中国科学院微生物研究所,中国微生物学会
文章信息
- 黄倩妮, 陶媛美慧, 黄虞远, 濮吉, 罗雪莲, 金东, 杨晶, 徐建国. 2022
- HUANG Qianni, TAO Yuanmeihui, HUANG Yuyuan, PU Ji, LUO Xuelian, JIN Dong, YANG Jing, XU Jianguo.
- 一株新型旱獭源性宽谱大肠杆菌噬菌体的分离鉴定及基因组分析
- Characterization and complete genomic analysis of a novel broad host-range Escherichia coli phage isolated from Marmota himalayana
- 微生物学报, 62(9): 3503-3517
- Acta Microbiologica Sinica, 62(9): 3503-3517
-
文章历史
- 收稿日期:2022-01-16
- 修回日期:2022-03-31
- 网络出版日期:2022-04-14
2. 中国疾病预防控制中心传染病预防控制所, 传染病预防控制国家重点实验室, 北京 102206;
3. 中国医学科学院, 未知微生物发现和功能研究院外创新单元, 北京 100005;
4. 南开大学公共卫生和健康研究院, 天津 300071
2. State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Prevention and Control, Chinese Center for Disease Control and Prevention, Beijing 102206, China;
3. Research Units of Discovery of Unknown Bacteria and Function, Chinese Academy of Medical Sciences, Beijing 100005, China;
4. Research Institute of Public Health, Nankai University, Tianjin 300071, China
大肠杆菌(Escherichia coli)是人类和动物的肠道菌群中最常见的细菌类群,包含无害正常菌群(normal flora)和引起人类感染的致病性大肠杆菌,根据其致病性可分为肠内致病型大肠杆菌以及肠外致病型大肠杆菌。目前研究最广泛的包括肠致病性大肠杆菌(Enteropathogenic E. coli,EPEC)、肠出血性大肠杆菌(enterohemorrhagic E. coli,EHEC)、肠产毒素性大肠杆菌(enterotoxigenic E. coli,ETEC)、肠侵袭性大肠杆菌(enteroinvasive E. coli,EIEC)、肠聚集性大肠杆菌(enteroaggregative E. coli,EAEC)、扩散粘附性大肠杆菌(diffusely adherent E. coli,DAEC)、新生儿脑膜炎相关大肠杆菌(neonatal meningitis-associated E. coli,NMEC)和尿路致病性大肠杆菌(uropathogenic E. coli,UPEC)等,可引起腹泻、脑膜炎、尿路感染甚至是溶血性尿毒症综合征(hemolytic uremic syndrome,HUS)等[1]临床症状。目前抗生素是治疗大肠杆菌感染性疾病的主要药物。因抗生素的滥用,大肠杆菌的耐药率不断上升,其多重耐药(multiple drug resistance,MDR)和广泛耐药(extreme drug resistant,XDR)菌株近期均有报道[2–3]。据2020年中国细菌耐药监测报告[4],大肠杆菌在医院临床革兰氏阴性分离菌中分离率排名第一(占革兰阴性菌29.7%),对三代头孢和喹诺酮的平均耐药率分别为51.6%和50.7%,耐碳青霉烯类大肠杆菌也偶见报道。耐药菌株的出现给临床治疗增加了难度,而噬菌体疗法是当前防治耐药大肠杆菌感染最有潜力的代替方法之一。
Cepko等[5]发现噬菌体能显著降低EAEC菌株在感染小鼠模型的体内定殖;Easwaran等[6]在石斑鱼模型中利用噬菌体治疗多重耐药大肠杆菌的感染,与对照组(70%)相比,实验组石斑鱼的存活率(100%)显著提高;Alomari等[7]发现益生菌联合噬菌体疗法可刺激犊牛免疫机制,在24–48 h内可完全消除大肠杆菌所引起的腹泻症状。早在2017年,世界卫生组织(WHO)就把大肠杆菌列为优先开发和研究新型抗菌剂的病原体[8]。近期,美国食品和药物管理局(FDA)批准了噬菌体疗法的临床试验,用于扩大准入的临床研究新药(eIND)[9–10]。噬菌体因其广阔的应用前景受到科学家的广泛关注。
课题组前期从喜马拉雅旱獭的肠道内容物中分离并测序了125株大肠杆菌,并证实旱獭分离源大肠杆菌携带多种毒力基因[11]。本研究以其中一株旱獭分离源的大肠杆菌为宿主菌,从喜马拉雅旱獭的肠道内容物样本中分离并鉴定了一株新型大肠杆菌噬菌体vB_EcoM_TH18,并对其生物学特性及基因组进行了系统研究。
1 材料与方法 1.1 材料 1.1.1 分离样本及菌株喜马拉雅旱獭肠内容物样本(50份)于2018年采集自青海省玉树藏族自治州。噬菌体裂解谱实验涉及155株菌,包括从125株旱獭分离源大肠杆菌[11]中随机遴选的94株分离株、37株国内不同来源E. coli菌株、3株E. coli O157:H7菌株(Sakai/Xuzhou21/EDL933)、4株沙门氏菌(Salmonella enteritidis、S. typhimurium、S. choleraesuis和S. paratyphi)以及17株临床耐药菌株,包括6株志贺氏菌(Shigella sonnei、S. flexneri 1a、S. flexneri 2a、S. flexneri 2b和S. flexneri Xv)、2株肺炎克雷伯杆菌(Klebsiella pneumoniae)、3株鲍曼不动杆菌(Acinetobacter baumannii)、3株屎肠球菌(Enterococcus faecium)和3株金黄色葡萄球菌(Staphylococcus aureus)。
旱獭分离源大肠杆菌及E. coli O157:H7菌株为本实验室分离鉴定并保存。国内来源E. coli菌株及沙门氏菌株分别由中国疾病预防控制中心传染病所熊衍文研究员和闫梅英研究员提供或惠赠。
1.1.2 主要试剂和仪器试剂:LB肉汤(Luria Bertani Broth),OXOID公司;细菌琼脂粉,BIODEE公司;聚乙二醇(PEG8000),Biotopped公司;氯化钠和氯化钙,AMRESCO公司;七水硫酸镁,BEIJING LABLEAD公司;Tris-HCl (pH=7.5),北京索莱宝科技有限公司;0.22 μm和0.45 μm微孔过滤器,Millipore公司;QIAamp MinElute Virus Spin Kit,QIAGEN公司。
仪器:核酸电泳仪,Bio-Rad公司;梯度PCR仪,Sensoquest公司;透射电镜,Hitachi公司。
1.2 噬菌体的富集与分离纯化将旱獭肠内容物样本混于SM缓冲液(100 mmol/L NaCl、8.1 mmol/L MgSO4·7H2O、50 mmol/L Tris-HCl)中,4 ℃、80 r/min过夜培养,使用0.45 μm过滤器过滤。取20 mL滤液加入1 mL对数生长期(OD600=0.4–0.5)的大肠杆菌、10 mL LB液体培养基,37 ℃振荡过夜,0.22 μm过滤器过滤,所得的上清液即为噬菌体原液。利用双层琼脂平板法[12]分离纯化噬菌体。取500 μL不同浓度的噬菌体原液稀释液平铺于LB平板(添加10 mmol/L CaCl2和2 mmol/L MgSO4)并倒置于37 ℃孵箱过夜培养,观察噬菌斑的形成情况。若有,则挑取清亮、边缘清晰的单个噬菌斑,置于SM缓冲液中,4 ℃静置过夜,使噬菌斑充分解离。重复以上步骤并挑取单个噬菌斑并增殖纯化5次以上,以获得噬斑形态大小均一的噬菌体。
1.3 噬菌体透射电镜的观察取20 μL纯化后的噬菌体裂解液吸附于铜网上,自然沉淀15 min,加入一滴2%的磷钨酸(PTA)负染10 min,利用透射电镜(TEM)观察并拍摄噬菌体的形态特征。
1.4 最佳感染复数(multiplicity of infection,MOI)及一步生长曲线的测定感染复数是指噬菌体浓度(PFU/mL)与细菌浓度(CFU/mL)的比值。将噬菌体与对数生长期的大肠杆菌按不同比例(1 000、100、10、1、0.1和0.01)混合,效价最高的比例即为噬菌体的最佳感染复数。
将对数期的大肠杆菌与噬菌体以最佳感染复数的比例混合,37 ℃孵育30 min后,离心1 min收集菌体,用LB液体培养基洗涤3次后,重悬于50 mL LB液体培养基中,37 ℃、180 r/min培养,每隔10 min检测1次培养液的效价,重复3次平行实验。绘制一步生长曲线,确定噬菌体的潜伏期和爆发期并计算裂解量。
裂解量计算公式:裂解量=裂解末期噬菌体效价/感染初期宿主菌浓度。
1.5 噬菌体pH稳定性测定LB液体培养基调节至不同的pH值后,高压灭菌。取100 μL噬菌体裂解液分别加入900 μL不同pH的LB液体培养基中,37 ℃孵育1 h后测定效价。
1.6 噬菌体宿主裂解谱及成斑效率的测定利用点板试验(spot test assay)初步测定噬菌体宿主裂解谱。取500 μL不同受试菌液平铺于双层琼脂平板,静置5 min,取5 μL噬菌体裂解液滴于平板上,静置至裂解液被吸收,过夜培养后观察有无噬菌斑的形成。检测噬菌体对受试菌株的裂解效率,并计算对受试菌株的成斑效率(efficiency of plating,EOP)[13],EOP检测方法参考文献[14]中所描述的步骤进行。EOP的计算公式为试验菌的空斑形成单位(PFU/mL)与宿主菌的空斑形成单位(PFU/mL)的比值。EOP≥0.5表示噬菌体对该受试菌株的裂解活性高,0.1≤EOP < 0.5表示裂解活性为中等,EOP < 0.1表示裂解活性低[15]。
1.7 噬菌体全基因组的提取以及生物学信息分析利用QIAamp MinElute Virus Spin Kit提取噬菌体DNA,并委托北京诺禾致源生物信息科技有限公司完成建库和测序。基于NCBI在线工具BLASTn (https://blast.ncbi.nlm.nih.gov/Blast)进行全基因组序列相似性比对,使用RAST 2.0[16] (http://rast.nmpdr.org/)进行注释,并利用GeneMark[17] (http://opal.biology.gatech.edu/GeneMark/)进行手动验证;基于耐药基因数据库(CARD)[18]和毒力因子数据库(VFDB)[19]检测耐药基因和毒力基因。通过tRNAscan-SE[20] (http://lowelab.ucsc.edu/tRNAscan-SE/)预测tRNA基因。使用CGView Server[21] (http://stothard.afns.ualberta.ca/cgview_server/)绘制基因组图谱。通过RAxML (v8.2.X)[22] (http://www.exelixis-lab.org/web/software/raxml/)和Mega-X (v.10.0.5)[23] (http://www.megasoftware. net)构建系统发育进化树。使用Easyfig 2.2.5[24] (http://mjsull.github.io/Easyfig/)绘制全基因组共线性比对图。通过CoreGenes 5.0软件[25]计算基因共享率。使用使用PHACTS软件[26]确定噬菌体生活方式。PHASTER软件[27]预测整合酶相关基因及其附着位点。
2 结果与分析 2.1 噬菌体的形态特征噬斑形态呈边缘光滑、无晕环、透亮、圆形,直径约为1.0 mm (图 1A)。在透射电镜下,噬菌体头部呈六边形,为典型的正20面体结构,狭窄的颈连接头部与尾部,尾部有收缩性,其头部直径为(90±5) nm,尾部长度为(115±5) nm (图 1B)。依据国际病毒分类委员会(International Committee on Taxonomy of Viruses,ICTV)公布的最新国际病毒分类方法[28],该噬菌体属有尾噬菌体目(Caudovirales) 肌尾噬菌体科(Myoviridae)。将本研究获得的大肠杆菌噬菌体,命名为vB_EcoM_TH18。
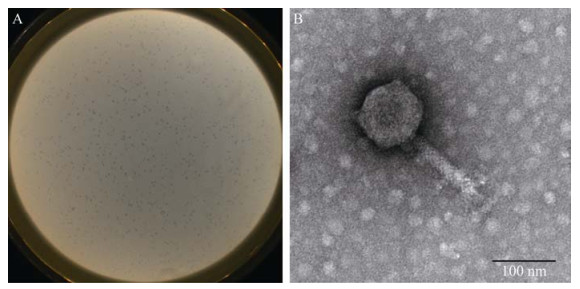
|
| 图 1 噬菌体vB_EcoM_TH18的形态特征 Figure 1 Morphology of phage vB_EcoM_TH18. A: plaques morphology; B: morphology by TEM. |
2.2 噬菌体最佳感染复数与一步生长曲线
当噬菌体与细菌的混合比例为1:1时,噬菌体效价最高,可达到1×108 PFU/mL,因此噬菌体最佳感染复数为1 (图 2A)。
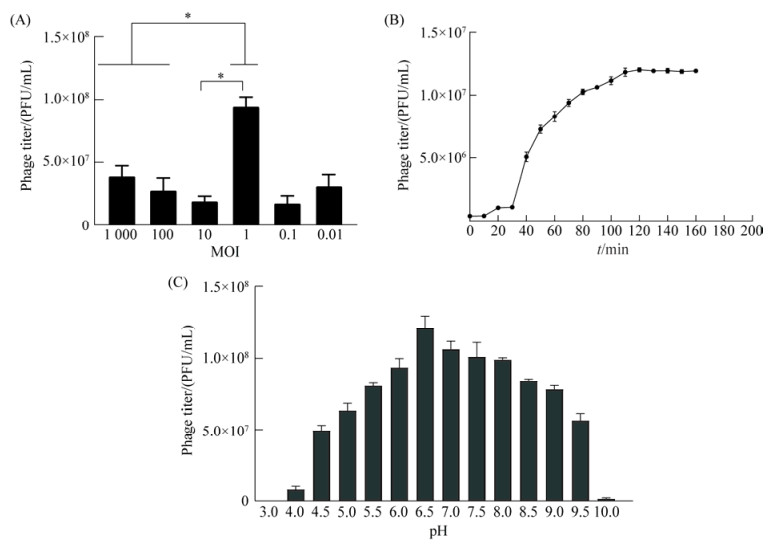
|
| 图 2 噬菌体vB_EcoM_TH18生物学特性 Figure 2 Biological characteristics of phage vB_EcoM_TH18. A: determination of optimal multiplicity of infection (MOI) of phage; B: one-step growth curve of phage; C: the influence of various pH on activity of phage. Each point represents the means SD from three replicate experiments. *: P < 0.05. |
利用实验数据绘制一步生长曲线。如图 2B所示,噬菌体感染宿主菌后的10 min内的效价无明显变化,潜伏期约为10 min;在20 min后噬菌体的效价快速上升,30 min进入爆发期,爆发期持续时间约为90 min,第110 min进入平台期,最高效价可达1.2×107 PFU/mL,裂解量为15 PFU/cell。
2.3 噬菌体pH稳定性vB_EcoM_TH18在pH 4.5–9.5范围内能维持较好的活性,效价维持稳定。当pH为6.5时,噬菌体的活性最高,效价达到1.27×108 PFU/mL。随着pH值的升高或降低,噬菌体的效价下降明显;过酸或过碱时,噬菌体会完全失活(图 2C)。
2.4 噬菌体宿主裂解谱及成斑效率为确定噬菌体的宿主裂解谱范围,如表 1所示,点板试验结果观察到vB_EcoM_TH18可裂解E. coli (n=73)及志贺杆菌(Shigella sonnei) (n=2)菌株,而其他菌属菌株均未观察到裂解现象。成斑试验结果显示,该噬菌体可有效裂解55.7% (73株)的E. coli菌株,包括8种不同致病型别及6种血清型。噬菌体对不同细菌菌株的裂解效率不同。研究发现,vB_EcoM_TH18对其中55% E. coli菌株具有高裂解活性,对32% E. coli菌株具有中等裂解活性,对13% E. coli菌株低裂解活性较低。而对100% Shigella sonnei菌株具有高裂解活性。研究发现噬菌体的裂解效率与细菌的致病型和血清型无关联。
| Bacterial | Pathotypes/ Serotypes | Spot test positive number/Tested number | Isolation source | EOP | ||
| EOP≥0.5 (n=42) |
0.1≤EOP < 0.5 (n=23) |
EOP < 0.1 (n=10) |
||||
| Escherichia coli | EHECa | 1/2 | Marmot | 1 | ||
| Escherichia coli | EPECb | 7/12 | Marmot/Clinical/ Environment/Food | 5 | 1 | 1 |
| Escherichia coli | EAECc | 9/12 | Marmot/Clinical/ Environment/Food | 3 | 4 | 2 |
| Escherichia coli | ETECd | 7/13 | Marmot | 3 | 3 | 1 |
| Escherichia coli | STECe | 12/33 | Marmot/Clinical/ Environment/Food | 9 | 1 | 2 |
| Escherichia coli | EIECf | 7/8 | Marmot | 4 | 2 | 1 |
| Escherichia coli | UPECg | 4/8 | Marmot | 1 | 2 | 1 |
| Escherichia coli | NMECh | 5/9 | Marmot | 3 | 2 | |
| Escherichia coli | ONTi | 13/20 | Marmot | 7 | 6 | |
| Escherichia coli | O157:H7 | 1/3 | Human | 1 | ||
| Escherichia coli | O113:H6 | 1/2 | Marmot | 1 | ||
| Escherichia coli | O16:H14 | 2/2 | Marmot | 1 | 1 | |
| Escherichia coli | O175:H28 | 1/1 | Marmot | 1 | ||
| Escherichia coli | O51:H1 | 2/2 | Marmot | 1 | 1 | |
| Escherichia coli | O8:H7 | 1/1 | Marmot | 1 | ||
| Shigella sonnei | – | 2/2 | Human | 2 | ||
| a: enterohemorrhagic E. coli; b: enteropathogenic E. coli; c: enteroaggregative E. coli; d: enterotoxigenic E. coli; e: shiga toxin-producing E. coli; f: enteroinvasive E. coli; g: uropathogenic E. coli; h: neonatal meningitis-associated E. coli; i: O-untypable. | ||||||
对国内不同来源的37株E. coli菌株进行成斑试验,所试验的菌株均进行耐药表型实验[29]。如图 3所示,vB_EcoM_TH18可裂解40.5% (n=15)来自8个不同省份的临床、食品、环境等来源的E. coli菌株。研究发现,vB_EcoM_TH18对其中73.3% E. coli菌株具有高裂解活性,其中就包含5株对氨苄西林、氨苄西林-舒巴坦、阿奇霉素、萘啶酮酸、四环素、头孢噻肟和甲氧苄啶-磺胺甲恶唑抗生素产生耐药性的E. coli菌株。研究表明,噬菌体裂解效率与细菌耐药表型无关联。
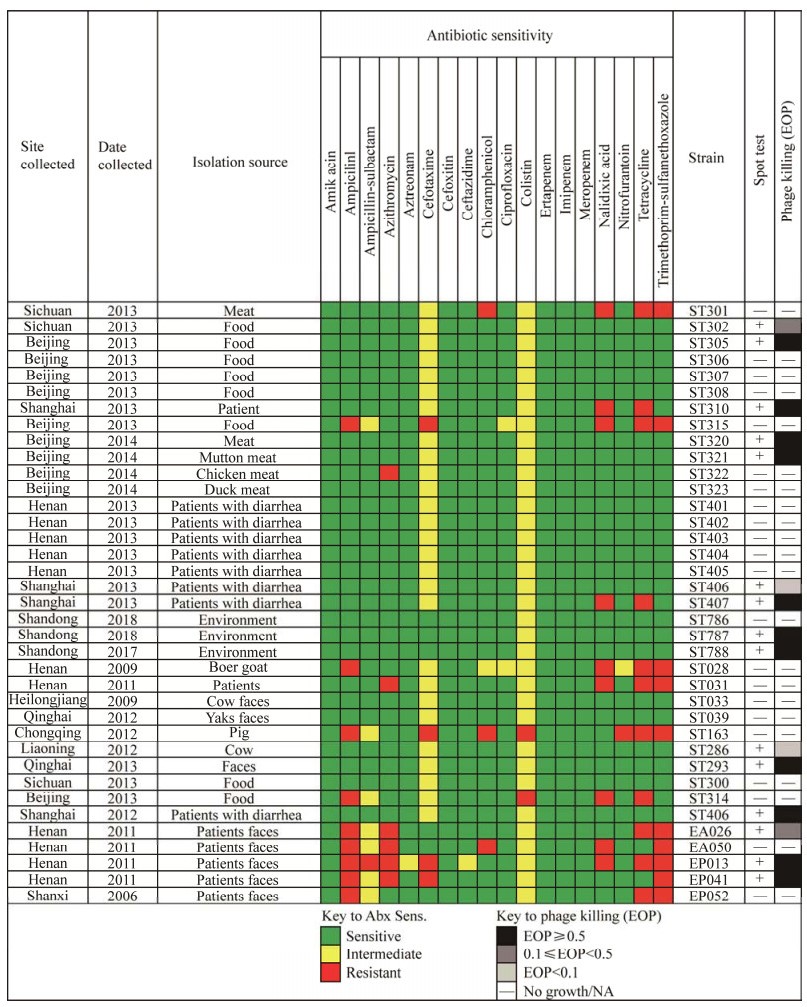
|
| 图 3 国内不同来源大肠杆菌菌株的抗生素敏感性及噬菌体成斑效率 Figure 3 Data for antibiotic sensitivity and phage killing (EOP) of E. coli isolated from different sources. |
2.5 噬菌体的全基因组特征
测序结果显示,vB_EcoM_TH18的基因组为线性双链DNA (dsDNA),长度为133 882 bp (GenBank登录号为ON075799),GC含量为39.95%,低于大肠杆菌的GC含量均值50.55% (基于NCBI数据库1 600余个基因组数据统计)。通过RAST在线注释,结果见图 4,共注释到210个蛋白质编码序列(CDS),其中129个(61.4%)在反义链上转录,其余在正义链上转录。基因组共预测到13个tRNA基因,位于69 818和75 486位点之间,未预测到毒力和耐药基因。通过PHACTs和PHASTER软件进行分类和预测,未预测到整合酶相关的基因,结果提示vB_EcoM_TH18为1株裂解性噬菌体。
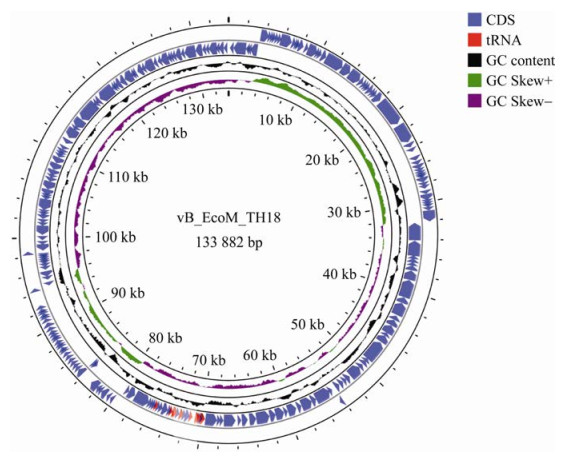
|
| 图 4 噬菌体vB_EcoM_TH18基因组图谱 Figure 4 The genome map of phage vB_EcoM_TH18. |
vB_EcoM_TH18的基因组中共有175个CDS (83.3%)预测为假定蛋白(hypothetical protein),预测为已知功能蛋白的CDS仅有35个(16.7%),分为6个模块:编码形态结构、核苷酸复制与代谢、DNA包装、宿主菌裂解、溶原调节和代谢相关酶(图 5)。在形态结构模块中,CDS 82编码尾纤维蛋白,在感染初期可特异性识别细菌表面受体[30–33]。核苷酸复制与代谢模块中,CDS 5编码抗终止子因子,参与噬菌体独特的基因调控形式,介导基因表达[34]。核酸内切酶(CDS 90、CDS 191)、核酸外切酶(CDS 189)、厌氧和需氧核糖核苷酸还原酶大亚基(CDS 175、CDS 176、CDS 177、CDS 178、CDS 180、CDS 181)以及胸苷酸合酶(CDS 184)共同合成用于噬菌体DNA合成的脱氧核糖核苷酸[35],此外核酸内切酶中包含HNH型归巢核酸内切酶(CDS90),该酶可介导内含子水平转移从而参与各个感染时期不同转录类别[36–37];DNA连接酶(CDS 203)、DNA聚合酶(CDS 34、CDS 36)、DNA引物酶以及DNA解旋酶(CDS 24、CDS 33)参与合成噬菌体DNA[35];RNA连接酶2 (CDS 18)用于连接RNA,参与封闭dsRNA/DNA杂交种的缺口[38]。终止酶大亚基(CDS 89、CDS 91、CDS 92)是DNA转移到空衣壳的必要条件,启动或终止噬菌体DNA包装反应[39]。CDS 172编码细胞内溶素(endolysin),可水解细胞壁肽聚酶,以释放子代噬菌体[40]。CDS 45、CDS 49编码ea22蛋白,可能有利于维持溶原状态,抑制噬菌体的裂解并对细菌的存活产生影响[41]。
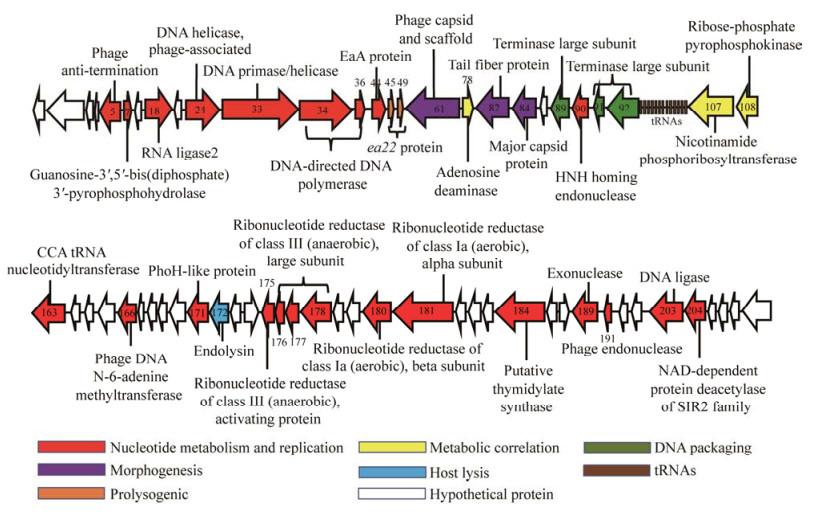
|
| 图 5 噬菌体基因组中35个功能基因结构图谱 Figure 5 Schematic diagram of the 35 functional CDSs of phage. The predicted CDSs are indicated as arrowheads and the number in arrows represents the serial number of CDSs. |
2.6 系统进化发育分析和比较基因组学
经过BLASTn比对,vB_EcoM_TH18的基因组与Avunavirus属内的Escherichia phage Av-05 (GenBank登录号:KM190144)具有较高的相似性(95.17%),覆盖率为75%。比较两者的基因组大小及tRNA含量,均有差异[42]。基于两者保守且有进化意义的序列(主要衣壳蛋白和终止酶大亚基)进行比较,其同源性分别为98.03%和99.62%。
为进一步明确vB_EcoM_TH18的分类学位置,基于全基因组序列、主要衣壳蛋白和终止酶大亚基序列分别构建系统发育树。3个进化树结果(图 6)均表明TH18与Av-05处于在同一进化分支上,两者具有共同的进化起源。
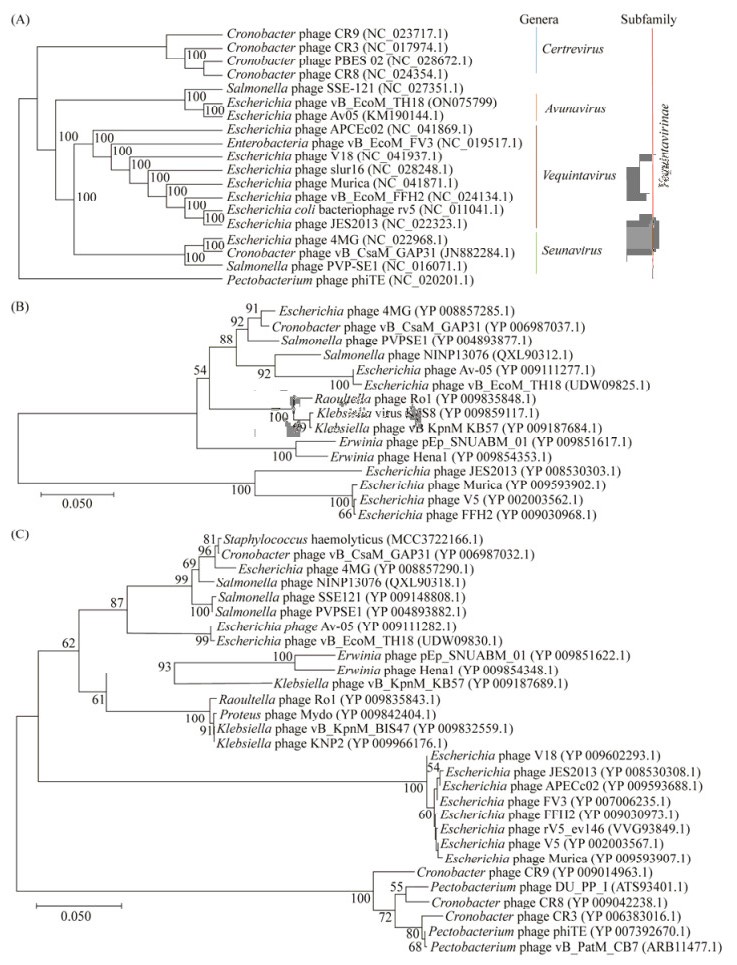
|
| 图 6 噬菌体vB_EcoM_TH18系统发育关系分析 Figure 6 Phylogenetic relationship of phage. A: phylogenetic tree of whole genomic sequence; B: phylogenetic tree of major capsid protein; C: phylogenetic tree of terminase large subunit. The numbers of nodes are the results of 1 000 bootstrap replicates and the scale bar represents the genetic distance. |
利用Easyfig比较TH18与Av-05基因组的差异,如图 7所示,灰色色调颜色越深,则表明二者相互连线的编码序列相似度越高。结果提示噬菌体TH18的基因组长度大于噬菌体Av-05。两者基因组中大部分CDS具有较高的同源性,而连线的空白处提示存在相似性较低的CDS,相同基因位点处及两者整体基因组排列存在差异。特别是CoreGenes软件计算出两者的基因共享率仅为76%,并且提示部分基因是噬菌体TH18独有的,如endolysin及NAD- dependent protein deacetylase of SIR2 family。

|
| 图 7 vB_EcoM_TH18与噬菌体Av-05基因组比对图 Figure 7 Genome comparison of phage vB_EcoM_TH18 and Av-05. The orange arrows represent CDS and the grey bars indicate the similarity of sequences, the intensity of grey indicates the degree of sequence similarity. |
综上所述,vB_EcoM_TH18不同于噬菌体Av-05,是肌尾噬菌体科Vequintavirinae亚科Avunavirus属的一株新型噬菌体。
3 讨论与结论本研究以实验前期研究保存的一株旱獭大肠杆菌(E. coli HT2012160)为宿主菌,从喜马拉雅旱獭肠内容物中分离出一株新型大肠杆菌噬菌体。依据最新命名规则,将该噬菌体命名为vB_EcoM_TH18,其中vB代表病毒噬菌体(Viruses bacteriophage);Eco代表大肠杆菌;M代表肌尾噬菌体科。
噬菌体的尾纤维蛋白可特异性识别宿主菌表面的受体,TH18与Av-05的尾纤维蛋白虽高度同源(92.07%),但二者的宿主裂解谱[43]不完全相同:TH18不裂解沙门氏菌,相反,Av-05可裂解多种血清型沙门氏菌。分析发现两者尾纤维蛋白的序列中有10个碱基位点存在差异,可能是造成两者裂解谱差异的关键。噬菌体TH18的宿主裂解谱相较噬菌体myPSH1131[12]宽,可有效裂解8种致病型、6种血清型E. coli菌株、国内不同来源的耐药E. coli菌株及临床分离来源的宋内志贺氏菌。噬菌体具有高度宿主特异性,通常只能感染单种或亚种细菌,这是目前噬菌体疗法面临的障碍[44]。然而,仅少部分噬菌体具有较宽的宿主裂解谱范围,可感染不同门、科、属的细菌[45–47]。大肠杆菌和志贺氏菌作为同科内不同属的成员,我们有理由认为噬菌体TH18可感染肠杆菌科内的不同属、种细菌。结合EOP试验结果发现,TH18是一株裂解能力较强的宽谱噬菌体。同时结合噬菌体生物学特性,发现该噬菌体生长潜伏期短,具有耐酸碱特性,极具应用和开发价值。
基因组中未发现毒力和耐药相关基因,以及整合酶相关基因簇。尽管注释到有促溶源特性(prolysogenic)的ea22蛋白[48],但ea22蛋白是否表达并如何调控噬菌体裂解周期的机制尚不明确。经丝裂霉素C诱导裂解实验,证实该噬菌体是一株裂解性噬菌体(步骤未展示)。噬菌体TH18在基因组的安全性,可保证其在后续生产和应用中的安全性。
综上所述,vB_EcoM_TH18的生物学特性及基因组学的研究结果表明,该噬菌体具有作为抗菌剂应用的潜力。
致谢
感谢中国疾病预防控制中心传染病所新腹泻病室樊粉霞老师对本论文提供技术指导和帮助;感谢中国疾病预防控制中心传染病所新病原室杨茜博士生和刘倩博士生为本文涉及的国内大肠杆菌菌株提供来源信息。
| [1] | Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clinical Microbiology Reviews, 2013, 26(4): 822-880. DOI:10.1128/CMR.00022-13 |
| [2] | Aliabadi S, Jauneikaite E, Müller-Pebody B, Hope R, Vihta KD, Horner C, Costelloe CE. Exploring temporal trends and risk factors for resistance in Escherichia coli-causing bacteraemia in England between 2013 and 2018: an ecological study. Journal of Antimicrobial Chemotherapy, 2021, 77(3): 782-792. |
| [3] | Feldman SF, Temkin E, Wullfhart L, Nutman A, Schechner V, Shitrit P, Shvartz R, Schwaber MJ, Andremont A, Carmeli Y. A nationwide population-based study of Escherichia coli bloodstream infections: incidence, antimicrobial resistance and mortality. Clinical Microbiology and Infection, 2021. |
| [4] | 全国细菌耐药检测网(CARSS). 2020年全国细菌耐药监测报告(简要版). http://www.carss.cn/Report/Details?aId=808. 访问时间2021年. |
| [5] | Cepko LCS, Garling EE, Dinsdale MJ, Scott WP, Bandy L, Nice T, Faber-Hammond J, Mellies JL. Myoviridae phage PDX kills enteroaggregative Escherichia coli without human microbiome dysbiosis. Journal of Medical Microbiology, 2020, 69(2): 309-323. DOI:10.1099/jmm.0.001162 |
| [6] | Easwaran M, De Zoysa M, Shin HJ. Application of phage therapy: synergistic effect of phage EcSw (ΦEcSw) and antibiotic combination towards antibiotic-resistant Escherichia coli. Transboundary and Emerging Diseases, 2020, 67(6): 2809-2817. DOI:10.1111/tbed.13646 |
| [7] | Alomari MMM, Dec M, Nowaczek A, Puchalski A, Wernicki A, Kowalski C, Urban-Chmiel R. Therapeutic and prophylactic effect of the experimental bacteriophage treatment to control diarrhea caused by E. coli in newborn calves. ACS Infectious Diseases, 2021, 7(8): 2093-2101. DOI:10.1021/acsinfecdis.1c00010 |
| [8] | World Health Organization. GLOBAL Priority List Of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua¼12017. Accessed year: 2021. |
| [9] | Furfaro LL, Payne MS, Chang BJ. Bacteriophage therapy: clinical trials and regulatory hurdles. Frontiers in Cellular and Infection Microbiology, 2018, 8: 376. DOI:10.3389/fcimb.2018.00376 |
| [10] | Cystic Fibrosis News Today. FDA Clears Clinical Trial of AP-PA02 Phage Therapy for P. aeruginosa Infections. https://cysticfibrosisnewstoday.com/2020/10/20/fda-clears-phase-1b-2a-cf-clinical-trial-ap-pa02-phage-therapy-against-pseudomonas-aeruginosa-infections/Accessed year: 2021. |
| [11] | Lu S, Jin D, Wu SS, Yang J, Lan RT, Bai XN, Liu S, Meng Q, Yuan XJ, Zhou J, Pu J, Chen Q, Dai H, Hu YY, Xiong YW, Ye CY, Xu JG. Insights into the evolution of pathogenicity of Escherichia coli from genomic analysis of intestinal E. coli of Marmota himalayana in Qinghai-Tibet plateau of China. Emerging Microbes & Infections, 2016, 5(12): e122. |
| [12] | Manohar P, Tamhankar AJ, Lundborg CS, Ramesh N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS One, 2018, 13(10): e0206278. DOI:10.1371/journal.pone.0206278 |
| [13] | Khan Mirzaei M, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One, 2015, 10(3): e0118557. DOI:10.1371/journal.pone.0118557 |
| [14] | Lomelí-Ortega CO, Martínez-Sández AJ, Barajas- Sandoval DR, Reyes AG, Magallón-Barajas F, Veyrand-Quíros B, Gannon L, Harrison C, Michniewski S, Millard A, Quiroz-Guzmán E. Isolation and characterization of vibriophage vB_Vc_SrVc9: an effective agent in preventing Vibrio campbellii infections in brine shrimp nauplii (Artemia franciscana). Journal of Applied Microbiology, 2021, 131(1): 36-49. DOI:10.1111/jam.14937 |
| [15] | Wei BD, Cong C, Zhang LC, Zheng L, Chen L, Yu W, Xu YP. Complete genome analysis of the newly isolated Shigella sonnei phage vB_SsoM_Z31. Archives of Virology, 2021, 166(9): 2597-2602. DOI:10.1007/s00705-021-05121-y |
| [16] | Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics, 2008, 9: 75. DOI:10.1186/1471-2164-9-75 |
| [17] | Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Research, 1999, 27(19): 3911-3920. DOI:10.1093/nar/27.19.3911 |
| [18] | Jia BF, Raphenya AR, Alcock B, Waglechner N, Guo PY, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research, 2016, 45(D1): D566-D573. |
| [19] | Chen LH, Zheng DD, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Research, 2015, 44(D1): D694-D697. |
| [20] | Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 1997, 25(5): 955-964. DOI:10.1093/nar/25.5.955 |
| [21] | Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics, 2004, 21(4): 537-539. |
| [22] | Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 2014, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
| [23] | Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096 |
| [24] | Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics, 2011, 27(7): 1009-1010. DOI:10.1093/bioinformatics/btr039 |
| [25] | Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Applied and Environmental Microbiology, 2013, 79(24): 7696-7701. DOI:10.1128/AEM.02411-13 |
| [26] | McNair K, Bailey BA, Edwards RA. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics, 2012, 28(5): 614-618. DOI:10.1093/bioinformatics/bts014 |
| [27] | Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang YJ, Wishart DS. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Research, 2016, 44(W1): W16-W21. DOI:10.1093/nar/gkw387 |
| [28] | Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Research, 2018, 46(D1): D708-D717. DOI:10.1093/nar/gkx932 |
| [29] | Pan YY, Hu B, Bai XN, Yang X, Cao LJ, Liu Q, Sun H, Li J, Zhang J, Jin D, Xiong YW. Antimicrobial resistance of non-O157 shiga toxin-producing Escherichia coli isolated from humans and domestic animals. Antibiotics: Basel, Switzerland, 2021, 10(1): 74. |
| [30] | Wilson JH, Luftig RB, Wood WB. Interaction of bacteriophage T4 tail fiber components with a lipopolysaccharide fraction from Escherichia coli. Journal of Molecular Biology, 1970, 51(2): 423-434. DOI:10.1016/0022-2836(70)90152-X |
| [31] | Van De Putte P, Cramer S, Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage Mu. Nature, 1980, 286(5770): 218-222. DOI:10.1038/286218a0 |
| [32] | Grundy FJ, Howf MM. Involvement of the invertible G segment in bacteriophage Mu tail fiber biosynthesis. Virology, 1984, 134(2): 296-317. DOI:10.1016/0042-6822(84)90299-X |
| [33] | Le S, He XS, Tan YL, Huang GT, Zhang L, Lux R, Shi WY, Hu FQ. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS One, 2013, 8(7): e68562. DOI:10.1371/journal.pone.0068562 |
| [34] | Yang HQ, Ma YF, Wang YT, Yang HX, Shen W, Chen XZ. Transcription regulation mechanisms of bacteriophages: recent advances and future prospects. Bioengineered, 2014, 5(5): 300-304. DOI:10.4161/bioe.32110 |
| [35] | Kropinski AM, Waddell T, Meng JC, Franklin K, Ackermann HW, Ahmed R, Mazzocco A, Yates J 3rd, Lingohr EJ, Johnson RP. The host-range, genomics and proteomics of Escherichia coli O157: H7 bacteriophage rV5. Virology Journal, 2013, 10: 76. DOI:10.1186/1743-422X-10-76 |
| [36] | Sitbon E, Pietrokovski S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends in Biochemical Sciences, 2003, 28(9): 473-477. DOI:10.1016/S0968-0004(03)00170-1 |
| [37] | Gu JM, Liu XH, Yang M, Li Y, Sun CJ, Lu R, Song J, Zhang QM, Lei LC, Feng X, Du CT, Yu H, Yang YJ, Han WY. Genomic characterization of lytic Staphylococcus aureus phage GH15: providing new clues to intron shift in phages. The Journal of General Virology, 2013, 94(Pt 4): 906–915. |
| [38] | Zhelkovsky AM, McReynolds LA. Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase-engineering a thermostable ATP independent enzyme. BMC Molecular Biology, 2012, 13: 24. DOI:10.1186/1471-2199-13-24 |
| [39] | Cui ZH, Xu ZW, Wei YL, Zhang Q, Qin KH, Ji XL. Characterization and genome analysis of a novel mu-like phage VW-6B isolated from the napahai plateau wetland of China. Current Microbiology, 2021, 78(1): 150-158. DOI:10.1007/s00284-020-02277-9 |
| [40] | Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiology, 2012, 7(10): 1147-1171. DOI:10.2217/fmb.12.97 |
| [41] | Dydecka A, Bloch S, Necel A, Topka G, Węgrzyn A, Tong JG, Donaldson LW, Węgrzyn G, Nejman- Faleńczyk B. The ea22 gene of lambdoid phages: preserved prolysogenic function despite of high sequence diversity. Virus Genes, 2020, 56(2): 266-277. DOI:10.1007/s11262-020-01734-8 |
| [42] | Amarillas L, López-Cuevas O, León-Félix J, Castro-Del Campo N, Gerba CP, Chaidez C. Genomic analysis of broad-host-range enterobacteriophage av-05. Genome Announcements, 2015, 3(3): e00282-15. |
| [43] | López-Cuevas O, Castro-Del Campo N, León-Félix J, González-Robles A, Chaidez C. Characterization of bacteriophages with a lytic effect on various Salmonella serotypes and Escherichia coli O157: H7. Canadian Journal of Microbiology, 2011, 57(12): 1042-1051. DOI:10.1139/w11-099 |
| [44] | Kim S, Kim SH, Rahman M, Kim J. Characterization of a Salmonella Enteritidis bacteriophage showing broad lytic activity against Gram-negative enteric bacteria. Journal of Microbiology: Seoul, Korea, 2018, 56(12): 917-925. |
| [45] | Malki K, Kula A, Bruder K, Sible E, Hatzopoulos T, Steidel S, Watkins SC, Putonti C. Bacteriophages isolated from Lake Michigan demonstrate broad host-range across several bacterial Phyla. Virology Journal, 2015, 12: 164. DOI:10.1186/s12985-015-0395-0 |
| [46] | Buttimer C, Hendrix H, Oliveira H, Casey A, Neve H, McAuliffe O, Ross RP, Hill C, Noben JP, O'Mahony J, Lavigne R, Coffey A. Things are getting hairy: enterobacteria bacteriophage vB_PcaM_CBB. Frontiers in Microbiology, 2017, 8: 44. |
| [47] | Van Zyl LJ, Nemavhulani S, Cass J, Cowan DA, Trindade M. Three novel bacteriophages isolated from the East African Rift Valley soda lakes. Virology Journal, 2016, 13(1): 204. DOI:10.1186/s12985-016-0656-6 |
| [48] | Tong JG, Nejman-Faleńczyk B, Bloch S, Węgrzyn A, Węgrzyn G, Donaldson LW. Ea22 proteins from lambda and shiga toxin-producing bacteriophages balance structural diversity with functional similarity. ACS Omega, 2020, 5(21): 12236-12244. DOI:10.1021/acsomega.0c00894 |
 2022, Vol. 62
2022, Vol. 62




