZHANG B, CHEN L, JIN JY, ZHONG N, CAI X, ZOU SP, ZHOU HY, LIU ZQ, ZHENG YG. Strengthening the (R)-pantoate pathway to produce D-pantothenic acid based on systematic metabolic analysis[J]. Food Bioscience, 2021, 43: 101283 DOI:10.1016/j.fbio.2021.101283. |
| [6] |
田开仁, 薛二淑, 宋倩倩, 乔建军, 李艳妮. CRISPR-dCas9调控基因转录的研究进展[J]. 中国生物工程杂志, 2018, 38(7): 94-101.
DOI:10.13523/j.cb.20180713 TIAN KR, XUE ES, SONG QQ, QIAO JJ, LI YN. The research progress of CRISPR-dCas9 in transcriptional regulation[J]. China Biotechnology, 2018, 38(7): 94-101 (in Chinese). |
| [7] | STACHLER AE, SCHWARZ TS, SCHREIBER S, MARCHFELDER A. CRISPRi as an efficient tool for gene repression in archaea[J]. Methods, 2020, 172: 76-85 DOI:10.1016/j.ymeth.2019.05.023. |
| [8] | BATIANIS C, KOZAEVA E, DAMALAS SG, MARTÍN-PASCUAL M, VOLKE DC, NIKEL PI, MARTINS DOS SANTOS VAP. An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida[J]. Microbial Biotechnology, 2020, 13(2): 368-385 DOI:10.1111/1751-7915.13533. |
| [9] | FANG LX, FAN J, LUO SL, CHEN YR, WANG CY, CAO YX, SONG H. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids[J]. Nature Communications, 2021, 12: 4976 DOI:10.1038/s41467-021-25243-w. |
| [10] | LI N, SHAN XY, ZHOU JW, YU SQ. Identification of key genes through the constructed CRISPR-dcas9 to facilitate the efficient production of O-acetylhomoserine in Corynebacterium glutamicum[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 978686 DOI:10.3389/fbioe.2022.978686. |
| [11] | LI B, ZHANG B, WANG P, CAI X, TANG YQ, JIN JY, LIANG JX, LIU ZQ, ZHENG YG. Targeting metabolic driving and minimization of by-products synthesis for high-yield production of D-pantothenate in Escherichia coli[J]. Biotechnology Journal, 2022, 17(1): e2100431 DOI:10.1002/biot.202100431. |
| [12] |
罗玉常, 窦文芳, 张晓梅, 史劲松, 许正宏. 谷氨酸棒杆菌ilvE基因的敲除对相关氨基酸合成的影响[J]. 生物技术通报, 2012(11): 185-191.
LUO YC, DOU WF, ZHANG XM, SHI JS, XU ZH. Effect of ilvE gene knockout on amino acids synthesis of Corynebacterium glutamicum[J]. Biotechnology Bulletin, 2012(11): 185-191 (in Chinese). |
| [13] | STAUFFER LT, STAUFFER GV. Role for the leucine-responsive regulatory protein (Lrp) as a structural protein in regulating the Escherichia coli gcvTHP operon[J]. Microbiology, 1999, 145(3): 569-576 DOI:10.1099/13500872-145-3-569. |
| [14] | GUO L, DIAO WW, GAO C, HU GP, DING Q, YE C, CHEN XL, LIU J, LIU LM. Engineering Escherichia coli lifespan for enhancing chemical production[J]. Nature Catalysis, 2020, 3(3): 307-318 DOI:10.1038/s41929-019-0411-7. |
| [15] | YANG YT, BENNETT GN, SAN KY. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli[J]. Metabolic Engineering, 2001, 3(2): 115-123 DOI:10.1006/mben.2000.0166. |
| [16] | GOSSET G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate: sugar phosphotransferase system[J]. Microbial Cell Factories, 2005, 4(1): 14 DOI:10.1186/1475-2859-4-14. |
| [17] | LAI SJ, ZHANG Y, LIU SW, LIANG Y, SHANG XL, CHAI X, WEN TY. Metabolic engineering and flux analysis of Corynebacterium glutamicum for L-serine production[J]. Science China Life Sciences, 2012, 55(4): 283-290 DOI:10.1007/s11427-012-4304-0. |
| [18] | LIU SY, DAI ZW, COOPER DE, KIRSCH DG, LOCASALE JW. Quantitative analysis of the physiological contributions of glucose to the TCA cycle[J]. Cell Metabolism, 2020, 32(4): 619-628.e21 DOI:10.1016/j.cmet.2020.09.005. |
| [19] | SHIMADA T, FUJITA N, YAMAMOTO K, ISHIHAMA A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources[J]. PLoS One, 2011, 6(6): e20081 DOI:10.1371/journal.pone.0020081. |
| [20] | JAHAN N, MAEDA K, MATSUOKA Y, SUGIMOTO Y, KURATA H. Development of an accurate kinetic model for the central carbon metabolism of Escherichia coli[J]. Microbial Cell Factories, 2016, 15(1): 112 DOI:10.1186/s12934-016-0511-x. |
| [21] | SINGHA TK, GULATI P, MOHANTY A, KHASA YP, KAPOOR RK, KUMAR S. Efficient genetic approaches for improvement of plasmid based expression of recombinant protein in Escherichia coli: a review[J]. Process Biochemistry, 2017, 55: 17-31 DOI:10.1016/j.procbio.2017.01.026. |
| [22] | HECHT A, GLASGOW J, JASCHKE PR, BAWAZER LA, MUNSON MS, COCHRAN JR, ENDY D, SALIT M. Measurements of translation initiation from all 64 codons in E. coli[J]. Nucleic Acids Research, 2017, 45(7): 3615-3626 DOI:10.1093/nar/gkx070. |
| [23] | GAO C, WANG SH, HU GP, GUO L, CHEN XL, XU P, LIU LM. Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning[J]. Biotechnology and Bioengineering, 2018, 115(3): 661-672 DOI:10.1002/bit.26486. |
 2023, Vol. 63
2023, Vol. 63




