中国科学院微生物研究所,中国微生物学会
文章信息
- 李倩, 朱奎. 2022
- LI Qian, ZHU Kui.
- 耐药细菌中交互敏感现象研究进展
- Advance in collateral sensitivity in drug-resistant bacteria
- 微生物学报, 62(5): 1688-1697
- Acta Microbiologica Sinica, 62(5): 1688-1697
-
文章历史
- 收稿日期:2021-11-07
- 修回日期:2022-01-26
- 网络出版日期:2022-02-21
抗菌药物的不合理使用导致细菌耐药性问题日益严重,是全球高度关注的公共卫生问题之一。2014年报道全球每年大约70万人因细菌耐药性而死亡,如果不加以控制,到2050年细菌耐药性每年将导致1 000万人死亡,并使全球GDP损失超过100万亿美元[1–4]。为遏制细菌耐药性的发生和发展,现已经提出了多种应对策略。目前,新型抗菌化合物及增效剂的研发仍是对抗耐药菌的主要策略[5]。例如,一种广谱抗菌药物增效剂SLAP-S25能与多类抗菌药物协同使用,从而恢复临床常见革兰阴性耐药菌对抗菌药物的敏感性[6–7]。同时,宿主导向的抗菌治疗和抗菌药物替代物等也是目前的研究热点和难点[8–12]。虽然这些策略将为长期应对细菌耐药性提供理论依据及技术支持,但大多数项目仍处于临床前研究,无法迅速治疗耐药病原菌感染。因此,迫切需要找到新的应对策略以减缓耐药菌导致的临床上无药可用的窘境[13]。
耐药菌的交互敏感性(collateral sensitivity,也称附带敏感性)是细菌耐药性进化过程中的一种常见表型,是细菌改变其遗传物质或者生理生化特性来适应某一类抗菌药物压力时产生对另一类或者几类抗菌药物敏感性升高的现象[14]。交互敏感性的研究将为耐药菌防控策略提供新的突破口。采用交互敏感的抗菌策略可以通过利用药物之间的特定相互作用来增加耐药病原菌对现有抗菌药物的敏感性。因此,利用细菌的交互敏感性不仅可以在菌株已经产生耐药性后实现有效治疗,还可以提高现有抗菌药物的疗效,延长其使用寿命,实现临床上迅速、高效的治疗策略[15–16]。此外,深入研究细菌交互敏感机制将有利于进一步揭示耐药菌株的产生与形成规律,为靶向耐药菌的新型抗菌药物提供理论基础[17]。在本文中,我们探讨了耐药细菌中交互敏感性的最新研究进展,重点介绍了交互敏感性的概念、表型以及分子机制,以期为研发新的抗菌策略提供新思路。
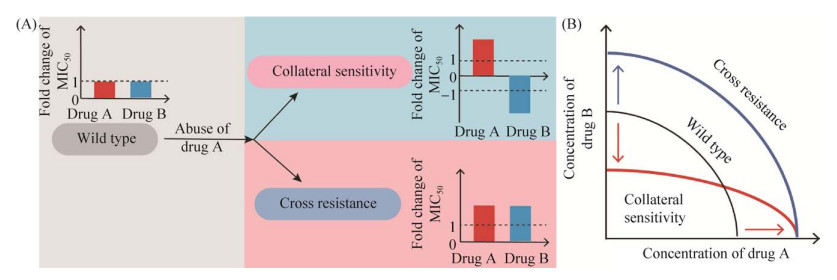
1 交互敏感性细菌在抗菌药物持续使用时会通过改变自身遗传特性等方式进化出耐药性,通常表现为合成抗菌药物水解酶、抗菌靶点突变、合成外排泵蛋白和改变自身代谢途径等。复杂的耐药机制和抗菌机制之间产生了关联和矛盾,形成交互敏感和交叉耐药(cross resistance)。例如,质子动力势的强弱与外排泵蛋白的活性紧密相关,当细菌通过大量表达外排泵活性蛋白产生耐药性时,又不可避免地使依赖质子动力势的药物进入胞内的量增多,这种生理功能的矛盾性导致细菌产生交互敏感(图 1A)。另外,cfr耐药基因会使抗菌靶点发生甲基化修饰从而使作用在此位点的5类抗菌药物(噁唑烷酮类、截短侧耳素类、链阳霉素类、林可胺类以及酰胺醇类)出现交叉耐药的现象[18]。交互敏感性可以指导研发设计新型抗菌药或合理的联合用药方案来对抗耐药菌。研究表明,74%的实验室耐药菌株都表现出对一类或多类药物的敏感性增强[19–21]。因此,在细菌耐药性的进化过程中,交互敏感性是一种常见且值得关注的现象。
|
| 图 1 交互敏感性与交叉耐药性模式图 Figure 1 Scheme of collateral sensitivity and cross resistance in bacteria. A: scheme of cross-resistance and collateral sensitivity; B: the change of antibiotic sensitivity. |
交互敏感性的概念最早是用来解释癌细胞在长期化疗后出现的对药物敏感性变化[22]。细菌中交互敏感性的描述最早报道于1952年,Bryson和Szybalski观察到大肠杆菌菌株在获得氯霉素抗性后对多黏菌素B变得敏感[23]。近年来,在细菌耐药性发展迅速而新型抗菌药物逐年减少的危机下,关于耐药菌交互敏感性表型及机制的研究逐渐成为热点[13, 24]。在抗菌药物的选择性压力下细菌出现耐药性突变频率大大增加,这种突变往往不会给细菌造成很大适应性代价,并且突变菌株可在特定抗菌药物或者无抗菌药物压力时存活并持续传播相关基因。但某些耐药性突变可能会造成细菌自身结构或者生理功能的改变,使其对另一类药物趋向于敏感,产生交互敏感现象(图 1B)。因此,揭示耐药细菌交互敏感性的产生及形成规律将有助于预判和逆转细菌的耐药性,制定合适的给药方案,提高耐药细菌的防控效率[25–26]。
2 交互敏感研究进展 2.1 交互敏感现象研究概述交互敏感是细菌耐药性进化过程中一种常见的现象,在大肠杆菌[27]、沙门菌[28]、铜绿假单胞菌[29–30]、鲍曼不动杆菌[31]、金黄色葡萄球菌[30]和其他多种病原菌中均有报道[32–34]。2009年,研究人员发现实验室菌株及临床分离大肠杆菌菌株都存在交互敏感现象,并对交互敏感现象进行更深入的探究。梳理菌株的交互敏感和交叉耐药网络是当前广泛采用的研究方法,可以为临床长期使用药物治疗提供数据参考[35]。例如,使用交叉耐药网络中的药物进行治疗时即可预测到耐药性后期发展轨迹,以及时调整用药策略,避免诱导出高水平耐药及无效治疗的可能性。相反,基于交互敏感网络的药物组合的顺序治疗可以减少高水平耐药发展的可能性,并且可以预见到一类抗菌药物耐药后的治疗方案。这些研究表明,交叉耐药和交互敏感网络可为治疗最初选择抗菌药物及预测耐药性发展轨迹奠定基础。除此之外,还可通过菌株基因组进化分析推测交互敏感产生机制[36–37]。例如,通过构建数学模型模拟计算来判断葡萄球菌及粪肠球菌等菌株的交互敏感性[38–40],并以此来判断菌株的耐药发展方向。
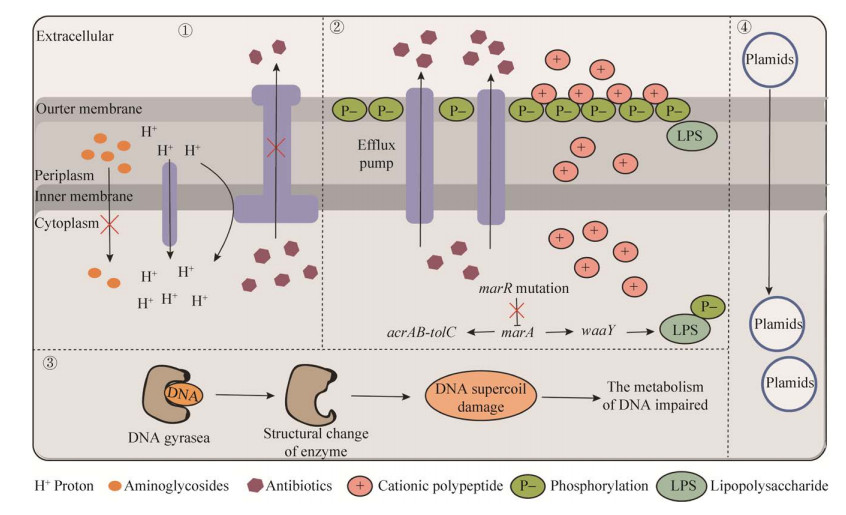
2.2 交互敏感机制研究进展目前,关于交互敏感机制的研究尚处于起步阶段(图 2)。在细菌遗传物质改变介导的交互敏感性方面取得了一些进展,可以总结为3点:(1) 耐药基因片段介导的交互敏感;(2) 基因突变引起的交互敏感;(3) 耐药质粒导致的交互敏感。此外,还有一些其他机制介导交互敏感的研究,如转录重编程导致细菌中其他生物学功能发生改变[41–42]。
|
| 图 2 细菌交互敏感机制图 Figure 2 Molecular mechanism of collateral sensitivity in bacteria. Modes of action of collateral sensitivity of ①: aminoglycoside antibiotics resistant bacteria to other antibiotics; ②: MDR bacteria to antimicrobial peptides; ③: quinolone antibiotics resistant bacteria to other antibiotics; ④: plasmid-mediated resistance. |
首先,耐药基因除了可以引起细菌产生耐药表型外,还可以引起其他生物学效应,导致细菌产生交互敏感现象(表 1)。例如,氨基糖苷类抗菌药物常需要利用细菌的质子动力势(proton- motive force,PMF)进入胞内,发挥抗菌作用。因此,氨基糖苷类抗菌药物耐药的大肠杆菌可以通过降低PMF使氨基糖苷类药物的胞内累积量减少而实现耐药;但PMF降低会导致质子依赖的外排泵的活性降低,导致其他多类抗菌药外排减少,胞内抗菌药物含量升高,进而导致细菌生长被抑制[43]。相应地,细菌表达四环素耐药基因tet增加外排泵蛋白表达,四环素外排增多,引起四环素耐药;但高效的外排泵反而提高了胞外氨基糖苷类抗菌药物进入菌体内的效率,从而引起细菌对氨基糖苷类抗菌药物的敏感性增加[44]。
| Species | Resistance | Collateral sensitivity |
Mechanism | In vitro/vivo | References |
| E. coli | Aminoglycosides | β-lactam antibiotics | Reduction of PMF in aminoglycoside-resistant isolates diminish the activity of PMF-dependent major efflux pumps, leading to susceptibility to β-lactam antibiotics | In vitro | [43] |
| Mecillinam | Cefotaxime | Three non-synonymous of CTX-M-15 increase resistance against mecillinam that confer susceptibility to cefotaxime | In vitro/vivo | [46] | |
| Multi-antibiotics | Cationic antimicrobial peptide | marR mutant increase the expression of efflux pump inducing multi-resistance, while it facilitates the interaction of antimicrobial peptides via modulation of the LPS phosphorylation pathway | In vitro/vivo | [47] | |
| Azithromycin | Colistin | Plsmid-mediated collateral sensitivity | In vitro/vivo | [48–49] | |
| P. aeruginosa | Ciprofloxacin | Gentamycin | nfxB mutation in ciprofloxacin resistant bacteria up-regulates the transporer protein (MexC) that confer susceptibility to gentamycin | In vitro/vivo | [50] |
| Gentamycin | Penicillin | Unclear | In vitro/vivo | [51] | |
| Ceftazidime | Tobramycin | Unclear | In vitro/vivo | [52] | |
| MRSA | Methicillin | Meloxicillin- piperacillin-tazobactam (ME/PI/TZ) | ME/PI/TZ disrupt PBP2α to recover sensitive to β-lactam antibiotics | In vitro/vivo | [53] |
此外,基因突变也是产生交互敏感现象的重要机制。耐药基因以及其他基因突变均有可能导致基因功能改变,促进细菌产生交互敏感现象(表 1)。例如,喹诺酮类耐药菌株通过耐药基因突变改变自身的DNA拓扑异构酶的结构,减弱了酶与喹诺酮类药物的亲和力产生耐药[45]。但耐药菌DNA超螺旋结构的改变,从根本上影响了整个基因组的转录,最终使细菌无法应对其他抗菌药造成的损害而死亡。同时,在探究了产广谱β-内酰胺酶CTX-M-15大肠杆菌的交互敏感机制时发现,具有交互敏感表型菌株中的CTX-M-15都有3个错义突变,导致其对美洛西林或哌拉西林-他唑巴坦的耐药性增加,但突变的水解酶与几种头孢菌素药物亲和力减弱,因而细菌恢复对头孢菌素药物的敏感性[46]。此外,细菌可通过调节细菌外膜脂多糖(lipopolysaccharide,LPS)来实现对抗菌肽的交互敏感。具体来讲,marR基因突变上调了AcrAB-ToIC外排泵的表达,增加对多类抗菌药物的耐药性;marR基因突变同时上调了LPS磷酸化激酶WaaY,LPS的磷酸化增加了细菌外膜表面负电荷,增强了靶向外膜的阳离子抗菌肽的敏感性[47]。
最新研究表明耐药质粒可以介导耐药菌交互敏感性的形成(表 1)。2021年首次发现水平转移的耐药质粒可以导致大肠杆菌产生交互敏感现象[48–49],证明了耐药质粒转移到大肠杆菌内后对一些抗菌药物敏感性升高。该研究结果不仅为治疗质粒介导的耐药菌株提供了新思路,还将以往从基因片段的角度揭示耐药菌交互敏感性形成机制拓展到耐药质粒,为后继相关机制的研究提供了新的方向。我们课题组前期通过构建交互敏感网络,发现耐万古霉素的屎肠球菌(VREfm)和对新型海洋源抗菌化合物equisetin耐药的金黄色葡萄球菌都表现出交互敏感现象[40]。此外,我们证明磷酸化的VanR通过结合相似的启动子序列,共同调控万古霉素耐药基因簇与核糖体保护蛋白MsrC的表达,介导VREfm对截短侧耳素类药物的交互敏感[54]。
2.3 交互敏感性在临床菌株中的研究进展近年来,随着对交互敏感现象的研究越来越深入,研究对象逐渐从实验室菌株逐渐拓展到临床菌株。如何将交互敏感规律应用于临床菌株来控制耐药菌的进化,提高现有抗菌药的使用效率,延长其使用寿命是解决问题的关键。利用交互敏感现象指导临床用药有以下几方面优势:(1) 可以用相对较低剂量的抗菌药物抑制病原菌生长,阻断病原菌感染进程;(2) 基于交互敏感性的治疗策略可以优化抗菌药治疗组合,提高治疗慢性感染的成功率[55–56];(3) 利用交互敏感抗菌药物组合实现周期性循环用药,逆转病原菌的耐药性,提高现有抗菌药物的使用效率[57–59]。
基于耐药菌交互敏感性设计的药物组合可以实现相对较低剂量的抗菌药物防控耐药病原菌。例如,美罗培南耐药的大肠杆菌可与呋喃妥因表现出交互敏感,对呋喃妥因的MIC下降了64倍,较低剂量的呋喃妥因就能抑制耐药大肠杆菌的生长[60]。同时,通过构建交互敏感网络筛选出的抗菌药物组合可以治疗临床慢性感染,延缓甚至逆转耐药菌的进化方向。研究表明,由铜绿假单胞菌引起纤维性囊肿病人在已经出现对药物耐药的情况下,使用通过构建交互敏感网络筛选的抗菌药物组合,可以清除喹诺酮类药物耐药的绿脓杆菌亚群,实现有效治疗[50]。此外,利用交互敏感药物组合实现循环用药来治疗耐药菌的感染,并可根据使用的药物组合控制和预测耐药性发展方向,提高现有抗菌药物的使用效率。以临床分离大肠杆菌为研究对象,对常用的23种抗菌药物进行交互敏感药物组合筛选,获得几百种组合并最终确定庆大霉素和头孢呋辛可实现循环用药治疗[21]。之后,基于交互敏感的药物组合被陆续报道可以有效控制临床分离菌株,包括MRSA和致病性大肠杆菌等病原菌[61–62]。这些研究进一步证明了交互敏感性指导的给药方案在治疗临床感染中潜力巨大。
相较于交互敏感现象在实验室条件下的深入研究及其揭示的不同耐药性机制和抗菌药物作用之间的新联系,临床上开展交互敏感性的研究和应用仍处于初级阶段,还需更多的临床数据来指导药物组合的设计和合理用药[63–65]。但须强调,基于耐药病原菌交互敏感性的药物组合及联合用药策略将为未来临床合理用药提供理论基础和技术支撑,是破解临床上面对耐药菌感染而出现无药可用窘境的新突破口。
3 总结与展望尽管目前在交互敏感性的研究上取得了长足的进步,但仍有两个方面亟待开展深入研究。(1) 耐药细菌交互敏感性的分子机制尚不清楚。已有许多研究表明,交互敏感现象广泛存在于大肠杆菌、金黄色葡萄球菌、铜绿假单胞菌和表皮葡萄球菌等细菌中,但其形成机制的研究大多是集中在细菌遗传物质方面的研究,具体机制还有待进一步阐明。由于能产生交互敏感现象的菌种较多,而菌种之间差异非常大,且产生交互敏感的药物组合和机制各不相同,所以进一步阐明其具体机制存在一定难度。阐明交互敏感分子机制,揭示病原菌耐药进化的规律,高效预测细菌耐药发生发展方向,从而制定合理给药方案或靶向设计新药,提高耐药病原菌的治疗效率。(2)如何利用交互敏感设计抗菌药物组合。虽然关于耐药菌交互敏感的研究已经从实验室菌株拓展到临床菌株,但总体研究还处于初级阶段。基于交互敏感机制设计的药物组合在临床使用需要更加严谨的临床试验研究的数据支持。目前还缺乏评价这类药物组合药效的临床用药标准和评价体系,仍需要更系统的前瞻性临床治疗研究来检验其提高疗效或降低耐药发生率的能力。
综上所述,在细菌耐药性日益严重而新型抗菌药物匮乏的现状下,利用耐药菌交互敏感性策略将为防控耐药菌提供新的方向。期待早日阐明耐药菌的交互敏感性现象的产生机制及形成规律,为合理用药和治疗耐药病原菌感染提供新思路和强有力的技术支持。
| [1] | Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrobial Agents and Chemotherapy, 2009, 53(12): 5046-5054. DOI:10.1128/AAC.00774-09 |
| [2] | Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian GB, Dong BL, Huang XH, Yu LF, Gu DX, Ren HW, Chen XJ, Lv LC, He DD, Zhou HW, Liang ZS, Shen JZ. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases, 2016, 16(2): 161-168. DOI:10.1016/S1473-3099(15)00424-7 |
| [3] | He T, Wang R, Liu DJ, Walsh TR, Zhang R, Lv Y, Ke YB, Ji QJ, Wei RC, Liu ZH, Shen YB, Wang G, Sun LC, Lei L, Lv ZQ, Li Y, Pang MD, Wang LY, Sun QL, Fu YL, Song HW, Hao YX, Shen ZQ, Wang SL, Chen GX, Wu CM, Shen JZ, Wang Y. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nature Microbiology, 2019, 4(9): 1450-1456. DOI:10.1038/s41564-019-0445-2 |
| [4] | Kim L. The science of antibiotic discovery. Cell, 2020, 181(1): 29-45. DOI:10.1016/j.cell.2020.02.056 |
| [5] | Liu Y, Ding SY, Dietrich R, Märtlbauer E, Zhu K. A biosurfactant-inspired heptapeptide with improved specificity to kill MRSA. Angewandte Chemie International Edition, 2017, 56(6): 1486-1490. DOI:10.1002/anie.201609277 |
| [6] | Song MR, Liu Y, Huang XY, Ding SY, Wang Y, Shen JZ, Zhu K. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nature Microbiology, 2020, 5(8): 1040-1050. DOI:10.1038/s41564-020-0723-z |
| [7] | Song MR, Zhu K. A broad-spectrum antibiotic adjuvant SLAP-S25: one stone many birds. Microbial Cell, 2020, 7(8): 215-217. DOI:10.15698/mic2020.08.726 |
| [8] | Li Y, Liu F, Zhang J, Liu X, Xiao P, Bai H, Chen S, Wang D, Sung SHP, Kwok RTK, Shen J, Zhu K, Tang BZ. Efficient killing of multidrug-resistant internalized bacteria by AIEgens in vivo. Advanced Science, 2021, 8(9): 2001750. DOI:10.1002/advs.202001750 |
| [9] | Liu Y, Jia Y, Yang K, Li R, Xiao X, Zhu K, Wang Z. Metformin restores tetracyclines susceptibility against multidrug resistant bacteria. Advanced Science, 2020, 7(12): 1902227. DOI:10.1002/advs.201902227 |
| [10] | Yang ZQ, Huang YL, Zhou HW, Zhang R, Zhu K. Persistent carbapenem-resistant Klebsiella pneumoniae: a Trojan horse. The Lancet Infectious Diseases, 2018, 18(1): 22-23. DOI:10.1016/S1473-3099(17)30627-8 |
| [11] | Cui YF, Wang SL, Ding SY, Shen JZ, Zhu K. Toxins and mobile antimicrobial resistance genes in Bacillus probiotics constitute a potential risk for One Health. Journal of Hazardous Materials, 2020, 382: 121266. DOI:10.1016/j.jhazmat.2019.121266 |
| [12] |
Liu XY, Mao CS, Zhu K. Progress on host-acting antibacterial drugs. Chinese Journal of Animal Infectious Diseases, 2021, 29(4): 52-60.
(in Chinese) 刘晓晔, 毛畅思, 朱奎. 宿主导向抗菌药物的研究进展. 中国动物传染病学报, 2021, 29(4): 52-60. |
| [13] | Hall MD, Handley MD, Gottesman MM. Is resistance useless? Multidrug resistance and collateral sensitivity. Trends in Pharmacological Sciences, 2009, 30(10): 546-556. DOI:10.1016/j.tips.2009.07.003 |
| [14] | Pál C, Papp B, Lázár V. Collateral sensitivity of antibiotic-resistant microbes. Trends in Microbiology, 2015, 23(7): 401-407. DOI:10.1016/j.tim.2015.02.009 |
| [15] | Barbosa C, Trebosc V, Kemmer C, Rosenstiel P, Beardmore R, Schulenburg H, Jansen G. Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Molecular Biology and Evolution, 2017, 34(9): 2229-2244. DOI:10.1093/molbev/msx158 |
| [16] | Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science, 2016, 351(6268): aad3292. DOI:10.1126/science.aad3292 |
| [17] | Fodor A, Abate BA, Deák P, Fodor L, Gyenge E, Klein MG, Koncz Z, Muvevi J, Ötvös L, Székely G, Vozik D, Makrai L. Multidrug resistance (MDR) and collateral sensitivity in bacteria, with special attention to genetic and evolutionary aspects and to the perspectives of antimicrobial peptides-a review. Pathogens: Basel, Switzerland, 2020, 9(7): 522. |
| [18] | Vester B. The cfr and cfr-like multiple resistance genes. Research in Microbiology, 2018, 169(2): 61-66. DOI:10.1016/j.resmic.2017.12.003 |
| [19] | Colclough A, Corander J, Sheppard SK, Bayliss SC, Vos M. Patterns of cross-resistance and collateral sensitivity between clinical antibiotics and natural antimicrobials. Evolutionary Applications, 2019, 12(5): 878-887. DOI:10.1111/eva.12762 |
| [20] | Munck C, Gumpert HK, Nilsson Wallin AI, Wang HH, Sommer MOA. Prediction of resistance development against drug combinations by collateral responses to component drugs. Science Translational Medicine, 2014, 6(262): 262ra156. |
| [21] | Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Science Translational Medicine, 2013, 5(204): 132. |
| [22] | Hutchison DJ. Cross resistance and collateral sensitivity studies in cancer chemotherapy. Advances in Cancer Research, 1963, 7: 235-250. |
| [23] | Bryson V, Szybalski W. Microbial selection. Science, 1952, 116(3003): 45-51. DOI:10.1126/science.116.3003.45 |
| [24] | Maltas J, Wood KB. Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biology, 2019, 17(10): e3000515. DOI:10.1371/journal.pbio.3000515 |
| [25] | Podnecky NL, Fredheim EGA, Kloos J, Sørum V, Primicerio R, Roberts AP, Rozen DE, Samuelsen Ø, Johnsen PJ. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nature Communications, 2018, 9: 3673. DOI:10.1038/s41467-018-06143-y |
| [26] | Horinouchi T, Suzuki S, Kotani H, Tanabe K, Sakata N, Shimizu H, Furusawa C. Prediction of cross-resistance and collateral sensitivity by gene expression profiles and genomic mutations. Scientific Reports, 2017, 7: 14009. DOI:10.1038/s41598-017-14335-7 |
| [27] | Bollenbach T. Antimicrobial interactions: mechanisms and implications for drug discovery and resistance evolution. Current Opinion in Microbiology, 2015, 27: 1-9. DOI:10.1016/j.mib.2015.05.008 |
| [28] | Ju XY, Zhu MJ, Han JZ, Lu ZX, Zhao HZ, Bie XM. Combined effects and cross-interactions of different antibiotics and polypeptides in Salmonella bredeney. Microbial Drug Resistance: Larchmont, N Y, 2018, 24(10): 1450-1459. DOI:10.1089/mdr.2017.0367 |
| [29] | Jansen G, Mahrt N, Tueffers L, Barbosa C, Harjes M, Adolph G, Friedrichs A, Krenz-Weinreich A, Rosenstiel P, Schulenburg H. Association between clinical antibiotic resistance and susceptibility of Pseudomonas in the cystic fibrosis lung. Evolution, Medicine, and Public Health, 2016, 2016(1): 182-194. DOI:10.1093/emph/eow016 |
| [30] | Barbosa C, Römhild R, Rosenstiel P, Schulenburg H. Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. eLife, 2019, 8: e51481. DOI:10.7554/eLife.51481 |
| [31] | García-Quintanilla M, Carretero-Ledesma M, Moreno-Martínez P, Martín-Peña R, Pachón J, McConnell MJ. Lipopolysaccharide loss produces partial colistin dependence and collateral sensitivity to azithromycin, rifampicin and vancomycin in Acinetobacter baumannii. International Journal of Antimicrobial Agents, 2015, 46(6): 696-702. DOI:10.1016/j.ijantimicag.2015.07.017 |
| [32] | Harrison EM, Ba XL, Coll F, Blane B, Restif O, Carvell H, Köser CU, Jamrozy D, Reuter S, Lovering A, Gleadall N, Bellis KL, Uhlemann AC, Lowy FD, Massey RC, Grilo IR, Sobral R, Larsen J, Rhod Larsen A, Vingsbo Lundberg C, Parkhill J, Paterson GK, Holden MTG, Peacock SJ, Holmes MA. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nature Microbiology, 2019, 4(10): 1680-1691. DOI:10.1038/s41564-019-0471-0 |
| [33] | Lozano-Huntelman NA, Singh N, Valencia A, Mira P, Sakayan M, Boucher I, Tang S, Brennan K, Gianvecchio C, Fitz-Gibbon S, Yeh P. Evolution of antibiotic cross-resistance and collateral sensitivity in Staphylococcus epidermidis using the mutant prevention concentration and the mutant selection window. Evolutionary Applications, 2020, 13(4): 808-823. DOI:10.1111/eva.12903 |
| [34] | Flanagan JN, Kavanaugh L, Steck TR. Burkholderia multivorans exhibits antibiotic collateral sensitivity. Microbial Drug Resistance, 2020, 26(1): 1-8. DOI:10.1089/mdr.2019.0202 |
| [35] | Maltas J, Krasnick B, Wood KB. Using selection by nonantibiotic stressors to sensitize bacteria to antibiotics. Molecular Biology and Evolution, 2020, 37(5): 1394-1406. DOI:10.1093/molbev/msz303 |
| [36] | De Evgrafov MCR, Faza M, Asimakopoulos K, Sommer MOA. Systematic investigation of resistance evolution to common antibiotics reveals conserved collateral responses across common human pathogens. Antimicrobial Agents and Chemotherapy, 2020, 65(1): e01273-e01220. |
| [37] | Yu X, Zheng BW, Xiao F, Jin Y, Guo LH, Xu H, Luo QX, Xiao YH. Effect of short-term antimicrobial therapy on the tolerance and antibiotic resistance of multidrug-resistant Staphylococcus capitis. Infection and Drug Resistance, 2020, 13: 2017-2026. DOI:10.2147/IDR.S254141 |
| [38] | Sun Y, Lu H, Zhang X, Wu Q, Bi W, Liu H, Cao J, Zhou T. Phenotype and genotype alteration during adaptive evolution of Enterococcus faecalis to antimicrobials. Infection, Genetics and Evolution, 2018, 62: 80-85. DOI:10.1016/j.meegid.2018.03.029 |
| [39] | Kavanaugh LG, Flanagan JN, Steck TR. Reciprocal antibiotic collateral sensitivity in Burkholderia multivorans. International Journal of Antimicrobial Agents, 2020, 56(1): 105994. DOI:10.1016/j.ijantimicag.2020.105994 |
| [40] | Chen S, Liu D, Zhang Q, Guo P, Ding SY, Shen JZ, Zhu K, Lin WH. A marine antibiotic kills multidrug-resistant bacteria without detectable high-level resistance. ACS Infectious Diseases, 2021, 7(4): 884-893. DOI:10.1021/acsinfecdis.0c00913 |
| [41] | Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, Hrtyan M, Busa-Fekete R, Bogos B, Méhi O, Csörgő B, Pósfai G, Fekete G, Szappanos B, Kégl B, Papp B, Pál C. Bacterial evolution of antibiotic hypersensitivity. Molecular Systems Biology, 2013, 9: 700. DOI:10.1038/msb.2013.57 |
| [42] | Nichol D, Rutter J, Bryant C, Hujer AM, Lek S, Adams MD, Jeavons P, Anderson ARA, Bonomo RA, Scott JG. Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nature Communications, 2019, 10: 334. DOI:10.1038/s41467-018-08098-6 |
| [43] | Azimi L, Rastegar Lari A. Collateral sensitivity between aminoglycosides and beta-lactam antibiotics depends on active proton pumps. Microbial Pathogenesis, 2017, 112: 122-125. DOI:10.1016/j.micpath.2017.09.049 |
| [44] | Merlin TL, Davis GE, Anderson WL, Moyzis RK, Griffith JK. Aminoglycoside uptake increased by Tet gene expression. Antimicrobial Agents and Chemotherapy, 1989, 33(9): 1549-1552. DOI:10.1128/AAC.33.9.1549 |
| [45] | Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern WV. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. The Journal of Antimicrobial Chemotherapy, 1996, 38(3): 443-455. DOI:10.1093/jac/38.3.443 |
| [46] | Rosenkilde CEH, Munck C, Porse A, Linkevicius M, Andersson DI, Sommer MOA. Collateral sensitivity constrains resistance evolution of the CTX-M-15 β-lactamase. Nature Communications, 2019, 10: 618. DOI:10.1038/s41467-019-08529-y |
| [47] | Lázár V, Martins A, Spohn R, Daruka L, Grézal G, Fekete G, Számel M, Jangir PK, Kintses B, Csörgő B, Nyerges Á, Györkei Á, Kincses A, Dér A, Walter FR, Deli MA, Urbán E, Hegedűs Z, Olajos G, Méhi O, Bálint B, Nagy I, Martinek TA, Papp B, Pál C. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nature Microbiology, 2018, 3(6): 718-731. DOI:10.1038/s41564-018-0164-0 |
| [48] | Herencias C, Rodríguez-Beltrán J, León-Sampedro R, Valle AAD, Palkovičová J, Cantón R, Millán ÁS. Collateral sensitivity associated with antibiotic resistance plasmids. eLife, 2021, 10: e65130. DOI:10.7554/eLife.65130 |
| [49] | MacLean RC, San Millan A. The evolution of antibiotic resistance. Science, 2019, 365(6458): 1082-1083. DOI:10.1126/science.aax3879 |
| [50] | Imamovic L, Ellabaan MMH, Dantas Machado AM, Citterio L, Wulff T, Molin S, Krogh Johansen H, Sommer MOA. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell, 2018, 172(1/2): 121-134.e14. |
| [51] |
Chang BX, Ma Z, Qu Y, Lu ZX, Lü FX, Zhao HZ, Zhang C, Bie XM. Effect of Pseudomonas aeruginosa on the rule of cross-resistance and collateral sensitivity against different antibiotics. Journal of Nanjing Agricultural University, 2019, 42(5): 940-945.
(in Chinese) 常冰雪, 马志, 曲岩, 陆兆新, 吕凤霞, 赵海珍, 张充, 别小妹. 铜绿假单胞菌对不同抗生素交叉耐药性和附属敏感性产生规律的影响. 南京农业大学学报, 2019, 42(5): 940-945. |
| [52] | Hernando-Amado S, Sanz-García F, Martínez JL. Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Science Advances, 2020, 6(32): eaba5493. DOI:10.1126/sciadv.aba5493 |
| [53] | Gonzales PR, Pesesky MW, Bouley R, Ballard A, Biddy BA, Suckow MA, Wolter WR, Schroeder VA, Burnham CA, Mobashery S, Chang M, Dantas G. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nature Chemical Biology, 2015, 11(11): 855-861. DOI:10.1038/nchembio.1911 |
| [54] | Li Q, Chen S, Zhu K, Huang XL, Huang YC, Shen ZQ, Ding SY, Gu DX, Yang QW, Sun HL, Hu FP, Wang H, Cai JC, Ma B, Zhang R, Shen JZ. Collateral sensitivity to pleuromutilins in vancomycin-resistant Enterococcus faecium. Nature Communications, 2022 (Accepted). |
| [55] | Fuentes-Hernandez A, Plucain J, Gori F, Pena-Miller R, Reding C, Jansen G, Schulenburg H, Gudelj I, Beardmore R. Using a sequential regimen to eliminate bacteria at sublethal antibiotic dosages. PLoS Biology, 2015, 13(4): e1002104. DOI:10.1371/journal.pbio.1002104 |
| [56] | Liu JF, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science, 2020, 367(6474): 200-204.57. DOI:10.1126/science.aay3041 |
| [57] | Roemhild R, Andersson DI. Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathogens, 2021, 17(1): e1009172. DOI:10.1371/journal.ppat.1009172 |
| [58] | Suzuki S, Horinouchi T, Furusawa C. Acceleration and suppression of resistance development by antibiotic combinations. BMC Genomics, 2017, 18(1): 328. DOI:10.1186/s12864-017-3718-2 |
| [59] | Copp JN, Pletzer D, Brown AS, Van Der Heijden J, Miton CM, Edgar RJ, Rich MH, Little RF, Williams EM, Hancock REW, Tokuriki N, Ackerley DF. Mechanistic understanding enables the rational design of salicylanilide combination therapies for Gram- negative infections. mBio, 2020, 11(5): e02068-20. |
| [60] | Roemhild R, Linkevicius M, Andersson DI. Molecular mechanisms of collateral sensitivity to the antibiotic nitrofurantoin. PLoS Biology, 2020, 18(1): e3000612. DOI:10.1371/journal.pbio.3000612 |
| [61] | Rodriguez De Evgrafov M, Gumpert H, Munck C, Thomsen TT, Sommer MO. Collateral resistance and sensitivity modulate evolution of high-level resistance to drug combination treatment in Staphylococcus aureus. Molecular Biology and Evolution, 2015, 32(5): 1175-1185. DOI:10.1093/molbev/msv006 |
| [62] | Varela MC, Roch M, Taglialegna A, Long SW, Saavedra MO, Rose WE, Davis JJ, Hoffman LR, Hernandez RE, Rosato RR, Rosato AE. Carbapenems drive the collateral resistance to ceftaroline in cystic fibrosis patients with MRSA. Communications Biology, 2020, 3: 599. DOI:10.1038/s42003-020-01313-5 |
| [63] | Kim S, Lieberman TD, Kishony R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. PNAS, 2014, 111(40): 14494-14499. DOI:10.1073/pnas.1409800111 |
| [64] | Mitchison DA. Prevention of drug resistance by combined drug treatment of tuberculosis. Handbook of Experimental Pharmacology, 2012(211): 87-98. |
| [65] | Apjok G, Boross G, Nyerges Á, Fekete G, Lázár V, Papp B, Pál C, Csörgő B. Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Molecular Biology and Evolution, 2019, 36(8): 1601-1611. DOI:10.1093/molbev/msz109 |
 2022, Vol. 62
2022, Vol. 62