中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王冰, 吴红霞, 仇华吉, 孙元. 2021
- Bing Wang, Hongxia Wu, Hua-Ji Qiu, Yuan Sun. 2021
- 病毒工厂:病毒复制的关键场所
- Viral factory: a critical site for virus replication
- 微生物学报, 61(7): 1873-1881
- Acta Microbiologica Sinica, 61(7): 1873-1881
-
文章历史
- 收稿日期:2020-08-09
- 修回日期:2020-10-20
- 网络出版日期:2021-02-10
病毒是专性的胞内寄生生物,必须依靠细胞才能够进行复制。病毒在复制过程中,其基因组会发生突变,能够在感染细胞中产生新的基因组序列。大多数病毒的突变是致死性的,但也会产生一些新的病毒与细胞因子的相互作用,并可能产生新的复制方式[1]。因此,病毒在持续不断进化的过程中,可能会形成病毒工厂(viral factories, VFs)组装信号。VFs可在空间上促进协调病毒基因组的复制和组装,最大限度地提高细胞资源的使用效率,还可以为病毒提供逃避宿主细胞抗病毒天然免疫应答的场所[2–4]。虽然一些病毒可在细胞核内建立VFs[5–7],但目前研究较为充分的是在细胞质中的VFs。VFs具有2个重要特征:招募复制结构周围的病毒和细胞组件;大多数VFs招募和使用线粒体。随着基因组学、蛋白质组学和显微技术的进步,研究者已经在VFs形成研究方面取得了一定进展。本综述将重点介绍VFs的结构,以及参与其形成、内部大分子运输的关键细胞因子。
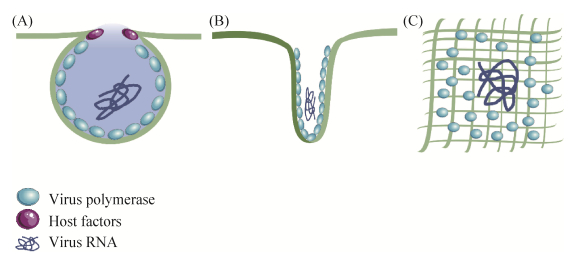
1 复制复合物的形成和结构在感染早期,病毒聚合酶及辅助因子与细胞膜相互作用,形成功能性复制复合物(replication complexes,RCs)[8]。这些复合体形成的病毒基因组具有多个拷贝,并将其转移到组装位点。病毒能够在细胞内膜和细胞质膜组装RCs,但后者较为少见。甲病毒(Alphavirus)的基因组为RNA,为披膜病毒科(Togaviridae)成员[9],该病毒能够利用质膜的胞质面组装RCs[9–12],随后,这些复合体被内化并运输至溶酶体,并在其中继续复制。披膜病毒的复制酶必须通过质膜来组装RCs,但目前该机制尚不清楚。痘苗病毒(vaccinia virus,VV)是痘病毒科(Poxviridae)的代表成员[13],该病毒能够与粗面内质网(rough endoplasmic reticulum,RER)囊池形成“微核”[14]。除上述情况外,大多数在胞浆中复制的病毒会形成3类膜结构:单层膜球体结构、管状立方形膜结构和由寡聚物形成的平面排列结构(图 1)。
|
| 图 1 病毒在细胞质复制形成的3种膜结构模式图 Figure 1 Models of membranous structures formed by viruses that replicated in the cytoplasm. A: spherules; B: cubic membranes; C: oligomeric arrays. |
球体结构由病毒非结构蛋白、病毒RNA和细胞辅助因子组成,为单层膜结构,通向胞浆的开口较小。该类球体由线粒体、内质网、溶酶体、过氧化物酶体、高尔基体或叶绿体等各种细胞器中的内陷形成[2, 15–16]。由于颈状连接限制了多余分子进入,所以能够保护新合成的病毒基因组不被降解,同时也控制了病毒基因组的输出。典型的球体是由芜菁黄花叶病毒(turnip yellow mosaic virus,TYMV)在叶绿体[17–18]、雀麦花叶病毒(brome mosaic virus,BMV)在内质网衍生的膜[19–20],以及塞姆利基森林病毒(semliki forest virus)和弗洛克豪斯病毒(flock house virus,FHV)在线粒体中组装的结构[21–23]。
布尼亚病毒(bunyamwera virus,BUNV)在高尔基体膜上组装非典型小球结构。BUNV是一种负链RNA病毒,是布尼亚病毒科(Bunyaviridae)的代表成员[24–26]。BUNV球体通常附着在圆柱形结构上[2];这些管状分子的开口通向细胞质,并随时间生长。研究表明,虽然球体是病毒基因组复制的场所,但圆柱体能够存储和保护复制的病毒RNA,同时等待运输到附近高尔基膜中的组装位点[27]。
膜的微管泡结构(membranous tubulovesicular structure,TBS)也经常存在于病毒感染的细胞中[2]。这种膜结构很可能起源于内质网,由病毒、RNA和DNA诱导形成。它们由高度弯曲的折叠脂质双层膜组成,为病毒粒子的复制及组装搭建了一个平台。研究表明,TBS间的连接通道可以促进分子从病毒RC到细胞质或细胞核的运输;外表面的孔隙能够控制物质进入,TBS高度弯曲的特性可能有助于病毒的出芽[28]。在感染严重急性呼吸综合征冠状病毒(severe acute respiratory syndrome coronavirus,SARS-CoV)的细胞中,可以观察到膜及双膜小泡(double membrane vesicles,DMV)与新生病毒粒子接触[29–30]。最初认为DMV是潜在的病毒复制部位[30],但通过结构分析,没有发现DMV与细胞质相连通[31]。此外,病毒复制酶和RNA复制中间产物——双链RNA的超微结构分析表明,TBS是病毒基因组复制的位点,而DMV可能具有其他作用,如存储复制的病毒RNA并使其逃避宿主免疫应答[30–32]。对于多种正链RNA病毒,如丙型肝炎病毒(hepatitis C virus,HCV)和脊髓灰质炎病毒(poliovirus)而言,DMV是其复制位点[33]。
登革热病毒(dengue virus,DENV)为黄病毒科(Flaviviridae)成员,是一种非常重要的人类病原体[34],它能够组装一个由内质网衍生的带有球状颗粒的网络[35–36]。这些复杂网络的结构分析结果表明,由ER衍生的含有RC的球体和附近ER膜中的组装位点之间存在联系[35]。
2 细胞器招募病毒聚合酶的表达和RCs的组装通常能够募集细胞器[37]。一些病毒会建立大型VFs[2],这些结构的直径为几微米[38]。在感染早期,BUNV能够组建VFs,该结构招募高尔基体、RER囊池和线粒体并能够改变自身形状、大小和位置,且进一步转变为圆形的膜网络。带有非典型球体的RC结构将高尔基体与RER[2]、新生病毒和线粒体相连[38]。因此,这些带有RC的“病毒管”也充当了工厂细胞器之间的物理连接,可能促进了物质交换。
某些病毒不是通过单个大型工厂进行复制,而是通过在局部募集细胞器和细胞因子来构建多个微型工厂,从而合成子代病毒粒子。Alphavirus和RUBV属于该种情况,它们均是通过修饰的溶酶体,即病毒复制细胞器进行复制[39–40]。RUBV复制细胞器周围存在大量细胞器,例如线粒体和RER[27]。尽管细胞器处于复制细胞器的外周,但它仍与细胞质进行物质信息交流[27]。RER通常围绕在复制细胞器外周,并且在这两个区室之间存在恒定的物质交换:RER将新合成的病毒蛋白运送至复制细胞器,而后者则将病毒基因组和亚基因组RNA转移到RER以继续蛋白合成[41]。不仅是RER,线粒体、RUBV装配细胞器和高尔基体也是如此[27]。
如上所述,多种VFs均能招募线粒体,它们可能是为病毒组装提供能量[42],且在某些情况下,它们还可以充当复制细胞器或用于病毒装配[42–46]。除此之外,线粒体可以为病毒复制提供其他宿主复制因子。p32是一种线粒体基质蛋白,能够参与多种凋亡途径[47–48],在感染RUBV和转染RUBV复制子的细胞中,该蛋白离开线粒体并存在于复制细胞器中[37]。研究者在RUBV复制细胞器的内膜中检测到了p32,还发现了病毒聚合酶和dsRNA分子[27]。p32参与线粒体募集并与RUBV衣壳结合[49],这种相互作用能够促进病毒复制,虽然机制未知,但研究者推测可能与RUBV亚基因组RNA的转录有关[50–51]。除RUBV外,p32还与其他病毒相互作用,例如人类免疫缺陷病毒1型(human immunodeficiency virus type 1,HIV-1)、单纯疱疹病毒(herpes simplex virus,HSV)、腺病毒、爱泼斯坦-巴尔病毒(epstein-barr virus)和丙肝病毒(hepatitis C virus,HCV),这种相互作用有利于病毒复制[52]。线粒体蛋白对番茄丛矮化病毒(tombusvirus)的复制具有重要作用,这些蛋白质包括MSP1 ATPase和MAM33,后者参与氧化磷酸化并与p33病毒复制酶相互作用;内部线粒体膜蛋白MDM38与病毒RNA结合并促进病毒复制;TUM1蛋白同样能够与p33相互作用[15]。
鉴定感染细胞中线粒体募集和修饰的因素可为解析VFs的形成提供重要线索。迄今为止,唯一确定的与VFs形成有关的信号传导途径是聚集体形成途径[2, 53]。错误折叠的蛋白质在细胞内聚集时,胞内会形成异常蛋白聚集体[54]。VFs和聚集体均位于微管组织中心(microtubular organizing center,MTOC),能够募集伴侣蛋白和线粒体,并构建由波形蛋白细丝形成的笼状结构[3]。疱疹病毒(herpes virus)、腺病毒和猿猴病毒40 (Simian vacuolating virus 40,SV40)可能利用早幼粒细胞白血病核小体(promyelocytic leukemia nuclear bodies, PML-NB),即一种与异常蛋白聚集体类似的结构,可作为组装核工厂的场所[55]。近期研究表明,其他多种胞内物质参与了多瘤病毒(polyomavirus)核工厂的形成[56]。病毒可能通过劫持聚集体形成途径以聚集病毒复制和形态发生所需的众多元件,并避免被细胞免疫系统所识别;或者,病毒大分子复合物可能被细胞误认为是错误折叠的蛋白质,从而引发聚集体的形成过程[3]。
3 VFs在病毒复制到形态发生过程中的结构变化VFs具有动态结构,能够不断变化以适应病毒需求,但目前仍然未知病毒如何协调这些变化。本部分将以BUNV、痘病毒(poxvirus)和巨型病毒(mimivirus)为例,对VFs动态变化过程进行叙述。BUNV在复制时能够建立紧凑的圆形结构,该结构在感染后期发生显著改变——线粒体和RER将远离核周区域,且带有病毒后代的分泌小泡向细胞表面移动[38],这和新生病毒颗粒的组装及成熟过程相吻合。而在VV感染过程中,其VFs会发生数次形态变化[2]。在病毒基因组复制过程中,VV首先组装带有附着线粒体的细胞质微核[14];随后形成一个类似聚集体的结构[57],其中未成熟的病毒利用非典型膜重塑机制进行组装[58]。随后,病毒在高尔基体完成最终的成熟过程和囊膜形成过程[59]。
与痘病毒类似,巨型病毒在感染细胞的胞质中完成整个病毒复制周期[60]。病毒感染细胞4 h后,即能观察到巨型病毒形成的VFs,它们是由病毒核心——能够将病毒DNA释放到细胞质中——诱导形成[60]。在感染后期,VFs融合形成外观与之前完全不同的单个结构,病毒在其中开始组装,并且可以观察到处于各个组装阶段的病毒粒子[61]。病毒感染细胞8 h后,VFs中存在成熟及未成熟病毒粒子,但病毒感染细胞10 h后,仅有成熟病毒粒子[62]。巨型病毒由DNA、病毒核心、衣壳和囊膜构成。工厂外不存在膜结构,但是由于成熟的病毒粒子具有囊膜,所以子代病毒粒子的组装也可能需要招募细胞膜结构[63]。
4 大分子复合物的移动促进病毒在VFs的组装在VFs内部,为了能将病毒基因组复制与成熟病毒粒子的组装和释放联系起来,必须能有效运输细胞和病毒大分子。因此,病毒需要控制细胞的运输系统。病毒可利用细胞骨架和相关的运动蛋白以及宿主膜系统来进行细胞内移动[64–66]。植物病毒能够编码运动蛋白(movement protein,MP),该蛋白通过植物胞间连丝使病毒基因组在细胞间运输[67]。除MP外,其他病毒蛋白也参与大分子在植物细胞内的运动,包括某些在病毒复制中起作用的蛋白[68]。例如,TMV的RCs定位于微丝并沿微丝运输[69–70]。病毒沿肌动蛋白丝运动时需要肌球蛋白——与微丝相关的分子马达的参与。另外,动物病毒还能利用囊泡进行运输[65, 71–72],如非洲猪瘟病毒(African swine fever virus,ASFV)。在BUNV的复制细胞器中已经检测到肌动蛋白和肌球蛋白[38]。用抑制上述蛋白质的药物处理细胞后的结果表明,肌动蛋白-肌球蛋白复合物介导了病毒基因组从RC运出[38]。
目前,细胞内病毒成分运输的许多问题仍然未知,例如,复制的基因组如何与复制细胞器中的病毒聚合酶分离,病毒如何在不同的细胞内区室中移动,或者放线菌素网络是否介导细胞内膜中病毒复合物的短距离运动。
5 结语和展望VFs是病毒入侵细胞后,在二者相互作用下产生的一种特殊结构。VFs是病毒完成复制的场所[2],且具有协助病毒逃避宿主天然免疫的功能,但其在形态方面存在较大差异,且结构也不尽相同,有些病毒还能在细胞内特定位点形成与其自身复制相关的特征性结构,如单股负链RNA病毒及狂犬病病毒(rabies virus,RV)在细胞内形成的内基氏小体(negri bodies,NB)[73],它不具有膜结构但是仍能通过聚集病毒蛋白来促进其复制。
病毒不仅能够通过调节宿主细胞周期的促进自身复制[4, 74],还能募集宿主细胞器并对其进行改造从而促进子代病毒的合成[14]。但募集及改造过程十分复杂,并涉及多种类型的相互作用,包括病毒核酸与宿主蛋白、病毒蛋白与宿主蛋白及病毒蛋白与宿主膜结构间的相互作用等,且这些相互作用处于动态变化之中,在病毒入侵细胞后的特定时间和部位产生,且作用时间短暂。毫无疑问,这种时空特异性及短暂的分子间相互作用将对解析VFs形成机制造成困难。因此,即使目前蛋白组学、基因组学等技术及分子间相互作用技术研究已经有了长足的进步,但仍难解析VFs的形成机制。
近年来,随着高分辨率电子显微镜的发展,针对VFs的形态学研究已经有了长足的进步,且通过对病毒不同复制阶段的VFs进行观察,研究者可以直观地了解到VFs的动态变化过程及不同成分的空间分布情况。但是,VFs不同组分在空间上的靠近或接触仍不能充分说明它们之间存在相互作用,还需要更为直接的证据对其进行验证。因此,仅依靠该技术仍难以解析VFs形成过程及机制。
由于VFs的形成需要经过复杂的生物学过程,因此单个研究人员或研究团队很难对其完整的形成机制进行全面解析,尤其是在科学研究日趋细化的今天。解析VFs形成过程需要不同领域间的相互合作及学科交叉。了解VFs形成过程不仅有助于加深人们对病毒复制的理解,其应用价值也不言而喻,是否可以通过阻断VFs形成而抑制病毒复制并研发新型抗病毒制剂?这无疑是一个可行且有应用前景的方向。
| [1] | Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiology and Molecular Biology Reviews, 2012, 76(2): 159-216. DOI:10.1128/MMBR.05023-11 |
| [2] | Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biology of the Cell, 2005, 97(2): 147-172. DOI:10.1042/BC20040058 |
| [3] | Netherton CL, Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Current Opinion in Virology, 2011, 1(5): 381-387. DOI:10.1016/j.coviro.2011.09.008 |
| [4] | Nevers Q, Albertini AA, Lagaudrière-Gesbert C, Gaudin Y. Negri bodies and other virus membrane-less replication compartments. Biochimica et Biophysica Acta, 2020, 1867(12): 118831. DOI:10.1016/j.bbamcr.2020.118831 |
| [5] | Peng L, Ryazantsev S, Sun R, Zhou ZH. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure, 2010, 18(1): 47-58. DOI:10.1016/j.str.2009.10.017 |
| [6] | Chang L, Godinez WJ, Kim IH, Tektonidis M, de Lanerolle P, Eils R, Rohr K, Knipe DM. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(21): E136-E144. DOI:10.1073/pnas.1103411108 |
| [7] | Erickson KD, Garcea RL. Viral replication centers and the DNA damage response in JC virus-infected cells. Virology, 2019, 528: 198-206. DOI:10.1016/j.virol.2018.12.014 |
| [8] | Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nature Reviews Microbiology, 2008, 6(5): 363-374. DOI:10.1038/nrmicro1890 |
| [9] | Pietilä MK, Hellström K, Ahola T. Alphavirus polymerase and RNA replication. Virus Research, 2017, 234: 44-57. DOI:10.1016/j.virusres.2017.01.007 |
| [10] | Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. Journal of Virology, 2010, 84(22): 11679-11695. DOI:10.1128/JVI.01441-10 |
| [11] | Spuul P, Balistreri G, Kääriäinen L, Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. Journal of Virology, 2010, 84(15): 7543-7557. DOI:10.1128/JVI.00477-10 |
| [12] | Risco C, Sanmartín-Conesa E, Tzeng WP, Frey TK, Seybold V, de Groot RJ. Specific, sensitive, high-resolution detection of protein molecules in eukaryotic cells using metal-tagging transmission electron microscopy. Structure, 2012, 20(5): 759-766. DOI:10.1016/j.str.2012.04.001 |
| [13] | Cantu F, Cao S, Hernandez C, Dhungel P, Spradlin J, Yang ZL. Poxvirus-encoded decapping enzymes promote selective translation of viral mRNAs. PLoS Pathogens, 2020, 16(10): e1008926. DOI:10.1371/journal.ppat.1008926 |
| [14] | Tolonen N, Doglio L, Schleich S, Krijnse Locker J. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Molecular Biology of the Cell, 2001, 12(7): 2031-2046. DOI:10.1091/mbc.12.7.2031 |
| [15] | Nagy PD, Pogany J. Chapter 4-global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Advances in Virus Research, 2010(76): 123-177. |
| [16] | Doyle N, Hawes PC, Simpson J, Adams LH, Maier HJ. The porcine deltacoronavirus replication organelle comprises double-membrane vesicles and zippered endoplasmic reticulum with double-membrane spherules. Viruses, 2019, 11(11): 1030. DOI:10.3390/v11111030 |
| [17] | Prod'homme D, Le Panse S, Drugeon G, Jupin I. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology, 2001, 281(1): 88-101. DOI:10.1006/viro.2000.0769 |
| [18] | Moriceau L, Jomat L, Bressanelli S, Alcaide-Loridan C, Jupin I. Identification and molecular characterization of the chloroplast targeting domain of turnip yellow mosaic virus replication proteins. Frontiers in Plant Science, 2017(8): 2138. |
| [19] | Schwartz M, Chen JB, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Molecular Cell, 2002, 9(3): 505-514. DOI:10.1016/S1097-2765(02)00474-4 |
| [20] | Sibert BS, Navine AK, Pennington J, Wang XF, Ahlquist P. Cowpea chlorotic mottle bromovirus replication proteins support template-selective RNA replication in Saccharomyces cerevisiae. PLoS ONE, 2018, 13(12): e0208743. DOI:10.1371/journal.pone.0208743 |
| [21] | Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO Journal, 1994, 13(18): 4204-4211. DOI:10.1002/j.1460-2075.1994.tb06740.x |
| [22] | Pietilä MK, van Hemert MJ, Ahola T. Purification of highly active alphavirus replication complexes demonstrates altered fractionation of multiple cellular membranes. Journal of Virology, 2018, 92(8): e01852-17. |
| [23] | Short JR, Speir JA, Gopal R, Pankratz LM, Lanman J, Schneemann A. Role of mitochondrial membrane spherules in flock house virus replication. Journal of Virology, 2016, 90(7): 3676-3683. DOI:10.1128/JVI.03080-15 |
| [24] | Elliott RM. Emerging viruses: the Bunyaviridae. Molecular Medicine, 1997, 3(9): 572-577. DOI:10.1007/BF03401814 |
| [25] | Walter CT, Barr JN. Recent advances in the molecular and cellular biology of Bunyaviruses. Journal of General Virology, 2011, 92(11): 2467-2484. DOI:10.1099/vir.0.035105-0 |
| [26] | Feng JJ, Wickenhagen A, Turnbull ML, Rezelj VV, Kreher F, Tilston-Lunel NL, Slack GS, Brennan B, Koudriakova E, Shaw AE, Rihn SJ, Rice CM, Bieniasz PD, Elliott RM, Shi XH, Wilson SJ. Interferon-stimulated gene (ISG)-expression screening reveals the specific antibunyaviral activity of ISG20. Journal of Virology, 2018, 92(13): e02140-17. |
| [27] | Fontana J, López-Iglesias C, Tzeng WP, Frey TK, Fernández JJ, Risco C. Three-dimensional structure of Rubella virus factories. Virology, 2010, 405(2): 579-591. DOI:10.1016/j.virol.2010.06.043 |
| [28] | Deng YR, Almsherqi ZA, Ng MML, Kohlwein SD. Do viruses subvert cholesterol homeostasis to induce host cubic membranes?. Trends in Cell Biology, 2010, 30(7): 371-379. |
| [29] | Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Rota PA, Bankamp B, Bellini WJ, Zaki SR. Ultrastructural characterization of SARS-coronavirus. Emerging Infectious Diseases, 2004, 10(2): 320-326. DOI:10.3201/eid1002.030913 |
| [30] | Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FGA, Koster AJ, Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biology, 2020, 18(6): e3000715. DOI:10.1371/journal.pbio.3000715 |
| [31] | Knoops K, Kikkert M, van den Worm SHE, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biology, 2008, 6(9): e226. DOI:10.1371/journal.pbio.0060226 |
| [32] | Knoops K, Bárcena M, Limpens RWAL, Koster AJ, Mommaas AM, Snijder EJ. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. Journal of Virology, 2012, 86(5): 2474-2487. DOI:10.1128/JVI.06677-11 |
| [33] | Kong LB, Aoyagi H, Yang ZB, Ouyang T, Matsuda M, Fujimoto A, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Suzuki T, Muramatsu M, Wakita T, Aizaki H. Surfeit 4 contributes to the replication of hepatitis C virus using double-membrane vesicles. Journal of Virology, 2020, 94(2): e00858-19. |
| [34] | Ferrari M, Zevini A, Palermo E, Muscolini M, Alexandridi M, Etna MP, Coccia EM, Fernandez-Sesma A, Coyne C, Zhang DD, Marques ETA, Olagnier D, Hiscott J. Dengue virus targets Nrf2 for NS2B3-mediated degradation leading to enhanced oxidative stress and viral replication. Journal of Virology, 2020, 94(24): e01551-20. |
| [35] | Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host & Microbe, 2009, 5(4): 365-375. |
| [36] | Eyre NS, Johnson SM, Eltahla AA, Aloi M, Aloia AL, McDevitt CA, Bull RA, Beard MR. Genome-wide mutagenesis of dengue virus reveals plasticity of the NS1 protein and enables generation of infectious tagged reporter viruses. Journal of Virology, 2017, 91(23): e01455-17. |
| [37] | Fontana J, Tzeng WP, Calderita G, Fraile-Ramos A, Frey TK, Risco C. Novel replication complex architecture in rubella replicon-transfected cells. Cellular Microbiology, 2007, 9(4): 875-890. DOI:10.1111/j.1462-5822.2006.00837.x |
| [38] | Fontana J, López-Montero N, Elliott RM, Fernández JJ, Risco C. The unique architecture of Bunyamwera virus factories around the Golgi complex. Cellular Microbiology, 2008, 10(10): 2012-2028. DOI:10.1111/j.1462-5822.2008.01184.x |
| [39] | Magliano D, Marshall JA, Bowden DS, Vardaxis N, Meanger J, Lee JY. Rubella virus replication complexes are virus-modified lysosomes. Virology, 1998, 240(1): 57-63. DOI:10.1006/viro.1997.8906 |
| [40] | Varjak M, Saul S, Arike L, Lulla A, Peil L, Merits A. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. Journal of Virology, 2013, 87(18): 10295-10312. DOI:10.1128/JVI.01105-13 |
| [41] | Lee JY, Bowden DS. Rubella virus replication and links to teratogenicity. Clinical Microbiology Reviews, 2000, 13(4): 571-587. DOI:10.1128/CMR.13.4.571 |
| [42] | Gregorczyk KP, Wyżewski Z, Szczepanowska J, Toka FN, Mielcarska MB, Bossowska-Nowicka M, Gieryńska M, Boratyńska-Jasińska A, Struzik J, Niemiałtowski MG, Szulc-Dąbrowska L. Ectromelia virus affects mitochondrial network morphology, distribution, and physiology in murine fibroblasts and macrophage cell line. Viruses, 2018, 10(5): 266. DOI:10.3390/v10050266 |
| [43] | Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biology, 2007, 5(9): e220. DOI:10.1371/journal.pbio.0050220 |
| [44] | Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology, 2008, 379(2): 294-305. DOI:10.1016/j.virol.2008.06.044 |
| [45] | Garzon S, Strykowski H, Charpentier G. Implication of mitochondria in the replication of Nodamura virus in larvae of the Lepidoptera, Galleria mellonella (L.) and in suckling mice.. Archives of Virology, 1990, 113(3/4): 165-176. |
| [46] | Lanman J, Crum J, Deerinck TJ, Gaietta GM, Schneemann A, Sosinsky GE, Ellisman MH, Johnson JE. Visualizing flock house virus infection in Drosophila cells with correlated fluorescence and electron microscopy. Journal of Structural Biology, 2008, 161(3): 439-446. DOI:10.1016/j.jsb.2007.09.009 |
| [47] | Itahana K, Zhang YP. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell, 2008, 13(6): 542-553. DOI:10.1016/j.ccr.2008.04.002 |
| [48] | Feichtinger RG, Oláhová M, Kishita Y, Garone C, Kremer LS, Yagi M, Uchiumi T, Jourdain AA, Thompson K, D'Souza AR, Kopajtich R, Alston CL, Koch J, Sperl W, Mastantuono E, Strom TM, Wortmann SB, Meitinger T, Pierre G, Chinnery PF, Chrzanowska-Lightowlers ZM, Lightowlers RN, DiMauro S, Calvo SE, Mootha VK, Moggio M, Sciacco M, Comi GP, Ronchi D, Murayama K, Ohtake A, Rebelo-Guiomar P, Kohda M, Kang DC, Mayr JA, Taylor RW, Okazaki Y, Minczuk M, Prokisch H. Biallelic C1QBP mutations cause severe neonatal-, childhood-, or later-onset cardiomyopathy associated with combined respiratory-chain deficiencies. AJHG, 2017, 101(4): 525-538. DOI:10.1016/j.ajhg.2017.08.015 |
| [49] | Claus C, Chey S, Heinrich S, Reins M, Richardt B, Pinkert S, Fechner H, Gaunitz F, Schäfer I, Seibel P, Liebert UG. Involvement of p32 and microtubules in alteration of mitochondrial functions by rubella virus. Journal of Virology, 2011, 85(8): 3881-3892. DOI:10.1128/JVI.02492-10 |
| [50] | Mohan KVK, Ghebrehiwet B, Atreya CD. The N-terminal conserved domain of rubella virus capsid interacts with the C-terminal region of cellular p32 and overexpression of p32 enhances the viral infectivity. Virus Research, 2002, 85(2): 151-161. DOI:10.1016/S0168-1702(02)00030-8 |
| [51] | Beatch MD, Everitt JC, Law LJ, Hobman TC. Interactions between rubella virus capsid and host protein p32 are important for virus replication. Journal of Virology, 2005, 79(16): 10807-10820. DOI:10.1128/JVI.79.16.10807-10820.2005 |
| [52] | Ghebrehiwet B, Lim BL, Kumar R, Feng X, Peerschke EI. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunological Reviews, 2001(180): 65-77. |
| [53] | Rojo G, Chamorro M, Salas ML, Viñuela E, Cuezva JM, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. Journal of Virology, 1998, 72(9): 7583-7588. DOI:10.1128/JVI.72.9.7583-7588.1998 |
| [54] | Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. Journal of Cell Biology, 1998, 143(7): 1883-1898. DOI:10.1083/jcb.143.7.1883 |
| [55] | Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. Journal of Structural Biology, 2000, 129(2/3): 278-287. |
| [56] | Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathogens, 2012, 8(4): e1002630. DOI:10.1371/journal.ppat.1002630 |
| [57] | Risco C, Rodríguez JR, López-Iglesias C, Carrascosa JL, Esteban M, Rodríguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. Journal of Virology, 2002, 76(4): 1839-1855. DOI:10.1128/JVI.76.4.1839-1855.2002 |
| [58] | Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host & Microbe, 2009, 6(1): 81-90. |
| [59] | Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. Journal of Virology, 1994, 68(1): 130-147. DOI:10.1128/jvi.68.1.130-147.1994 |
| [60] | Mutsafi Y, Zauberman N, Sabanay I, Minsky A. Vaccinia-like cytoplasmic replication of the giant mimivirus. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(13): 5978-5982. DOI:10.1073/pnas.0912737107 |
| [61] | Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun SY, Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biology, 2008, 6(5): e114. DOI:10.1371/journal.pbio.0060114 |
| [62] | de Castro IF, Volonté L, Risco C. Virus factories: biogenesis and structural design. Cellular Microbiology, 2013, 15(1): 24-34. DOI:10.1111/cmi.12029 |
| [63] | Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant mimivirus. Journal of Molecular Biology, 2005, 353(3): 493-496. DOI:10.1016/j.jmb.2005.08.060 |
| [64] | Boevink P, Oparka KJ. Virus-host interactions during movement processes. Plant Physiology, 2005, 138(4): 1815-1821. DOI:10.1104/pp.105.066761 |
| [65] | Greber UF, Way M. A superhighway to virus infection. Cell, 2006, 124(4): 741-754. DOI:10.1016/j.cell.2006.02.018 |
| [66] | Wang IH, Burckhardt CJ, Yakimovich A, Greber UF. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses, 2018, 10(4): 166. DOI:10.3390/v10040166 |
| [67] | Niehl A, Heinlein M. Cellular pathways for viral transport through plasmodesmata. Protoplasma, 2011, 248(1): 75-99. DOI:10.1007/s00709-010-0246-1 |
| [68] | Schoelz JE, Harries PA, Nelson RS. Intracellular transport of plant viruses: finding the door out of the cell. Molecular Plant, 2011, 4(5): 813-831. DOI:10.1093/mp/ssr070 |
| [69] | Harries P, Ding B. Cellular factors in plant virus movement: at the leading edge of macromolecular trafficking in plants. Virology, 2011, 411(2): 237-243. DOI:10.1016/j.virol.2010.12.021 |
| [70] | Heinlein M. Plant virus replication and movement. Virology, 2015(479-480): 657-671. |
| [71] | Iwasaki K, Omura T. Electron tomography of the supramolecular structure of virus-infected cells. Current Opinion in Structural Biology, 2010, 20(5): 632-639. DOI:10.1016/j.sbi.2010.08.007 |
| [72] | Morshed A, Dutta P, Kim MJ. Electrophoretic transport and dynamic deformation of bio-vesicles. Electrophoresis, 2019, 40(18/19): 2584-2591. |
| [73] | Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudrière-Gesbert C, Gaudin Y, Blondel D. Negri bodies are viral factories with properties of liquid organelles. Nature Communications, 2017, 8(1): 58. DOI:10.1038/s41467-017-00102-9 |
| [74] | Fan Y, Sanyal S, Bruzzone R. Breaking bad: how viruses subvert the cell cycle. Frontiers in Cellular and Infection Microbiology, 2018(8): 396. |
 2021, Vol. 61
2021, Vol. 61




