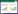中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 阮浩哲, 刘立明, 张伟国, 徐建中. 2021
- Haozhe Ruan, Liming Liu, Weiguo Zhang, Jianzhong Xu. 2021
- NADPH缺陷型Corynebacterium glutamicum重组菌株的构建及其性能分析
- Construction and performance analysis of NADPH-auxotrophic Corynebacterium glutamicum recombinant
- 微生物学报, 61(8): 2442-2456
- Acta Microbiologica Sinica, 61(8): 2442-2456
-
文章历史
- 收稿日期:2020-09-18
- 修回日期:2020-11-07
- 网络出版日期:2021-03-22
2. 江南大学食品科学与技术国家重点实验室, 江苏 无锡 214122
2. State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi 214122, Jiangsu Province, China
辅因子NADPH在细胞内分布广泛,通过参与800多个氧化还原反应来调节胞内氧化还原水平并影响众多基因表达、细胞功能、代谢途径、物质跨膜运输和胞内微环境[1]。胞内NADPH水平与目标代谢产物的合成之间的关系可分为以下几种类型:(1) 影响目标代谢产物产量:提高胞内NADPH水平,能促进胞内NADPH-依赖型产物的合成[2-3];(2) 影响底物转化率:调控胞内NADPH水平,能有效降低副产物积累,提高原料的转化率[4];(3) 影响目标代谢产物生产强度:增加胞内NADPH供给,满足中心碳代谢途径中关键酶对NADPH的需求,可实现目标产物高强度的生产[5];(4) 拓宽底物利用范围:促进胞内NADPH的再生,可提高菌体对糖蜜、羧基化合物等的利用[6-7];(5) 影响胞内微环境:改变NADPH水平会影响胞内NAD(H/+)状态和ATP含量,可调节胞内pH (pHi)和活性氧簇(ROS)的形成,提高细胞对酸胁迫和氧胁迫的适应力[8-10]。因此,对胞内NADPH水平的调控是菌种改造和发酵过程优化需考虑的一个重要指标。
胞内NADPH水平的调控策略可分为外源调控和内源调控。外源调控是指采用生化工程的方法,通过添加外源电子受体[8]、不同还原态碳源和NADP+前体物[11],改变溶氧[12]等实现对NADPH代谢的调控;而内源调控是基于细胞内NADPH合成与代谢的途径,通过代谢工程策略调节与NADP(H/+)代谢相关途径或酶活性。目前,内源调控是调节胞内NADPH水平的常用策略,具体思路可分为以下几个方面:(1) 调控磷酸戊糖(PP)途径:PP途径是多数微生物中主要的NADPH供给途径,由葡萄糖-6-磷酸脱氢酶(Zwf, 编码基因zwf)和6-磷酸葡萄糖酸脱氢酶(Gnd, 编码基因gnd)催化合成[1, 13];(2) 调控糖酵解(EMP)途径:将菌体内源的NAD+-依赖型甘油醛-3-磷酸脱氢酶(NAD+-GADPH)换成NADP+-GAPDH,可显著增加胞内NADPH水平,促进目标产物的合成[5, 14];(3) 调控三羧酸循环(TCA)途径:敲除琥珀酰-CoA合成酶,扰乱正常的TCA,强化流经苹果酸酶(MalE, 编码基因malE)的碳通量,可增加胞内NADPH的水平[15];(4) 调控转氢酶循环途径:调节E. coli中由膜结合吡啶核苷酸转氢酶PntAB (或mTH, 编码基因pntAB)或可溶性吡啶核苷酸转氢酶UdhA (或sTH)组成的转氢酶循环途径,调控NAD(H/+)与NADP(H/+)的相互转化水平,从而实现调节胞内NADPH水平[16]。C. glutamicum没有转氢酶循环途径,只有由丙酮酸/磷酸烯醇式丙酮酸羧化酶、MalE和苹果酸脱氢酶组成的类似转氢酶循环(transhydrogenase-like cycle)途径,MalE可将NADH转化为NADPH,提高胞内NADPH供给水平,促进目的产物的合成[17]。虽然上述代谢改造取得了比较显著的成果,但还存在以下不足:辅因子NADPH调控微生物细胞生理代谢功能的机制还不清晰,进而难以通过控制胞内NADPH水平实现目标产物合成途径的精细化调控。
本研究旨在通过阻断C. glutamicum胞内所有参与NADPH合成途径(图 1),构建1株NADPH缺陷型的重组菌株。该菌株可以作为有效的底盘细胞,用于比较不同的NADPH再生策略,进而可以直接筛选具有不同NADPH再生率的氧化还原酶,获得不同胞内NADPH水平的重组菌株,实现精确调控胞内NADPH的水平,为进一步阐明NADPH调控微生物细胞生理代谢功能的机制提供研究基础。

|
图 1 NADPH营养缺陷型C. glutamicum重组菌株的构建策略 Figure 1 Schematic representation of the NADPH auxtrophic C. glutamicum. NADPH and NADH metabolic pathway are shown in blue arrows and pink arrows, respectively. The red arrow and grey arrow represent the extrinsic routes.   |
1 材料和方法 1.1 试剂
D-葡萄糖酸、丙酮酸、α-酮戊二酸和草酰乙酸购自国药集团化学试剂有限公司。胰蛋白胨、酵母提取物购自英国Oxoid公司。质粒提取试剂盒、Taq DNA聚合酶、DNA Marker和蛋白质Marker购自南京诺唯赞生物科技有限公司。各种限制性内切酶购自美国ThermoFisher Scientific公司。
1.2 菌株、质粒与引物实验过程中涉及的菌株、质粒和引物如表 1所示,其中引物由苏州金唯智生物科技有限公司合成。
| C. glutamicum strains and plasmids | Characters | References |
| Strains | ||
| Lys-χ | L-lysine high-producing strain derived from C. glutamicum ATCC13032 by multiple rounds of random mutagenesis | Our Lab |
| Lys-χ ΔZ | Deletion of genes zwf in strain Lys-χ chromosome | This study |
| Lys-χ ΔZM | Deletion of genes zwf and malE in strain Lys-χ chromosome | This study |
| Lys-χ ΔZMICg | Deletion of genes zwf, malE and icdCg in strain Lys-χ chromosome | This study |
| Lys-χ ΔZMICg: : ISm, i.e., Lys-χ1 | Replacement of the natural icdCg gene with the Ptac-icdSm-rrnBT1T2 cassette in strain Lys-χ ΔZM chromosome | This study |
| Lys-χ1/PX-pntAB | Overexpression of E. coli pntAB under the promoter PX in strain Lys-χ1 (PX represents a series of promoters with different transcriptional activity, and the promoters used in this study are listed in Table 2) | This study |
| Plasmids | ||
| pK18mobsacB | Integration vector | Stratagene |
| pEC-XK99E | Overexpression plasmid with Kan resistance and promoter Ptrc | Stratagene |
| pK18mobsacB/∆zwf | Integration vector for deletion of zwf | [18] |
| pK18mobsacB/∆malE | Integration vector for deletion of malE | [18] |
| pK18mobsacB/∆icdCg | Integration vector for deletion of icdCg | [18] |
| pK18mobsacB/∆icdCg: : icdSm | Integration vector for replacement of the icdCg gene by the Ptac-icdSm-rrnBT1T2 cassette | [18] |
| pECM | Delection of lacIq in plasmid pEC-XK99E | This study |
| Ptrc-pntAB-gfp | Overexpression plasmid derived from pECM | This study |
| PX-pntAB-gfp | Overexpression plasmid derived from pECM, the promoter Ptrc was replaced by promoter PX | This study |
| Primer pairs | ||
| Zwf-F | GTGAGCACAAACACGAC | PCR verification of genes zwf, malE, icdCg and icdSm |
| Zwf-R | TTATGGCCTGCGCCAGGTGTG | |
| MalE-F | GCGTTCCACCCAAAACCT | |
| MalE-R | TGGACAGCTGCCTTGACT | |
| IcdCg-F | TTACTTCTTCAGTGCG | |
| IcdCg-R | ATGGCTAAGATCATCTG | |
| IcdSm-F | TGCCTGATAAGCCCGTT | |
| IcdSm-R | CAGAAAACTGACGGGTT | |
1.3 培养条件与培养基
37 ℃、100 r/min培养Escherichia coli,30 ℃、100 r/min培养C. glutamicum。在特定条件下,添加50 μg/mL或加25 μg/mL卡那霉素(Kan)用于筛选E. coli和C. glutamicum重组菌株。
Luria-Bertani (LB)培养基(g/L):蛋白胨10,酵母提取物5,NaCl 10,pH 7.0。LBG培养基:LB培养基添加5 g/L葡萄糖。Epo培养基:LBG培养基添加30 g/L甘氨酸,4 g/L异烟肼和1 g/L吐温-80。LBHIS培养基:LB培养基添加91 g/L山梨醇和18.5 g/L脑心浸出液。CgXII培养基(g/L):3-(N-吗啡啉)-丙磺酸42,(NH4)2SO4 20,尿素5,KH2PO4 1,K2HPO4·3H2O 1,MgSO4·7H2O 0.25,CaCl2 0.01,FeSO4·7H2O 0.01,MnSO4·H2O 0.01,ZnSO4·7H2O 0.01,NiCl·6H2O 0.0002,生物素0.0002,原儿茶酸0.00003。所有培养基利用20% (M/V) NaOH调节至pH 7.0,并于121 ℃灭菌20 min。
1.4 质粒和菌株方法 1.4.1 基因敲除质粒pK18mobsacB-∆zwf、pK18mobsacB-∆malE、pK18mobsacB-∆icdCg和pK18mobsacB-∆icdCg: : icdSm的构建:基因敲除质粒pK18mobsacB-∆zwf、pK18mobsacB-∆malE、pK18mobsacB-∆icdCg和pK18mobsacB-∆icdCg: : icdSm构建流程如图 2所示,具体构建方法参照杨汉昆等建立的方法进行[18]。
|
| 图 2 构建pK18mobsacB-∆zwf (A)、pK18mobsacB-∆malE (B)、pK18mobsacB-∆icdCg和pK18mobsacB-∆icdCg: : icdSm (C) Figure 2 The process of construction of pK18mobsacB-∆zwf (A), pK18mobsacB-∆malE (B), pK18mobsacB-∆icdCg and pK18mobsacB-∆icdCg: : icdSm (C). |
1.4.2 基因表达质粒PX-pntAB-gfp的构建:
根据National Center for Biotechnology Information中E. coli MG1655全基因组序列的PntAB基因序列设计引物(即pntAB-F: 5ʹ-CCGGAATTCGAAAGGA GATATACCATGCGAATTGGCATACCAA-3ʹ;pntAB- R: 5ʹ-CGTGAGCTCTTACAGAGCTTTCAG-3ʹ)。以E. coli MG1655基因组为模板,pntAB-F/pntAB-R为引物进行PCR获得pntAB基因片段。随后,采用限制性内切酶EcoR I和Sac I酶切质粒pECM和pntAB基因片段,胶回收后于22 ℃过夜酶连,经Kan抗性平板筛选和基因测序验证获得重组表达质粒pECM-pntAB (即Ptrc-pntAB)。随后,根据来源于Aequorea victoria的绿色荧光蛋白(GFP,编码基因gfp)氨基酸序列经密码子优化后,在其编码基因上游加入C. glutamicum SD识别序列并通过基因合成的方法连接到重组表达质粒Ptrc-pntAB中,从而获得目的重组质粒Ptrc-pntAB-gfp。为了获得不同pntAB基因表达强度的重组质粒,本实验选择将质粒pECM中的trc启动子通过融合PCR的方式替换成不同强度的启动子,从而获得重组表达质粒PX-pntAB-gfp。本实验所选用的启动子序列如表 2所示,其核苷酸序列由苏州金唯智生物科技有限公司合成。
| Promoters | Sequences (5ʹ→3ʹ) | References |
| Plac | TTTACACTTTATGCTTCCGGCTCGTATGTTG | [19] |
| Ptrp | TGTTGACAATTAATCATCGAACTAGTTAACTAGTACGCA | |
| Ptac | TGAGCTGTTGACAATTAATCATCGGCTCGTATATAATGTGTGGAATTGTGAGCGGATAACAATT | |
| PtacM | TGAGCTGTTGACAATTAATCATCGTGTGGTACCATGTGTGGAATTGTGAGCGGATAACAATT | |
| Pkan | CCGGAATTGCCAGCTGGGGCGCCCTCTGGTAAGGTTGGGAAGCCCTGCAA | |
| PPF104 | CAGCGTATTTGACCGATCCGGACACCTGGGATAATGTGTGGATTTGTCGG | |
| PlacM | TGAGCTGTTTACAATTAATCATCGTGTGGTACCATGTGTGGAATTG | [20] |
| Ptrc | TTGACAATTAATCATCCGGCTCGTAATG | |
| Psod | AGCGGTAACCATCACGGGTTCGGGTGCGAAAAACCATGCCATAACAGGAATGTTCCTTT CGAAAATTGAGGAAGCCTTATGCCCTACAACCCTACTTAGCTGCCAATTATTCCGGGCT TGTGACCCGCTACCCAATAAATAGGTGGGCTGAAAAATTTCGTTGCAATATCAACAAAA AGGCCTATCATTGGGAAGTGTCGCACCAAGTACTTTTGCGAAGCGCCATCTGACGGATT TTCAAAAGATGTATATGCTCGGTGCGGAAACCTACGAAAGGATTTTTTACCC |
|
| Ptuf | TGGCCGTTACCTGCGAATGTCCACAGGGTAGCTGGTAGTTGAAAATCAACGCCGTTG CCCTTAGGATTCAGTAACTGGCACATTTGTAATGCGCTAGATCTGTGTGCTCAGTCT TCCAGGCTGCTTATCACAGTGAAAGCAAACCAATTCGT GGCTGCGAAAGTCGTAGCCACCACGAAGTCCAGGAGGACATACA |
|
| Pgro | AGTTTGGCTGCCATGTGAATTTTTAGCACCCTCAACAGTTGAGTGCTGGCA CTCTCGGGGGTAGAGTGCCAAATAGGTTGTTTGACACACAGTTGTTCAC CCGCGACGACGGCTGTGCTGGAAACCCACAACCGGCACACACAAAATTTTTCTCAT |
[21] |
| Ppfk | TGGGTGATTGTTCCGGCGCGGGTGTTGTGATGGGTTTAATATGGAAGACA | [22] |
| PgapB | GCAGATACTGGAATCATTAACACCTTCCGCTTTGGGCTAATGTTGGGGGG | [22] |
| PgapA | GAATCCGCTGCAAAATCTTTGTTTCCCCGCTAAAGTTGGGGAC | |
| Ppgk | ACCCCGGGCTATTTTGTGTCTTTAATCAATACAATTGAATACCG | |
| Pgpm | TTTGCCGTATCTCGTGCGCAGAATTGCTTTTGAGGGAAAGATGGAGGAGA | |
| Peno | TTTCAACTGATTGCCTCATCGAAACAAGATTCGTGCAACAATTGGGTGTA | |
| Ppck | ACCTAAAGTTTTAACTAGTTCTGTATCTGAAAGCTACGCTAGGGGGCG | |
| PmalE | CATTGCGAAATTTTTGTTGAGCTACATATTTAGCTAGTGTTTTTGTTCCA | |
| PdapA | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTAACCTTGAACTCTA | [23] |
| PdapA-P4 | TAGGTTATTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTAACCTTGAACTCTA | |
| PdapA-R2 | TAGGTTCCTTCCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTAACCTTGAACTCTA | |
| PdapA-A16 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTATAATTGAACTCTA | |
| PdapA-A45 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGTAAAATTGAACTCTA | |
| PdapA-B6 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGCAACCATGAACTCTA | |
| PdapA-B27 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGAAACCATGAACTCTA | |
| PdapA-B31 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAAGGGAACCGTGAACTCTA | |
| PdapA-C2 | TAGGTTTTTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGATCGTAACCTTGAACTCTA | |
| PdapA-C13 | TAGGTTATTTGCGGGGTTGTTTAACCCCCAAATGAGGGAAGAACGTAACCTTGAACTCTA | |
| PdapA-C20 | TAGGTTCCTTCCGGGGTTGTTTAACCCCCAAATGAGGGAAGATGGTAACCTTGAACTCTA | |
| PH30 | CAAAAGCTGGGTACCAAAGTAACTTTTCGGTTAAGGTAGCGCATTCGTGGTGCCCGTGGCCCGGTTGGTTGGGCAGGAGTATATTGGGATCCA | [24] |
| PH36 | CAAAAGCTGGGTACCTCTATCTGGTGCCCTAAACGGGGGAATATTAACGGGCCCAGGGTGGTCGCACCTTGGTTGGTAGGAGTAGCATGGGATCCA |
1.4.3 NADPH营养缺陷型C. glutamicum重组菌株的构建
依次将上述重组质粒pK18mobsacB-∆zwf、pK18mobsacB-∆malE、pK18mobsacB-∆icdCg和pK18mobsacB-∆icdCg: : icdSm电转至C. glutamicum Lys-χ感受态细胞中,并筛选出目标重组菌株C. glutamicum Lys-χ Δzwf (即Lys-χ ΔZ)、C. glutamicum Lys-χ Δzwf ΔmalE (即Lys-χ ΔZM)、C. glutamicum Lys-χ Δzwf ΔmalE ΔicdCg (即Lys-χ ΔZMICg)和C. glutamicum Lys-χ Δzwf ΔmalE ΔicdCg: : icdSm (即Lys-χ ΔZMICg: : Ism或Lys-χ1)。目标重组菌株的具体筛选方法参照Wang等提出的方法进行[25]。
1.4.4 NADPH回补型C. glutamicum重组菌株的构建依次将上述重组基因表达质粒PX-pntAB-gfp电转至C. glutamicum Lys-χ1感受态细胞中,并筛选出目标重组菌株C. glutamicum Lys-χ1/PX-pntAB-gfp (即Lys-χ1/PX-pntAB-gfp)。目标重组菌株的具体筛选方法参照Xu等提出的方法进行[6]。
1.5 分析方法 1.5.1 NAD(P)+和NAD(P)H的检测和NAD(P)H/NAD(P)+的计算采用超声破碎法破碎细胞后,以酸性抽提液(0.5 mol/L HCl)提取氧化型吡啶核苷酸(NAD+和NADP+),以碱性抽提液(0.5 mol/L NaOH)提取还原型吡啶核苷酸(NADH和NADPH)。随后,借助从BioVision公司购置的定量分析试剂盒,利用酶循环法测定NAD(P)+和NAD(P)H的浓度并计算NADH/NAD+和NADPH/NADP+,其中以NAD/NADH Quantification Colorimeteric Kit特异性检测NAD+和NADH,以NADP/NADPH Quantification Colorimeteric Kit特异性检测NADP+和NADPH,具体步骤参照试剂盒说明书和我们前期建立的方法进行[6]。
1.5.2 绿色荧光蛋白(GFP)表达强度检测将活化好的种子转接到10 mL LBG液体培养基中培养10–12 h,吸取一定量的培养液4 ℃离心收集菌体。然后用磷酸盐(PBS)缓冲液(pH 7.4)清洗3遍收集菌体,并利用一定量的PBS缓冲液(pH 7.4)重悬菌体。最后,采用荧光激活细胞分选仪(Beckman Coulter, Inc., CA, USA)分析菌体绿色荧光强度,具体分析方法参考Yim等建立的方法进行[24]。
1.5.3 菌体生长情况的分析将发酵液离心10 min (12000 r/min),然后用dd H2O洗涤离心菌体3次,将菌体置于105 ℃烘干至恒重,最后计算并得到在本实验条件下C. glutamicum OD562与DCW (g/L)的关系。将定时取样的发酵液用0.25 mol/L的稀盐酸溶液稀释26倍后,用紫外分光光度计测定OD562,具体测定方法参照杨汉昆等建立的方法进行[18]。
1.5.4 葡萄糖含量及L-赖氨酸浓度的测定发酵液离心5 min (4 ℃,12000 r/min)后取上清并将其稀释100倍,通过生物传感分析仪SBA-40C测定发酵液中残留的葡萄糖含量(进样量25 µL)和L-赖氨酸浓度,具体测定方法参照杨汉昆等建立的方法[18]。
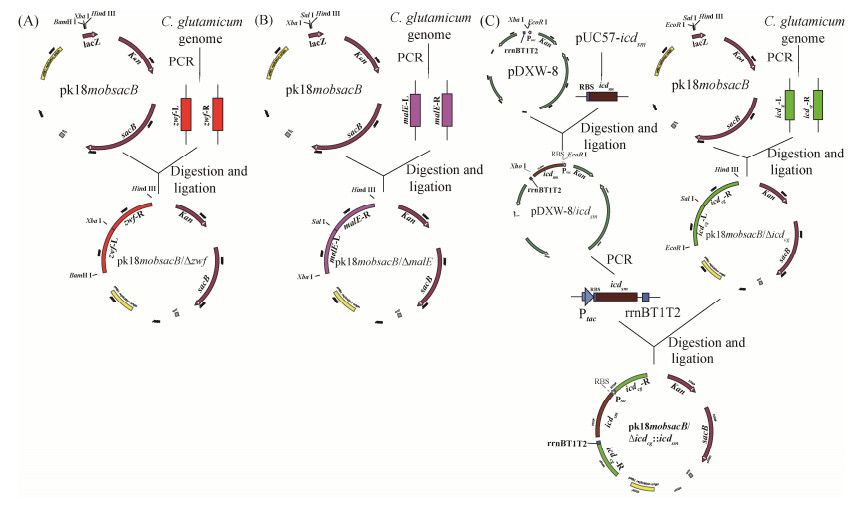
2 结果和分析 2.1 目的重组菌株的筛选与鉴定参照“材料和方法”中的方法,依次将目的重组质粒pK18mobsacB-∆zwf、pK18mobsacB-∆malE、pK18mobsacB-∆icdCg和pK18mobsacB-∆icdCg: : icdSm电转至C. glutamicum Lys-χ感受态细胞中,通过二次同源重组筛选出目标重组菌株。经过PCR验证,确定目标重组菌株(图 3)。从图 3可知,重组菌Lys-χ ΔZ、Lys-χ ΔZM和Lys-χ ΔZMICg中编码基因zwf、malE和icdCg都有缺失,而在重组菌Lys-χ ΔZMICg: : ISm中也存在来源于S. mutans的icdSm基因。这些结果表明,本研究所筛选的重组菌株为目的重组菌株。目的重组菌株的筛选与鉴定方法参照杨汉昆等建立的方法进行[18]。
|
| 图 3 目标重组菌株PCR验证 Figure 3 PCR analysis of the target recombinant strains. Lane 1, lane 2: the comparison between original and knock-out of gene zwf. Lane 3, lane 4: the comparison between original and knock-out of gene malE. Lane 5, lane 6: the comparison between original and knock-out of gene icdcg. Lane 7: the gene icdsm. |
2.2 改造Zwf影响菌体生长且显著影响L-赖氨酸合成
Zwf是PP途径中第一个限速酶,以NADP+为辅因子催化葡萄糖-6-磷酸形成6-磷酸葡萄糖酸内脂和NADPH (图 1)。为了构建NADPH营养缺陷型菌株,本研究首先选择失活Zwf,获得重组菌株C. glutamicum Lys-χ ΔZ。摇瓶发酵出发菌株Lys-χ和重组菌株Lys-χ ΔZ,发酵期间定时取样测定发酵液中菌体量。结果表明,重组菌株Lys-χ ΔZ表现出与出发菌株Lys-χ较差的菌体生长性能(图 4-A,B)。然而,与出发菌株Lys-χ相比,重组菌株Lys-χ ΔZ中L-赖氨酸产量显著下降(即1.8± 0.4 g/L),仅为出发菌株Lys-χ的9.1%,且单位菌体量下L-赖氨酸产量也显著降低[2.05 vs. 0.23 (g L-赖氨酸/g菌体)](图 4-C)。有趣的是,当以葡萄糖酸为底物时,重组菌株Lys-χ ΔZ中L-赖氨酸产量能够显著提高,达到15.4±1.3 g/L,为出发菌株Lys-χ的78.2%。然而,当以其他有机酸(如丙酮酸、α-酮戊二酸或草酰乙酸)为底物时(添加量与葡萄糖中“C”含量相当),L-赖氨酸产量并没有增加,甚至会降低L-赖氨酸产量(图 4-C)。

|
| 图 4 不同重组菌株在不同碳源下菌体生长和L-赖氨酸发酵情况 Figure 4 The cell growth at 24 h (A) and at 48 h (B) as well as the L-lysine production at 48 h (C) of different recombinant strains under the different carbone source. Glc: glucose; Gln: gluconate; Pyr: pyruvate; α-KG: α-ketoglutarate; OAA: oxaloacetate. |
为了确定出发菌株Lys-χ经过改造后是否改变了胞内NADPH水平,本文对出发菌株和重组菌株摇瓶发酵中期(即24 h后)胞内吡啶核苷酸进行了含量测定,具体数据如表 3所示。从表 3可以看出,重组菌株Lys-χ ΔZ胞内NADPH水平从出发菌株Lys-χ的4.18×10–4 nmol/(104细胞)下降到1.03×10–4 nmol/(104细胞),胞内NADPH/ NADP+降低了32.1%。值得指出的是,重组菌株Lys-χ ΔZ胞内另一种吡啶核苷酸(即NADH)相比出发菌株Lys-χ有显著的提高,从1.75×10–4 nmol/ (104细胞)提高到2.34×10–4 nmol/(104细胞),胞内NADH/NAD+升高了33.7% (表 3)。
| C. glutamicum | NADPH a | NADP+ a | NADPH/NADP+ | NADH a | NAD+ a | NADH/NAD+ |
| Lys-χ | 4.18 | 3.72 | 1.12 | 1.75 | 7.03 | 0.25 |
| Lys-χ ΔZ | 1.03 | 1.45 | 0.71 | 2.34 | 6.12 | 0.38 |
| Lys-χ ΔZM | 0.82 | 1.47 | 0.56 | 2.37 | 6.05 | 0.39 |
| Lys-χ ΔZMICg | ND | 1.22 | – | 1.62 | 7.04 | 0.23 |
| Lys-χ1 | ND | 1.25 | – | 2.93 | 5.66 | 0.52 |
| a: the unit is 10–4 nmol/(104 cell); ND: not detected; –: no computed data. | ||||||
2.3 协同改造Zwf、MalE和异柠檬酸脱氢酶(Icd)显著影响菌体生长和L-赖氨酸合成
C. glutamicum中除PP途径中Zwf和Gnd参与NADPH合成外,还有类似转氢酶循环途径中MalE和TCA循环中的Icd也参与NADPH的合成(图 1)[17]。为了进一步控制胞内NADPH的合成,本实验继续失活MalE和Icd。以葡萄糖为碳源时,在失活Zwf基础上继续失活MalE (即获得重组菌株Lys-χ ΔZM)对菌体生长影响不显著,但是在失活Zwf和MalE基础上继续失活Icd (即获得重组菌株Lys-χ ΔZMICg)菌体生长抑制明显(图 4-A,B)。需要指出的是,当以葡萄糖酸为碳源时,协同改造Zwf、MalE和Icd对菌体生长影响较小,而以其他有机酸(如丙酮酸、α-酮戊二酸和草酰乙酸)为碳源时,菌体生长受到显著影响(图 4-A)。此外,本研究发现将来源于变形链球菌(S. mutans)中的NAD+-Icd基因(icdSm)替换C. glutamicum自身NADP+-Icd基因(icdCg)获得重组菌株Lys-χ1,该重组菌株Lys-χ1在所有测试碳源中菌体生长都要优于重组菌株Lys-χ ΔZMICg (图 4-A,B)。在L-赖氨酸合成方面,以葡萄糖或其他有机酸(如丙酮酸、α-酮戊二酸和草酰乙酸)为碳源时,重组菌株Lys-χ ΔZMICg和Lys-χ1发酵液中检测不到L-赖氨酸的积累(图 4-C)。以葡萄糖酸为碳源时,重组菌株Lys-χ ΔZMICg [(5.9±1.0) g/L]和Lys-χ1 [(7.2±0.4) g/L]中L-赖氨酸产量有明显提高,但仍低于出发菌株Lys-χ [(15.4±1.4) g/L] (图 4-C)。进一步分析重组菌株Lys-χ ΔZMICg和Lys-χ1胞内吡啶核苷酸含量时发现,在重组菌株Lys-χ ΔZMICg和Lys-χ1胞内检测不到NADPH (表 3)。值得指出的是,当失活NADP+-Icd时胞内NADH水平和NADH/NAD+比例显著下降,而将NADP+-Icd替换成NAD+-Icd可显著提高胞内NADH水平和NADH/NAD+比例(表 3)。
综上所述,重组菌株Lys-χ ΔZMICg和Lys-χ1胞内都不积累NADPH,都是NADPH营养缺陷型C. glutamicum重组菌株。考虑到重组菌株Lys-χ1在菌体生长性能和L-赖氨酸发酵水平都要优于Lys-χ ΔZMICg,故本实验选择重组菌株Lys-χ1作为本研究的NADPH营养缺陷型C. glutamicum重组菌株并进行下一步研究。
2.4 不同强度启动子控制PntAB调节NADPH营养缺陷型C. glutamicum Lys-χ1中胞内NADPH的补给从图 4可知,NADPH营养缺陷型C. glutamicum Lys-χ1的菌株生长和L-赖氨酸合成都显著受到抑制,本研究推测可能是因为重组菌Lys-χ1胞内无法合成NADPH而破坏了菌体的正常代谢功能。为了证实上述推测,本实验通过采用不同强度启动子来控制来源于E. coli的PntAB在重组菌Lys-χ1胞内的表达水平,并分析不同重组菌株中胞内NADP(H/+)和NAD(H/+)水平。根据文献报道,本实验选择了32种不同强度的启动子(表 2)。为了进一步考查所选择的32种启动子在C. glutamicum中的表达强度,本实验将构建好的重组表达质粒PX-pntAB-gfp电转至重组菌Lys-χ1中,通过分析不同重组菌株中绿色荧光强度来比较不同启动子的活力。根据相对荧光强度,32种启动子分成5类,即超弱启动子(5种)、弱启动子(5种)、中等启动子(7种)、强启动子(9种)和超强启动子(6种)(图 5)。

|
| 图 5 不同启动子的转录强度分析 Figure 5 Promoter strength analysis of different pormoters by flow cytometry. |
由于PntAB和GFP都是在同一启动子下控制表达,因此理论上PntAB在不同启动子下也具有不同的转录水平,从而调控胞内不同的NADPH水平。为此,本实验分析了不同重组菌株中胞内NADP(H/+)和NAD(H/+)水平。从表 4可知,除了携带有启动子PdapA-B6和PdapA-C2控制的表达质粒外,携带有其余30种启动子控制的表达质粒的重组菌株都能恢复胞内NADPH的积累,同时改变胞内NAD(H/+)水平。此外,启动子的强弱、荧光强度和胞内NADPH水平三者呈线性关系,即不同强度的启动子控制的表达质粒因PntAB表达水平不同,重组菌株中胞内NADPH积累水平也不同,具体表现为携带有超强启动子控制的表达质粒的重组菌株胞内NADPH水平最高[如重组菌Lys-χ1/PtacM-pntAB-gfp胞内NADPH水平为19.34×10–4 nmol/(104细胞)],而携带有超弱启动子控制的表达质粒的重组菌株胞内NADPH水平最低[如重组菌Lys-χ1/PdapA-B27-pntAB-gfp胞内NADPH水平为0.28×10–4 nmol/(104细胞)](表 4)。需要指出的是,伴随着重组菌胞内NADPH水平的升高,胞内NADH水平则逐渐降低(表 4)。
| C. glutamicum | NADPH b | NADP+ b | NADH b | NAD+ b | Cell weight c | L-lysine c |
| Lys-χ1 | ND | 1.25 | 2.93 | 5.66 | 0.3±0.1 | ND |
| Lys-χ1/PdapA-B6-pntAB-gfp | ND | 1.32 | 2.92 | 5.62 | 0.5±0.2 | ND |
| Lys-χ1/PdapA-B27-pntAB-gfp | 0.28 | 1.41 | 2.84 | 5.68 | 4±0.2 | ND |
| Lys-χ1/PdapA-B31-pntAB-gfp | 0.32 | 1.54 | 2.81 | 5.68 | 4.5±0.1 | ND |
| Lys-χ1/PdapA-C2-pntAB-gfp | ND | 1.28 | 2.93 | 5.77 | 0.6±0.2 | ND |
| Lys-χ1/PdapA-C13-pntAB-gfp | 0.35 | 1.56 | 2.79 | 5.69 | 4.7±0.5 | ND |
| Lys-χ1/PdapA-P4-pntAB-gfp | 0.75 | 1.70 | 2.69 | 6.03 | 7.4±0.4 | 0.3±0.3 |
| Lys-χ1/PdapAR2-pntAB-gfp | 0.58 | 1.63 | 2.74 | 5.82 | 7.3±0.5 | ND |
| Lys-χ1/PgapB-pntAB-gfp | 0.83 | 1.49 | 2.66 | 6.05 | 6.7±0.8 | 0.5±0.2 |
| Lys-χ1/Ppck-pntAB-gfp | 1.02 | 1.45 | 2.53 | 6.3 | 7.9±0.5 | 1.8±0.1 |
| Lys-χ1/PmalE-pntAB-gfp | 0.93 | 1.55 | 2.6 | 6.16 | 7.7±0.3 | 1±0.3 |
| Lys-χ1/PdapA-pntAB-gfp | 1.88 | 2.35 | 2.43 | 6.45 | 9.9±1.0 | 8.5±0.5 |
| Lys-χ1/PgapA-pntAB-gfp | 4.34 | 3.81 | 1.98 | 7.26 | 9.8±0.7 | 20.4±1.7 |
| Lys-χ1/Ppgk-pntAB-gfp | 3.22 | 3.32 | 2.29 | 6.86 | 10.2±0.6 | 18.8±0.9 |
| Lys-χ1/Pgpm-pntAB-gfp | 3.61 | 3.28 | 2.08 | 7.07 | 9.8±1.1 | 19.8±1.5 |
| Lys-χ1/Peno-pntAB-gfp | 3.46 | 3.26 | 2.17 | 6.93 | 10.0±0.8 | 19.2±1.4 |
| Lys-χ1/Ppfk-pntAB-gfp | 3.29 | 3.27 | 2.22 | 6.62 | 9.9±0.3 | 19.0±2.3 |
| Lys-χ1/PPF104-pntAB-gfp | 2.45 | 2.72 | 2.34 | 6.58 | 9.5±1.2 | 13.2±1.5 |
| Lys-χ1/PdapA-A16-pntAB-gfp | 5.81 | 4.61 | 1.65 | 7.51 | 9.3±0.9 | 19.7±1.8 |
| Lys-χ1/PdapA-A45-pntAB-gfp | 4.93 | 4.04 | 1.91 | 7.32 | 9.8±1.6 | 20.7±0.3 |
| Lys-χ1/PdapA-C20-pntAB-gfp | 5.29 | 4.23 | 1.68 | 7.43 | 10.2±1.3 | 20.0±1.4 |
| Lys-χ1/Pgro-pntAB-gfp | 6.35 | 4.93 | 1.54 | 7.8 | 9.1±0.4 | 17.9±2.0 |
| Lys-χ1/Plac-pntAB-gfp | 5.16 | 4.20 | 1.78 | 7.52 | 9.8±1.3 | 20.4±2.1 |
| Lys-χ1/Ptrc-pntAB-gfp | 6.09 | 4.80 | 1.49 | 7.47 | 9.3±0.6 | 19.0±1.4 |
| Lys-χ1/Ptrp-pntAB-gfp | 5.30 | 4.23 | 1.66 | 7.43 | 9.8±0.4 | 20.2±1.7 |
| Lys-χ1/Ptac-pntAB-gfp | 5.87 | 4.59 | 1.59 | 7.48 | 9.9±1.0 | 19.7±1.3 |
| Lys-χ1/Pkan-pntAB-gfp | 7.68 | 5.73 | 1.39 | 7.98 | 9.1±0.7 | 15.6±1.2 |
| Lys-χ1/PlacM-pntAB-gfp | 15.68 | 7.16 | 0.83 | 8.27 | 7.5±0.8 | 5.9±0.6 |
| Lys-χ1/PtacM-pntAB-gfp | 19.34 | 7.61 | 0.57 | 8.43 | 5.4±0.4 | 2.4±0.7 |
| Lys-χ1/Psod-pntAB-gfp | 10.03 | 6.01 | 1.21 | 9.31 | 8.7±0.5 | 13.7±0.9 |
| Lys-χ1/Ptuf-pntAB-gfp | 13.42 | 6.74 | 1.03 | 8.76 | 8.4±0.8 | 9.3±0.6 |
| Lys-χ1/PH30-pntAB-gfp | 11.17 | 6.38 | 1.19 | 9.89 | 8.5±0.3 | 12±1.6 |
| Lys-χ1/PH36-pntAB-gfp | 15.10 | 6.93 | 0.89 | 8.22 | 7.8±0.9 | 6.2±0.7 |
| a: the strains list in this table were not ranked according to the level of promoters; b: the unit is 10–4 nmol/(104 cell); c: the unit is g/L; ND: not detected. | ||||||
2.5 NADPH营养缺陷型C. glutamicum Lys-χ1中异源表达PntAB可恢复胞内L-赖氨酸积累
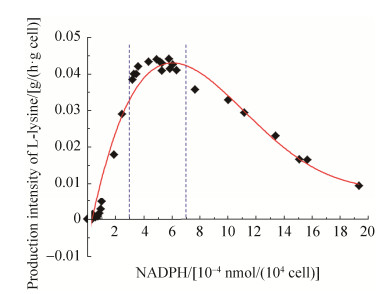
由表 4可知,NADPH营养缺陷型C. glutamicum Lys-χ1胞内NADP(H/+)和NAD(H/+)水平可通过不同强度启动子控制PntAB的表达来调节。前面研究发现,当失活胞内Zwf、MalE和Icd时,因胞内NADPH水平显著降低,从而会显著影响菌体生长和L-赖氨酸合成(图 4)。为此,本实验分析了通过不同强度启动子控制PntAB在NADPH营养缺陷型C. glutamicum Lys-χ1中的表达,考察胞内NADPH水平的变化对菌体生长和L-赖氨酸合成的影响。需要指出的是,尽管重组菌Lys-χ1/PtacM-pntAB-gfp的细胞内NADPH水平最高,但最终菌体量和L-赖氨酸产量并不是最高(表 4)。通过分析不同胞内NADPH浓度下重组菌株L-赖氨酸生产强度发现,菌体量和L-赖氨酸产量在胞内NADPH水平一定范围内会随着NADPH浓度的增加而增加,但是当超过NADPH阈值时,会随着胞内NADPH浓度的增加而降低(图 6)。从图 6中可以得知,当胞内NADPH浓度在(3–7)×10–4 nmol/(104 cell)时,细胞处于最大L-赖氨酸生产强度。结合表 4和图 5的结果可以发现,当以强启动子控制PntAB在NADPH营养缺陷型C. glutamicum Lys-χ1中的表达时,L-赖氨酸生产强度最大,其中以启动子PdapA-A16控制PntAB表达时L-赖氨酸生产强度最高(即重组菌Lys-χ1/PdapA-A16- pntAB-gfp),达到0.044 g/(h·g cell)。然而,当以弱启动子或超强启动子控制PntAB在重组菌Lys-χ1中的表达时,L-赖氨酸生产强度为最大值的1%–75%。例如,以弱启动子PdapA-R2控制PntAB表达的重组菌Lys-χ1/PdapA-R2-pntAB-gfp的L-赖氨酸生产强度为最大值的~1.3%,而超强启动子Psod控制PntAB表达的重组菌Lys-χ1/Psod- pntAB-gfp的L-赖氨酸生产强度为最大值的~74.3%。
|
| 图 6 不同强度启动子控制NADPH水平对L-赖氨酸生产强度的影响 Figure 6 The effect of intracellular NADPH level controlled by promoters with different transcriptional activity on production intensity of L-lysine. |
3 讨论
本研究首次以L-赖氨酸高产菌株C. glutamicum Lys-χ为出发菌株构建了1株NADPH营养缺陷型重组菌株C. glutamicum Lys-χ1。重组菌Lys-χ1可作为底盘细胞,用于比较不同的NADPH再生策略,获得不同胞内NADPH水平的重组菌株,进而实现精确调控胞内NADPH水平,为进一步阐明NADPH调控微生物细胞生理代谢功能的机制提供研究基础。
在C. glutamicum中,PP途径是NADPH的主要合成途径,其中Zwf和Gnd参与催化NADPH的合成[1, 13]。为了构建NADPH营养缺陷型菌株,本研究首先选择失活Zwf,获得重组菌株C. glutamicum Lys-χ ΔZ。研究发现,失活Zwf会显著降低L-赖氨酸的合成(图 4)。众多研究指出,合成1 mol的L-赖氨酸需要消耗4 mol NADPH[1, 5, 13-14]。由于PP途径被阻断,胞内NADPH水平不能满足L-赖氨酸的需求,因此重组菌Lys-χ ΔZ合成L-赖氨酸的能力显著降低。需要指出的是,Zwf的失活也会影响菌体的生长,这结果与Lindner等报道的不一致。Lindner等发现,失活E. coli中Zwf不影响菌体生长[26]。在E. coli中存在PntAB或UdhA组成的转氢酶循环途径,该途径可以调控NAD(H/+)与NADP(H/+)的相互转化水平,从而实现调节胞内NADPH水平[16]。然而,C. glutamicum中没有转氢酶循环途径,不能将胞内富余的NADH转化成NADPH,从而导致胞内NADPH匮乏,进而影响菌体生长[17]。当同时失活C. glutamicum中Zwf、MalE和IcdCg时,胞内检测不到NADPH的积累,同时菌体生长和L-赖氨酸合成都受到显著抑制(表 3)。另外,将重组菌Lys-χ ΔZM中自身的NADP-IcdCg替换成NAD-IcdSm时(即重组菌Lys-χ1),重组菌株中也检测不到胞内NADPH的积累(表 3)。这些结果表明,C. glutamicum中参与NADPH合成的酶为Zwf、MalE和IcdCg,改造这些酶会阻断胞内NADPH的积累。值得指出的是,当以葡萄糖酸为底物时,重组菌株中菌体生长和L-赖氨酸合成性能都有明显的改善,而以其他有机酸(如丙酮酸、α-酮戊二酸或草酰乙酸)为底物时则没有明显影响(图 4)。其原因可能是,葡萄糖酸在磷酸化酶的作用下形成6-磷酸葡萄糖酸,进而通过Gnd催化形成核酮糖-5-磷酸和NADPH,从而满足菌体生长和L-赖氨酸合成对NADPH的需求[26]。
PntAB催化NADH形成NADPH而不直接涉及碳代谢途径,从而调节胞内NADP(H/+)和NAD(H/+)水平[16]。研究发现,在NADPH营养缺陷型重组菌Lys-χ1中异源表达PntAB可以恢复重组菌中胞内NADPH的积累,并在一定程度上恢复菌体生长和L-赖氨酸合成(表 4)。此外,本研究发现在C. glutamicum中具有不同强度的启动子控制PntAB表达水平不同,使得重组菌株中胞内NADPH积累水平也不同。当携带有超强启动子控制的表达质粒的重组菌株胞内NADPH水平最高[如重组菌Lys-χ1/PtacM-pntAB-gfp胞内NADPH水平为19.34×10–4 nmol/(104细胞)],而携带有超弱启动子控制的表达质粒的重组菌株胞内NADPH水平最低[如重组菌Lys-χ1/PdapA-B27-pntAB-gfp胞内NADPH水平为0.28×10–4 nmol/(104细胞)](表 4)。然而,高细胞内NADPH水平,并不代表高菌体量和L-赖氨酸产量。在一定范围内胞内NADPH水平下,菌体量和L-赖氨酸产量会随着NADPH浓度的增加而增加,但是当超过NADPH阈值时,会随着胞内NADPH浓度的增加而降低(图 6)。这一结果表明,胞内过量的NADPH会抑制菌体生长和L-赖氨酸的合成,其原因可能是胞内过量的NADPH打破了细胞内氧化还原平衡[4, 27]。综上所述,本研究最终获得的NADPH营养缺陷型重组菌Lys-χ1可以作为底盘细胞,通过不同强度的启动子控制外源PntAB的表达水平来控制重组菌胞内NADPH水平,从而获得不同胞内NADPH水平的重组菌株,为进一步阐明NADPH调控微生物细胞生理代谢功能的机制提供研究基础。此外,有研究报道,利用E. coli的NADPH营养缺陷型菌株可以筛选具有高效且专一性的NADPH合成酶[28]。因此,NADPH营养缺陷型重组菌株C. glutamicum Lys-χ1在研究NADPH再生策略和对微生物细胞生理代谢功能的影响中具有很大优势。
| [1] | Xu JZ, Yang HK, Zhang WG. NADPH metabolism: a survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Critical Reviews in Biotechnology, 2018, 38(7): 1061-1076. DOI:10.1080/07388551.2018.1437387 |
| [2] | Qi HS, Li SS, Zhao SM, Huang D, Xia ML, Wen JP. Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis. PLoS ONE, 2014, 9(4): e93815. DOI:10.1371/journal.pone.0093815 |
| [3] | Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MMY, Arnold FH. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metabolic Engineering, 2011, 13(3): 345-352. DOI:10.1016/j.ymben.2011.02.004 |
| [4] | Bartek T, Blombach B, Zönnchen E, Makus P, Lang S, Eikmanns BJ, Oldiges M. Importance of NADPH supply for improved L-valine formation in Corynebacterium glutamicum. Biotechnology Progress, 2010, 26(2): 361-371. |
| [5] | Xu JZ, Han M, Zhang JL, Guo YF, Zhang WG. Metabolic engineering Corynebacterium glutamicum for the L-lysine production by increasing the flux into L-lysine biosynthetic pathway. Amino Acids, 2014, 46(9): 2165-2175. DOI:10.1007/s00726-014-1768-1 |
| [6] | Xu JZ, Ruan HZ, Yu HB, Liu LM, Zhang WG. Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production from mixed sugar. Microbial Cell Factories, 2020, 19(1): 39. DOI:10.1186/s12934-020-1294-7 |
| [7] | Xu Q, Xu X, Huang H, Li S. Efficient synthesis of (R)-2-chloro-1-phenylethol using a yeast carbonyl reductase with broad substrate spectrum and 2-propanol as cosubstrate. Biochemical Engineering Journal, 2015, 103: 277-285. DOI:10.1016/j.bej.2015.08.009 |
| [8] | Chen Y, Liu QG, Chen XC, Wu JL, Guo T, Zhu CJ, Ying HJ. Redirecting metabolic flux in Saccharomyces cerevisiae through regulation of cofactors in UMP production. Journal of Industrial Microbiology & Biotechnology, 2015, 42(4): 577-583. DOI:10.1007/s10295-014-1536-y |
| [9] | Soga N, Kinosita K Jr, Yoshida M Jr, Suzuki T Jr. Kinetic equivalence of transmembrane pH and electrical potential differences in ATP synthesis. Journal of Biological Chemistry, 2012, 287(12): 9633-9639. DOI:10.1074/jbc.M111.335356 |
| [10] | Kang SW, Lee S, Lee EK. ROS and energy metabolism in cancer cells: alliance for fast growth. Archives of Pharmacal Research, 2015, 38(3): 338-345. DOI:10.1007/s12272-015-0550-6 |
| [11] | Li N, Zhang YY, Ye Q, Zhang YZ, Chen Y, Chen XC, Wu JL, Bai JX, Xie JJ, Ying HJ. Effect of ribose, xylose, aspartic acid, glutamine and nicotinic acid on ethyl (S)-4-chloro-3-hydroxybutanoate synthesis by recombinant Escherichia coli. Bioresource Technology, 2012, 118: 572-575. DOI:10.1016/j.biortech.2012.02.102 |
| [12] | Singh R, Mailloux RJ, Puiseux-Dao S, Appanna VD. Oxidative stress evokes a metabolic adaptation that favors increased NADPH synthesis and decreased NADH production in Pseudomonas fluorescens. Journal of Bacteriology, 2007, 189(18): 6665-6675. DOI:10.1128/JB.00555-07 |
| [13] | Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C. Metabolic flux engineering of L-lysine production in Corynebacterium glutamicum-over expression and modification of G6P dehydrogenase. Journal of Biotechnology, 2007, 132(2): 99-109. DOI:10.1016/j.jbiotec.2007.05.026 |
| [14] | Martínez I, Zhu JF, Lin H, Bennett GN, San KY. Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metabolic Engineering, 2008, 10(6): 352-359. DOI:10.1016/j.ymben.2008.09.001 |
| [15] | Kind S, Becker J, Wittmann C. Increased lysine production by flux coupling of the tricarboxylic acid cycle and the lysine biosynthetic pathway-metabolic engineering of the availability of succinyl-CoA in Corynebacterium glutamicum. Metabolic Engineering, 2013, 15: 184-195. DOI:10.1016/j.ymben.2012.07.005 |
| [16] | Chou HH, Marx CJ, Sauer U. Transhydrogenase promotes the robustness and evolvability of E. coli deficient in NADPH production. PLoS Genetics, 2015, 11(2): e1005007. DOI:10.1371/journal.pgen.1005007 |
| [17] | Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Applied and Environmental Microbiology, 2011, 77(10): 3300-3310. DOI:10.1128/AEM.02972-10 |
| [18] |
Yang HK, Xu JZ, Zhang WG. Blocking NADPH biosynthesis affected growth and metabolites formation of Corynebacterium glutamicum. Food and Fermentation Industries, 2019, 45(10): 1-9.
(in Chinese) 杨汉昆, 徐建中, 张伟国. 阻断辅因子NADPH合成对谷氨酸棒杆菌生长及产物合成的影响. 食品与发酵工业, 2019, 45(10): 1-9. |
| [19] | 徐大庆. 黄色短杆菌载体系统的构建及其产L-缬氨酸代谢工程育种的初步研究. 江南大学博士学位论文, 2010. |
| [20] | Jiang Y, Qian FH, Yang JJ, Liu YM, Dong F, Xu CM, Sun BB, Chen B, Xu XS, Li Y, Wang RX, Yang S. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nature Communications, 2017, 8: 15179. DOI:10.1038/ncomms15179 |
| [21] | 叶菁. 木糖异构酶基因xylA在谷氨酸棒杆菌中的克隆与表达研究. 华中科技大学博士学位论文, 2013. |
| [22] | Pátek M, Nešvera J. Promoters and plasmid vectors of Corynebacterium glutamicum. //Yukawa H, Inui M (eds). Corynebacterium glutamicum: Biology and Biotechnology. Berlin, Heidelberg: Springer-Verlag, 2013: 51-88. |
| [23] | Vašicová P, Pátek M, Nešvera J, Sahm H, Eikmanns B. Analysis of the Corynebacterium glutamicum dapA promoter. Journal of Bacteriology, 1999, 181(19): 6188-6191. DOI:10.1128/JB.181.19.6188-6191.1999 |
| [24] | Yim SS, An SJ, Kang M, Lee J, Jeong KJ. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnology and Bioengineering, 2013, 110(11): 2959-2969. DOI:10.1002/bit.24954 |
| [25] | Wang LP, Yu HB, Xu JZ, Ruan HZ, Zhang WG. Deciphering the crucial roles of AraC-type transcriptional regulator Cgl2680 on NADPH metabolism and L-lysine production in Corynebacterium glutamicum. World Journal of Microbiology and Biotechnology, 2020, 36(6): 1-15. DOI:10.1007/s11274-020-02861-y |
| [26] | Lindner SN, Ramirez LC, Krüsemann JL, Yishai O, Belkhelfa S, He H, Bouzon M, Döring V, Bar-Even A. NADPH-auxotrophic E. coli: a sensor strain for testing in vivo regeneration of NADPH. ACS Synthetic Biology, 2018, 7(12): 2742-2749. DOI:10.1021/acssynbio.8b00313 |
| [27] | Xu JZ, Ruan HZ, Chen XL, Zhang F, Zhang WG. Equilibrium of the intracellular redox state for improving cell growth and L-lysine yield of Corynebacterium glutamicum by optimal cofactor swapping. Microbial Cell Factories, 2019, 18(1): 65. DOI:10.1186/s12934-019-1114-0 |
| [28] | Calzadiaz-Ramirez L, Calvó-Tusell C, Stoffel GMM, Lindner SN, Osuna S, Erb TJ, Garcia-Borràs M, Bar-Even A, Acevedo-Rocha CG. In vivo selection for formate dehydrogenases with high efficiency and specificity toward NADP+. ACS Catalysis, 2020, 10(14): 7512-7525. DOI:10.1021/acscatal.0c01487 |
 2021, Vol. 61
2021, Vol. 61