中国科学院微生物研究所,中国微生物学会
文章信息
- 孙新蕾, 吴长城, 谭文杰, 魏兰兰. 2022
- SUN Xinlei, WU Changcheng, TAN Wenjie, WEI Lanlan.
- 宏基因组测序在中枢神经系统感染性疾病诊断中的应用及研究进展
- Metagenomic next-generation sequencing in diagnosis of infectious diseases of central nervous system
- 微生物学报, 62(10): 3722-3731
- Acta Microbiologica Sinica, 62(10): 3722-3731
-
文章历史
- 收稿日期:2022-02-23
- 修回日期:2022-04-19
- 网络出版日期:2022-05-09
2. 中国疾病预防控制中心, 病毒病预防控制所, 北京 102206;
3. 深圳市第三人民医院, 广东 深圳 518112
2. National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China;
3. The Third People's Hospital of Shenzhen, Shenzhen 518112, Guangdong, China
中枢神经系统感染性疾病主要包括脑炎、脑膜炎和脊髓炎等。脑炎多由病毒感染引起,常见于儿童,可引起严重后遗症,其中最常见的症状包括发育迟缓(35.0%)、行为异常(18.0%)和智力缺陷(17.5%)等[1]。脑膜炎主要分为无菌性和细菌性两类。前者较为常见,通常是自限性的且预后良好;后者具有起病急、死亡率高且后遗症严重的特点,需及时明确感染病原体以对症治疗[2]。脊髓炎可引起髓鞘或轴突损伤,致使患者丧失感觉功能甚至瘫痪等[3]。近期针对我国急性脑膜炎或脑炎患者(acute meningitis or encephalitis,AME)持续10年的纵向监测结果显示:AME易感于儿童,病毒性病原体是造成患儿死亡的主要原因之一[4]。我国CNS感染患者中常见的病原体也是病毒。新发、再发和耐药病原体导致的病因不明及多重感染的病例数日益增加,进一步增大了临床病原诊断难度。因此,开发快速、灵敏且无偏倚的病原检测技术对中枢神经系统感染性疾病的精准诊疗具有重要意义。
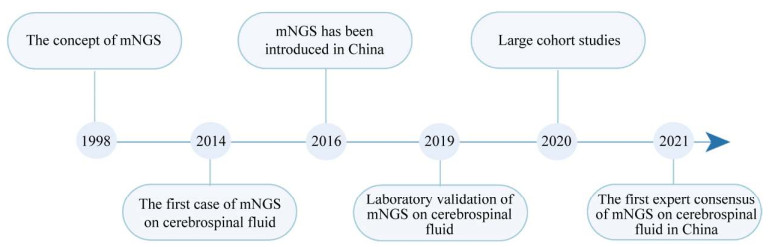
1 宏基因组测序宏基因组测序(metagenomic next-generation sequencing,mNGS)是一种基于下一代测序技术的新型病原检测技术。相较于传统检测技术,其能更加快速、准确和高通量地识别和分型病原[5]。mNGS检测目标覆盖了大量已知基因组序列的病原且可持续纳入新发现的病原。图 1概括了mNGS技术应用于临床诊断的发展历程并节选了国内外部分驱动该技术发展的关键事件。mNGS最初被应用于环境微生态研究[6]。2014年,Wilson等利用mNGS确定了一名病因不明、反复发热及免疫缺陷症的脑炎患者的病因是钩端螺旋体感染[7]。此事件正式拉开了mNGS应用于临床检测的序幕。2016年,mNGS技术在国内进入应用阶段[8]。2019年,Miller等开发一套适用于CNS感染的mNGS检测流程,其实验室也通过了临床实验室改进法案修正案(Clinical Laboratory Improvement Amendments,CLIA)的认证[9]。同年,Wilson等领衔的多中心脑脊液样本研究全面地揭示了mNGS在病原鉴定、耐药基因预测、疾病动态监测和演化分析中的潜在应用价值[10]。近期我国自主开展的多项队列研究再次证实了mNGS技术巨大的病原检测潜力,与传统方法相结合可有效地提高病原检出率[11–14]。2021年,吴钢等撰写了《中枢神经系统感染性疾病的脑脊液宏基因组学第二代测序应用专家共识》[15]。这是国内首篇关于脑脊液mNGS临床应用的专家共识,为CNS感染性疾病精准诊断提供了重要的帮助。
|
| 图 1 mNGS技术应用于中枢神经系统感染性疾病诊断的发展历程 Figure 1 Application of mNGS in the diagnosis of infectious diseases in central nervous system. |
2 mNGS实验及数据分析 2.1 mNGS实验流程与相关技术 2.1.1 样本处理
脑脊液(cerebrospinal fluid,CSF)是诊断CNS感染性疾病的主要样本。由于病原载量较低,脑脊液样本mNGS检测的灵敏性和特异性极易受背景菌和污染菌等的影响[16]。因此,待检患者临床样本的采集、运输及检测等全过程必须避免污染。若采集后4 h内进行检测,脑脊液样本可在2–8 ℃条件下运输和储存。若存储时长低于1周,可将样本储存于–20 ℃。预计保存时长超过1周的样本应储存于–70冰箱,运输过程需全程干冰。需长期保存的样本还应加入RNA稳定剂以抑制核酸降解。《中枢神经系统感染性疾病的脑脊液宏基因组学第二代测序应用专家共识》探讨了适用于mNGS检测的脑脊液样本采集要求,也具有重要的参考价值[15]。
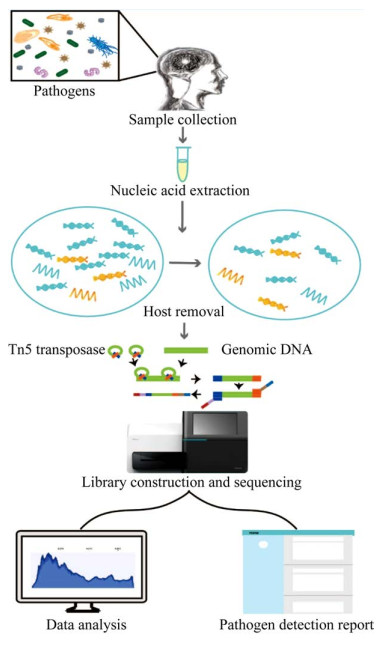
2.1.2 核酸提取病原核酸的有效提取是开展mNGS检测的前提,因此应设计预实验验证核酸提取方案是否有效。一般提取方法无法有效破壁的病原体,如结核杆菌和真菌等,珠打、煮沸等额外处理步骤可有效提升核酸提取率以避免假阴性检测结果。实验过程还应引入阳性和阴性对照组对整体流程进行质量控制(图 2)。阳性对照常使用已知病原微生物等的预混液;阴性对照可使用核酸提取试剂洗脱缓冲液等[17]。
|
| 图 2 mNGS检测流程示意图 Figure 2 mNGS workflow. |
2.1.3 人源核酸去除
脑脊液中人源核酸占比较高严重降低了病原的检出率,直接进行测序可能导致假阴性检测结果。因此在测序文库构建之前,人源核酸应被尽可能地去除[16]。常用的去人源DNA技术有:离心、过滤和差异裂解等。离心法操作简单且成本较低。Gu等采用16 000 r/min离心10 min去除脑脊液中人源细胞沉淀[18]。过滤法可以滤除颗粒较大的细胞和真菌等,适合于富集病毒。Kawada等采用0.45 µm过滤器去除脑脊液中的人源细胞[19]。差异裂解法可选择性裂解游离人源细胞DNA。Simner等在脑脊液中加入皂苷裂解人源细胞并用DNase消化游离的人源DNA[20]。RNA建库还需额外注意以下事项:总RNA需使用DNase处理以除去残留的DNA污染[9]。上述方案均存在一定的局限性,实际操作可考虑综合应用多种方案以获得理想的去宿主核酸效果。此外总RNA中约80%–90%为核糖体RNA[21],可使用探针杂交[22]和CRISPR- Cas9[23]等方法去除宿主核糖体RNA。
2.1.4 测序文库构建脑脊液测序文库构建包括核酸片段化、接头连接、扩增及纯化4个主要步骤。RNA需在文库构建前完成逆转录和二链合成步骤。核酸总量需满足构建测序文库的最低要求。总核酸一般采用酶切或机械打断方式进行片段化。其中转座酶打断的方式要求的起始核酸量较低,适用于脑脊液样本建库。Nextera XT DNA Library Preparation Kit等被广泛应用于构建脑脊液宏基因组测序文库[9–10]。文库质检合格后进行双端或单端测序。脑脊液样本宏基因组测序数据量通常不应低于20 000 000测序读长(reads)[24]。
2.2 数据分析mNGS数据分析主要包括质量控制、人源序列去除、序列组装、物种鉴定及耐药和毒力基因分析等。测序读长可用Fastp[25]和Trimmomatic[26]等工具进行质量控制。质控合格的reads可使用BWA[27]或Bowtie2[28]等工具将其比对到人类参考基因组。比对结果可通过Samtools[29]等工具滤除人源序列。剩余的reads使用SPAdes[30]及Megahit[31]等工具进行不依赖于参考基因的de novo组装,也可使用CLC Genomics Workbench (QIAGEN)等图形化分析工具进行参考基因组依赖的一致性序列(consensus sequence)组装。组装出的序列可借助BLAST[32]等工具与微生物数据库进行比对。常用的病原数据库有美国生物技术信息中心(National Center for Biotechnology Information,NCBI)的核酸序列数据库(Nucleotide Sequence Database,NT)和参考序列数据库(Reference Sequence Database,Refseq)、临床级微生物数据库(Food and Drug Administration-database for Regulatory Grade Microbial Sequences,FDA-ARGOS)、全球微生物数据中心(World Data Center for Microorganisms,WDCM)和基因组分类学数据库(Genome Taxonomy Database,GTDB)等。此外,Kraken等工具采用了基于Kmer匹配的算法[33],可快速根据原始测序数据鉴定样本中微生物的种属。耐药和毒力基因分析一般使用PathoFact[34]等工具,将组装序列与公共参考数据库比较进行注释。相关参考数据库包括毒力因子数据库(Virulence Factors Database,VFDB)和抗性基因数据库(Antibiotic Resistance Genes Database,ARDB)等。
当前mNGS数据分析尚缺乏标准化流程[35],病原微生物基因组数据库也不尽完善。同一测序数据不同分析流程的分析结果间可能存在一定差异。因此,在搭建生物信息分析平台时应进行充分地调试,严格引入质量控制体系以保证生成合理且稳定的病原鉴定结果[36]。最后,针对不同种类的病原应设置适应的阈值判断标准,报告解读时还须注意排除污染菌与试剂工程菌等干扰。数据分析相关流程的搭建也可参考其他相关综述[35–37]。
3 mNGS应用于CNS感染性疾病诊断研究进展 3.1 mNGS利于检出脑脊液中罕见和未知病原体mNGS在检测新发和罕见病原方面存在显著优势。基于mNGS技术,大量未知的引起人中枢神经系统感染的病原体被不断地鉴定出来,如松鼠博尔纳病毒[38]和伪狂犬病病毒等[39–42]。相较于传统检测方法,mNGS技术更有利于罕见病原的检测,如嗜冷杆菌[43]、解脲支原体[44]、广州管圆线虫[45–46]和猕猴α疱疹病毒1型[47]等均可被mNGS检出。此外,脑脊液mNGS检测为疑难危重CNS感染病患提供快速精准的诊疗依据,有利于协助临床医生合理地使用药物[48]。
3.2 mNGS在临床诊断中的重要应用价值和巨大发展前景近期多个研究中心开展的针对CNS感染性疾病的大规模队列研究,证实mNGS技术在临床诊疗和公共卫生领域具有重大应用价值。相对于传统检测方法,mNGS具有较高的敏感性[49],不易受抗生素影响且能在短时间内进行大量病原体的筛查鉴定[13]。其强大的病原检出能力可有效排除感染性病因,帮助自身免疫性和肿瘤等患者及时接受精准治疗[50]。针对特定病原体的队列研究表明,mNGS对结核性脑膜炎诊断的灵敏度明显高于分枝杆菌生长指示管培养、改良Ziehl-Neelsen染色和Xpert MTB/RIF方法,可作为一线脑脊液检测方法[51]。mNGS有助于隐球菌的鉴定,与传统检测方法结合也可显著提高检出率[52]。mNGS也能有效地检出传统检测方法不易发现的寄生虫[53]。对204名特发性脑膜炎、脑炎或脊髓炎患者进行急性感染性疾病的精确诊断研究,发现mNGS使感染诊断率提高了22%。mNGS与常规检测(培养、抗原检测和免疫原性检测)比较,阳性符合率为80%,阴性符合率为98%[10]。mNGS可进一步分析耐药基因和毒力因子有助于临床医生合理有效地用药[11],并能持续监测疾病进展和治疗效果从而帮助临床医生及时地调整治疗方案[13]。mNGS还被应用于CNS微生物群落探究,加强对人中枢神经系统的认识,有助于临床诊断和治疗CNS感染相关疾病[54]。mNGS联合其他病原检测技术,有利于临床精准诊疗,可提升CNS感染的诊断率[10, 55]。mNGS还被应用于预测和监测新突发传染病疫情[56]。
4 mNGS应用于CNS感染病原检测的局限性mNGS有利于CNS感染病原体的检出,但其广泛应用尚受诸多因素的限制。高宿主核酸占比严重影响病原的检出,现有去宿主核酸方法的效果还不够理想,更高效率的方法亟待开发。mNGS结果易受检测样本、试剂和实验室环境中背景微生物的干扰,严格遵守检测流程的质量控制程序非常重要。此外,定期检测常见背景微生物菌群也有利于避免假阳性检出结果。病原参考数据库方面也存在不足之处:当前部分微生物基因组序列中仍存在空白和错误;针对CNS感染的专用病原数据库亟待开发。相较于DNA病原,mNGS检测RNA病原的灵敏度尚存在一定的不足。预扩增方法可增加病原RNA的丰度,提升病原检出率。由于CNS感染性疾病的mNGS诊断尚缺乏标准的检测流程和规范的报告解读标准,未来该技术还需要不断地优化和完善。尽管mNGS在CNS感染性疾病诊疗中发挥了重要的作用,临床医生依然不能完全依赖于mNGS的病原检测结果,应结合患者病史和其他检测指标等信息筛选出主要致病病原以诊断病因[57]。当前,mNGS检测的成本还比较高,限制了其在CNS感染病原检测的广泛应用。随着mNGS技术的成熟,其检测成本将有所下降。
5 总结和展望mNGS在CNS感染性疾病诊疗中已展现出了巨大的优势和潜力,但临床实践中仍面临诸多挑战。微生物学家和临床医生等应携手一同优化和完善mNGS技术,推动该技术临床应用的规范化。需要特别说明的是,应用mNGS技术不应完全摒弃传统检测方法,只有充分结合并发挥各种方案的优势才能更好地服务于CNS感染性疾病的快速精准诊疗。我们相信基于mNGS技术的CNS感染性疾病诊疗将成为临床微生物学的重要研究前沿之一。mNGS技术将有力地推动CNS感染性疾病病原精准诊断并提升病患生存率。
致谢
感谢赵旻炅在作图和文献整理上的帮助和支持。感谢朱绪杰和李珍对于论文写作提出宝贵的修改意见。
| [1] | Khandaker G, Jung J, Britton PN, King C, Yin JK, Jones CA. Long-term outcomes of infective encephalitis in children: a systematic review and meta-analysis. Developmental Medicine and Child Neurology, 2016, 58(11): 1108-1115. DOI:10.1111/dmcn.13197 |
| [2] | Mount HR, Boyle SD. Aseptic and bacterial meningitis: evaluation, treatment, and prevention. American Family Physician, 2017, 96(5): 314-322. |
| [3] | West TW. Transverse myelitis—a review of the presentation, diagnosis, and initial management. Discovery Medicine, 2013, 16(88): 167-177. |
| [4] | Wang LP, Yuan Y, Liu YL, Lu QB, Shi LS, Ren X, Zhou SX, Zhang HY, Zhang XA, Wang X, Wang YF, Lin SH, Zhang CH, Geng MJ, Li J, Zhao SW, Yi ZG, Chen X, Yang ZS, Meng L, Wang XH, Cui AL, Lai SJ, Liu MY, Zhu YL, Xu WB, Chen Y, Yuan ZH, Li MF, Huang LY, Jing HQ, Li ZJ, Liu W, Fang LQ, Wu JG, Hay SI, Yang WZ, Gao GF. Etiological and epidemiological features of acute meningitis or encephalitis in China: a nationwide active surveillance study. The Lancet Regional Health-Western Pacific, 2022, 20: 100361. DOI:10.1016/j.lanwpc.2021.100361 |
| [5] | Forbes JD, Knox NC, Ronholm J, Pagotto F, Reimer A. Metagenomics: the next culture-independent game changer. Frontiers in Microbiology, 2017, 8: 1069. DOI:10.3389/fmicb.2017.01069 |
| [6] | Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology, 1998, 5(10): R245-R249. |
| [7] | Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu GX, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, De Risi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. The New England Journal of Medicine, 2014, 370(25): 2408-2417. DOI:10.1056/NEJMoa1401268 |
| [8] | Guan HZ, Shen A, Lv X, Yang XZ, Ren HT, Zhao YH, Zhang YX, Gong YP, Ni PX, Wu HL, Zhu YC, Cui LY. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. Journal of NeuroVirology, 2016, 22(2): 240-245. DOI:10.1007/s13365-015-0390-7 |
| [9] | Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, Stryke D, Pham E, Fung B, Bolosky WJ, Ingebrigtsen D, Lorizio W, Paff SM, Leake JA, Pesano R, De Biasi R, Dominguez S, Chiu CY. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Research, 2019, 29(5): 831-842. DOI:10.1101/gr.238170.118 |
| [10] | Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu GX, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, Berger A, Douglas V, Josephson SA, Chow FC, Fulton BD, De Risi JL, Gelfand JM, Naccache SN, Bender J, Dien Bard J, Murkey J, Carlson M, Vespa PM, Vijayan T, Allyn PR, Campeau S, Humphries RM, Klausner JD, Ganzon CD, Memar F, Ocampo NA, Zimmermann LL, Cohen SH, Polage CR, De Biasi RL, Haller B, Dallas R, Maron G, Hayden R, Messacar K, Dominguez SR, Miller S, Chiu CY. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. The New England Journal of Medicine, 2019, 380(24): 2327-2340. DOI:10.1056/NEJMoa1803396 |
| [11] | Fan SY, Wang XJ, Hu YF, Shi JP, Zou YL, Zhao WL, Qiao XD, Wang CJ, Chin JH, Liu L, Qin LZ, Wang SN, Li HF, Yue W, Zhang WH, Li XH, Ge Y, Wu HL, Chen WJ, Li YJ, Guan TJ, Li SY, Wu YH, Zhou GY, Liu Z, Piao YS, Zhang JZ, Ren CH, Cui L, Liu CY, Ren HT, Zhao YH, Feng S, Jiang HS, Wang JW, Bu H, Guo SG, Peng B, Cui LY, Li W, Guan HZ. Metagenomic next-generation sequencing of cerebrospinal fluid for the diagnosis of central nervous system infections: a multicentre prospective study. bioRxiv, 2019, Doi: 10.1101/658047. |
| [12] | Xing XW, Zhang JT, Ma YB, He MW, Yao GE, Wang W, Qi XK, Chen XY, Wu L, Wang XL, Huang YH, Du J, Wang HF, Wang RF, Yang F, Yu SY. Metagenomic next-generation sequencing for diagnosis of infectious encephalitis and meningitis: a large, prospective case series of 213 patients. Frontiers in Cellular and Infection Microbiology, 2020, 10: 88. DOI:10.3389/fcimb.2020.00088 |
| [13] | Zhang Y, Cui P, Zhang HC, Wu HL, Ye MZ, Zhu YM, Ai JW, Zhang WH. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. Journal of Translational Medicine, 2020, 18(1): 199. DOI:10.1186/s12967-020-02360-6 |
| [14] | Qian LY, Shi YJ, Li FQ, Wang YF, Ma M, Zhang YF, Shao YW, Zheng GH, Zhang GJ. Metagenomic next-generation sequencing of cerebrospinal fluid for the diagnosis of external ventricular and lumbar drainage-associated ventriculitis and meningitis. Frontiers in Microbiology, 2020, 11: 596175. DOI:10.3389/fmicb.2020.596175 |
| [15] |
Chinese Society of Infectious Diseases and Cerebrospinal Fluid Cytology. Expert consensus on clinical application of metagenomic next-generation sequencing of cerebrospinal fluid in the diagnosis of infectious diseases of the central nervous system. Chinese Journal of Neurology, 2021, 54(12): 1234-1240.
(in Chinese) 中华医学会神经病学分会感染性疾病与脑脊液细胞学学组. 中枢神经系统感染性疾病的脑脊液宏基因组学第二代测序应用专家共识. 中华神经科杂志, 2021, 54(12): 1234-1240. DOI:10.3760/cma.j.cn113694-20210730-00532 |
| [16] | Liu DL, Zhou HW, Xu T, Yang QW, Mo X, Shi DW, Ai JW, Zhang JJ, Tao Y, Wen DH, Tong YG, Ren LL, Zhang W, Xie SM, Chen WJ, Xing WL, Zhao JY, Wu YL, Meng XF, Ouyang C, Jiang Z, Liang ZK, Tan HQ, Fang Y, Qin N, Guan YL, Gai W, Xu SH, Wu WJ, Zhang WH, Zhang CT, Wang YC. Multicenter assessment of shotgun metagenomics for pathogen detection. EBioMedicine, 2021, 74: 103649. DOI:10.1016/j.ebiom.2021.103649 |
| [17] |
Branch of Laboratory Medicine. Expert consensus on clinical standardized application of metagenomics next-generation sequencing for detection of pathogenic microorganisms. Chinese Journal of Laboratory Medicine, 2020, 43(12): 1181-1195.
(in Chinese) 中华医学会检验医学分会. 高通量宏基因组测序技术检测病原微生物的临床应用规范化专家共识. 中华检验医学杂志, 2020, 43(12): 1181-1195. DOI:10.3760/cma.j.cn114452-20200903-00704 |
| [18] | Gu W, Deng XD, Marco L, Sucu YD, Shaun A, Doug S, Scot F, Allan G, Kevin R, Kelsey Z, Hannah S, Yu GX, Gurpreet I, Benjamin B, Chow ED, Amy B, Wilson MR, Candace W, Elaine H, Steve M, De Risi JL, Chiu CY. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nature Medicine, 2020, 27(1): 115-124. |
| [19] | Kawada JI, Okuno Y, Torii Y, Hayano S, Ando S, Kamiya Y, Kojima S, Ito Y. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Scientific Reports, 2016, 6(1): 33452. DOI:10.1038/srep33452 |
| [20] | Simner PJ, Miller HB, Breitwieser FP, Gabriel PM, Pardo CA, Salzberg SL, Sears CL, Thomas DL, Eberhart CG, Carroll KC. Development and optimization of metagenomic next-generation sequencing methods for cerebrospinal fluid diagnostics. Journal of Clinical Microbiology, 2018, 56(9): e00472-18. |
| [21] | O'Neil D, Glowatz H, Schlumpberge M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Current Protocols in Molecular Biology, 2013(SUPPL. 103): Chapter 4. |
| [22] | Petrova OE, Fernando GA, Claudia Z, Karin S. Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterial biofilm and mixed pathogen culture transcriptomes. Scientific Reports, 2017, 7(1): 41114. DOI:10.1038/srep41114 |
| [23] | Song LY, Xie KB. Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing. Microbiome, 2020, 8(1): 80. DOI:10.1186/s40168-020-00859-0 |
| [24] |
Clinical Microbiology Group, Branch of Laboratory Medicine, Chinese Medical Association, Clinical Microbiology Group, Branch of Microbiology and Immunology, Chinese Medical Association, Clinical Microbiology and Infection Branch, China Association For the Promotion of International Exchanges in Health Care. Metagenomic high throughput sequencing technology is applied to China's expert consensus on pathogen detection of infectious diseases. Chinese Journal of Laboratory Medicine, 2021, 44(2): 107-120.
(in Chinese) 中华医学会检验医学分会临床微生物学组, 中华医学会微生物学与免疫学分会临床微生物学组, 中国医疗保健国际交流促进会临床微生物与感染分会. 宏基因组高通量测序技术应用于感染性疾病病原检测中国专家共识. 中华检验医学杂志, 2021, 44(2): 107-120. DOI:10.3760/cma.j.cn114452-20201026-00794 |
| [25] | Chen SF, Zhou YQ, Chen YR, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics: Oxford, England, 2018, 34(17): i884-i890. DOI:10.1093/bioinformatics/bty560 |
| [26] | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics: Oxford, England, 2014, 30(15): 2114-2120. DOI:10.1093/bioinformatics/btu170 |
| [27] | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009, 25(14): 1754-1760. DOI:10.1093/bioinformatics/btp324 |
| [28] | Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 2009, 10(3): R25. DOI:10.1186/gb-2009-10-3-r25 |
| [29] | Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup 1GPDP. The sequence alignment/map format and SAMtools. Bioinformatics, 2009, 25(16): 2078-2079. DOI:10.1093/bioinformatics/btp352 |
| [30] | Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology, 2012, 19(5): 455-477. DOI:10.1089/cmb.2012.0021 |
| [31] | Li DH, Liu CM, Luo RB, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 2015, 31(10): 1674-1676. DOI:10.1093/bioinformatics/btv033 |
| [32] | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology, 1990, 215(3): 403-410. DOI:10.1016/S0022-2836(05)80360-2 |
| [33] | Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology, 2014, 15(3): R46. DOI:10.1186/gb-2014-15-3-r46 |
| [34] | De Nies L, Lopes S, Heintz-Buschart A, Laczny CC, May P, Wilmes P. PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. bioRxiv, 2020, Doi: 10.1101/2020.03.24.006148. |
| [35] | Chiu CY, Miller SA. Clinical metagenomics. Nature Reviews Genetics, 2019, 20(6): 341-355. DOI:10.1038/s41576-019-0113-7 |
| [36] |
Branch of Laboratory Medicine Chinese Medical Association. Expert consensus on the standardized management of bioinformatics analysis for the detection of pathogenic microorganisms in mNGS. Chinese Journal of Laboratory Medicine, 2021, 44(9): 799-807.
(in Chinese) 中华医学会检验医学分会. 宏基因组测序病原微生物检测生物信息学分析规范化管理专家共识. 中华检验医学杂志, 2021, 44(9): 799-807. |
| [37] | Li N, Cai QQ, Miao Q, Song ZS, Fang Y, Hu BJ. High-throughput metagenomics for identification of pathogens in the clinical settings. Small Methods, 2021, 5(1): 2000792. DOI:10.1002/smtd.202000792 |
| [38] | Hoffmann B, Tappe D, Höper D, Herden C, Boldt A, Mawrin C, Niederstraßer O, Müller T, Jenckel M, van der Grinten E, Lutter C, Abendroth B, Teifke JP, Cadar D, Schmidt-Chanasit J, Ulrich RG, Beer M. A variegated squirrel bornavirus associated with fatal human encephalitis. The New England Journal of Medicine, 2015, 373(2): 154-162. DOI:10.1056/NEJMoa1415627 |
| [39] | Ai JW, Weng SS, Cheng Q, Cui P, Li YJ, Wu HL, Zhu YM, Xu B, Zhang WH. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerging Infectious Diseases, 2018, 24(6): 1087-1090. DOI:10.3201/eid2406.171612 |
| [40] | Yang X, Guan HZ, Li C, Li Y, Wang SJ, Zhao XH, Zhao YY, Liu YM. Characteristics of human encephalitis caused by pseudorabies virus: a case series study. International Journal of Infectious Diseases, 2019, 87: 92-99. DOI:10.1016/j.ijid.2019.08.007 |
| [41] | Liu QY, Wang XJ, Xie CH, Ding SF, Yang HN, Guo SB, Li JX, Qin LZ, Ban FG, Wang DF, Wang C, Feng LX, Ma HC, Wu B, Zhang LP, Dong CX, Xing L, Zhang JW, Chen HC, Yan RQ, Wang XR, Li W. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clinical Infectious Diseases, 2020, 73(11): e3690-e3700. |
| [42] | Zhou YY, Nie C, Wen H, Long Y, Zhou MH, Xie ZC, Hong DJ. Human viral encephalitis associated with suid herpesvirus 1. Neurological Sciences, 2022, 43(4): 2681-2692. DOI:10.1007/s10072-021-05633-0 |
| [43] | Ortiz-Alcántara JM, Segura-Candelas JM, Garcés- Ayala F, Gonzalez-Durán E, Rodríguez-Castillo A, Alcántara-Pérez P, Wong-Arámbula C, González-Villa M, León-Ávila G, García-Chéquer AJ, Diaz-Quiñonez JA, Méndez-Tenorio A, Ramírez-González JE. Fatal Psychrobacter sp. infection in a pediatric patient with meningitis identified by metagenomic next-generation sequencing in cerebrospinal fluid. Archives of Microbiology, 2016, 198(2): 129-135. |
| [44] | Zhan CY, Chen LH, Hu LL. Neonatal Ureaplasma parvum meningitis complicated with subdural hematoma: a case report and literature review. BMC Infectious Diseases, 2021, 21(1): 268. DOI:10.1186/s12879-021-05968-1 |
| [45] | Feng L, Zhang AW, Que JL, Zhou HY, Wang HY, Guan YL, Shen CZ, Sun XS, Lai R, Peng FH, Feng HY, Chen L. The metagenomic next-generation sequencing in diagnosing central nervous system angiostrongyliasis: a case report. BMC Infectious Diseases, 2020, 20(1): 691. DOI:10.1186/s12879-020-05410-y |
| [46] | Xie M, Zhou Z, Guo SH, Li ZQ, Zhao H, Deng JS. Next-generation sequencing specifies Angiostrongylus eosinophilic meningoencephalitis in infants: two case reports. Medicine, 2019, 98(35): e16985. DOI:10.1097/MD.0000000000016985 |
| [47] |
Zhao L, Qi WJ, Du HJ, Zheng Y, Liu JY, Wang WL, Wang GX, Pan Y, Huang BY, Feng ZM, Zhang DT, Yang P, Chen YW, Li S, Ma CN, Han J, Wang QY, Tan WJ. Confirmation of the first human case with Macacine alphaherpesvirus 1 infection in China. International Journal of Virology, 2021(4): 278-282.
(in Chinese) 赵莉, 齐文杰, 杜海军, 郑阳, 刘景院, 王文玲, 王国兴, 潘阳, 黄保英, 冯兆民, 张代涛, 杨鹏, 陈艳伟, 李爽, 马春娜, 韩俊, 王全意, 谭文杰. 中国首例人感染猕猴α疱疹病毒1型病例的确诊. 国际病毒学杂志, 2021(4): 278-282. DOI:10.3760/cma.j.issn.1673-4092.2021.04.004 |
| [48] | Chen B, Chen Z, Yang YS, Cai GL, Xu XJ, Guan HZ, Ren HT, Tuo HZ. Next-generation sequencing combined with serological tests based pathogen analysis for a neurocysticercosis patient with a 20-year history: a case report. BMC Neurology, 2021, 21(1): 236. DOI:10.1186/s12883-021-02277-7 |
| [49] | Yan LP, Sun WW, Lu ZH, Fan L. Metagenomic next-generation sequencing (mNGS) in cerebrospinal fluid for rapid diagnosis of tuberculosis meningitis in HIV-negative population. International Journal of Infectious Diseases, 2020, 96: 270-275. DOI:10.1016/j.ijid.2020.04.048 |
| [50] | Ramachandran PS, Wilson MR. Metagenomics for neurological infections-expanding our imagination. Nature Reviews Neurology, 2020, 16(10): 547-556. DOI:10.1038/s41582-020-0374-y |
| [51] | Chen YX, Wang YQ, Liu XJ, Li W, Fu HY, Liu XY, Zhang X, Zhou XQ, Yang BZ, Yao J, Ma XL, Han LJ, Li H, Zheng LH. Comparative diagnostic utility of metagenomic next-generation sequencing, GeneXpert, modified Ziehl-Neelsen staining, and culture using cerebrospinal fluid for tuberculous meningitis: a multi-center, retrospective study in China. Journal of Clinical Laboratory Analysis, 2022, 36(4): e24307. |
| [52] | Xing XW, Zhang JT, Ma YB, Zheng N, Yang F, Yu SY. Apparent performance of metagenomic next-generation sequencing in the diagnosis of cryptococcal meningitis: a descriptive study. Journal of Medical Microbiology, 2019, 68(8): 1204-1210. DOI:10.1099/jmm.0.000994 |
| [53] | Hu ZL, Weng X, Xu CH, Lin Y, Cheng C, Wei HX, Chen W. Metagenomic next-generation sequencing as a diagnostic tool for toxoplasmic encephalitis. Annals of Clinical Microbiology and Antimicrobials, 2018, 17(1): 45. DOI:10.1186/s12941-018-0298-1 |
| [54] | Kang YY, Ji XC, Guo L, Xia H, Yang XF, Xie Z, Shi XD, Wu R, Feng DY, Wang C, Chen M, Zhang WL, Wei H, Guan YL, Ye K, Zhao G. Cerebrospinal fluid from healthy pregnant women does not harbor a detectable microbial community. Microbiology Spectrum, 2021, 9(3): e0076921. DOI:10.1128/Spectrum.00769-21 |
| [55] | Ge MM, Gan MY, Yan K, Xiao FF, Yang L, Wu BB, Xiao ML, Ba Y, Zhang R, Wang J, Cheng GQ, Wang LS, Cao Y, Zhou WH, Hu LY. Combining metagenomic sequencing with whole exome sequencing to optimize clinical strategies in neonates with a suspected central nervous system infection. Frontiers in Cellular and Infection Microbiology, 2021, 11: 671109. DOI:10.3389/fcimb.2021.671109 |
| [56] | Saha S, Ramesh A, Kalantar K, Malaker R, Hasanuzzaman M, Khan LM, Mayday MY, Sajib MSI, Li LM, Langelier C, Rahman H, Crawford ED, Tato CM, Islam M, Juan YF, de Bourcy C, Dimitrov B, Wang J, Tang J, Sheu J, Egger R, de Carvalho TR, Wilson MR, Saha SK, De Risi JL. Unbiased metagenomic sequencing for pediatric meningitis in Bangladesh reveals neuroinvasive chikungunya virus outbreak and other unrealized pathogens. bioRxiv, 2019, DOI: 10.1101/579532. |
| [57] | 毕铭辕, 汪春付, 连建奇, 孙永涛. 宏基因组测序在感染性疾病中的应用与反思. 中华临床感染病杂志, 2019, 12(5): 379-384. DOI:10.3760/cma.j.issn.1674-2397.2019.05.012 |
 2022, Vol. 62
2022, Vol. 62