中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王泽铭, 李传虹, 马巧丽, 李千雪, 魏亚茹, 赵吉, 于景丽, 希尼尼根. 2021
- Zeming Wang, Chuanhong Li, Qiaoli Ma, Qianxue Li, Yaru Wei, Ji Zhao, Jingli Yu, Xininigen. 2021
- 湿度盐度pH协同驱动锡林河景观疣微菌群空间异质性
- Moisture, salinity and pH co-driving spatial heterogeneity of Verrucomicrobial populations in Xilin River landscape
- 微生物学报, 61(6): 1728-1742
- Acta Microbiologica Sinica, 61(6): 1728-1742
-
文章历史
- 收稿日期:2021-02-28
- 修回日期:2021-05-11
- 网络出版日期:2021-05-14
2. 内蒙古农业大学兽医学院, 内蒙古 呼和浩特 010018
2. College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot 010018, Inner Mongolia Autonomous Region, China
Hedlund等在1997年将疣微菌门(Verrucomicrobia)划分为一个新的细菌类群[1]。疣微菌门包含丰祐菌纲(Opitutae)、斯巴达杆菌纲(Spartobacteria)、疣微菌纲(Verrucomicrobiae)等主要类群。疣微菌在富营养环境下可利用硫酸盐降解多糖[2];在寡营养条件下可通过化能自养代谢获取能量[3];在强酸环境条件下具有甲烷氧化功能,是阻碍甲烷排放的重要功能群[4-5]。Nixon等通过32个疣微菌全基因组分析证实Candidatus Marcellius具有固氮相关的nifHDKENB基因,其编码的固氮酶具有完整的三羧酸循环和糖酵解代谢途径[6]。丰祐菌纲成员存在固氮基因[7],Dunfield等[8]和Chiang等[9]证实subdivision 6具有好氧的甲烷氧化和固氮功能,这说明疣微菌部分成员具有生物固氮和减缓甲烷排放的双重环境效应,属于生态友好型环境微生物。疣微菌可通过植物-微生物相互作用促进矿物质的溶解增加土壤矿质营养,产生的植物激素等生物活性化合物有助于杀死病原生物[10],这说明疣微菌也是制备生态环保型菌肥和生物杀虫剂的重要原料。
疣微菌群广泛分布于全球土壤[2]、淡水[11]、海洋[12-13]等陆地及水域生态系统,在全球碳氮硫生物地球化学循环中发挥重要作用。南极洲、欧洲和美洲等181种土壤及波罗的海咸水表面均以斯巴达杆菌纲成员为主[2]。淡水生境和波罗的海咸水表面的疣微菌群分别占全细菌群落组成的19%[6]和12%[14]。湿度、pH、有机碳和全氮含量是影响秦岭南侧根际土壤疣微菌等细菌多样性的主要环境变量[15];盐度是影响页岩气井疣微菌群丰度的重要环境变量[6];土壤颗粒物组成是影响土壤酶活性及疣微菌相对丰度的重要因子[16];高海拔区淡水湿地疣微菌的相对丰度会随着磷含量的增加而增加[17-19],但富含腐殖质的湖泊中疣微菌的相对丰度会随着磷含量的增加而减少[20];海洋疣微菌群主要受盐度、硝酸盐等因素影响[21]。综上所述,湿度、pH、有机碳、全氮、全磷、盐度、硝酸盐、土壤颗粒组成等可能是影响疣微菌群组成和功能分化的关键因素。
锡林河流域是中国北方较具代表性的河流景观。受气候变化和人类活动的双重影响,锡林河景观呈破碎化趋势,生态系统脆弱[22]。微生物群落是景观功能及服务变化的最佳指示生物[23-24]。本研究以锡林河流域疣微菌群的景观异质性作为切入点,选择河床中心、河床边缘、牛轭湖床、低河漫滩、高河漫滩、低阶地和高阶地等景观要素,研究疣微菌群的景观空间异质性及环境驱动力问题。本研究的意义在于借助疣微菌群的空间分布规律建立指示景观生态安全的微生物指标体系,并为今后实现其甲烷氧化和生物固氮等生态友好功能奠定基础。
1 材料和方法 1.1 研究区概况及景观要素的选择锡林河流域(43°39′−44°36′N、115°33′−117°28′E)属于中温带干旱大陆性季风气候。该区年均温约2.6 ℃,年均降水量约300 mm,全年51%-89%的降水量集中在5-8月。代表性的植被包括水莎草(Juncellus serotinus)等水生植被、灯芯草(Juncus effusus)和鹅绒委陵菜(Potentilla anserina)等湿生植被、羊草(Leymus chinensis)和大针茅(Stipa grandis)等旱生植被;代表性的土壤包括沼泽土、沼泽化草甸土、草甸土和栗钙土等。研究区为放牧利用样地。
本研究在锡林河流域典型河段基于水文及微地形差异选取长期淹水的水生生境(R)、季节性淹水的湿生生境(F)、长期不淹水的旱生生境(T)等3种生境类型及河床、河漫滩、阶地等划分7个景观要素。河床景观要素包括河床中心(riverbed center,RC)、河床边缘(riverbed edge,RE)和牛轭湖床(oxbow lake-bed,RO);河漫滩景观要素包括低河漫滩(low floodplain,LF)和高河漫滩(high floodplain,HF);阶地景观要素包括低阶地(low terrace,LT)和高阶地(high terrace,HT),见图 1-A。在RC、RE和RO、LF和HF、LT和HT各景观要素中分别采集4个土壤样品(图 1-A)。土壤湿度(moisture)、盐度(salinity)、pH、氨态氮(ammonia)、硝态氮(nitrate)、总有机碳(total organic carbon,TOC)、全氮(total nitrogen,TN)、全磷(total phosphorus,TP)、土壤颗粒组成(sand,silt,clay)等环境变量的检测方法参考文献[25-26]。环境变量的检测结果见图 1-B。

|
| 图 1 基于生境和微地形差异划分锡林河流域不同景观要素(A)及环境变量(B)的陆向分布图 Figure 1 Landward distribution map of different landscape components (A) based on the difference of habitats and microtopography as well as environmental variables (B) in Xilin River basin. |
1.2 生物信息学分析
采用Excel 2010进行基础数据分析及饼形图、累积柱状图和折线图等基本图形的绘制。采用SPSS 24.0进行单因素方差分析及单个类群与单个环境变量的皮尔森相关性分析。基于R语言Vegan程序包进行热图分析。采用CANOCO 5.0 (Biometris-Plant Research International,Wageningen,Netherlands)对不同景观要素疣微菌群和多个环境变量进行多重相关的冗余分析(redundancy analysis,RDA)。通过CANOCO 5.0的变异权重分析(variation partitioning analysis,VPA)计算影响疣微菌群景观空间异质性的单个环境变量的解释度,通过R语言中的Random forest程序包进行随机森林分析(random forest analysis,RFA)找出相关性最大的环境变量组合,再基于双组及三组环境变量的VPA,找到解释度最高的环境变量组合及关键性的环境变量。
2 结果和分析 2.1 基于16S rRNA基因系统发育分析的疣微菌群分类学谱系及相对丰度分布特征锡林河流域水生的河床中心、河床边缘与牛轭湖床,湿生的低河漫滩与高河漫滩,旱生的河流低阶地与高阶地等7个景观要素共检测到来自35个门类的742类细菌(属水平)。隶属于疣微菌门的21类细菌属在旱生的低阶地景观要素中的累积相对丰度最高,在水生的河床中心景观要素中累积相对丰度次高,在湿生的河漫滩景观要素中累积相对丰度最低。不同景观要素中21类疣微菌群的累积相对丰度排序为LT>RC>HT>RO>其他3个景观要素(P<0.05)。
交替球菌属(Alterococcus)、丰祐菌属(Opitutus)、Xiphinematobacter、Chthoniobacter和Haloferula、突柄杆菌属(Prosthecobacter)等21个疣微菌属隶属于丰祐菌纲(Opitutae)、斯巴达杆菌纲(Spartobacteria)、疣微菌纲(Verrucomicrobiae)等5个纲,丰祐菌目(Opitutales)、Chthoniobacterales、疣微菌目(Verrucomicrobiales)等6个目,丰祐菌科(Opitutaceae)、Xiphinematobacteraceae、Chthoniobacteraceae、疣微菌科(Verrucomicrobiaceae)、DA101_soil_group和DEV007等7个科(图 2)。
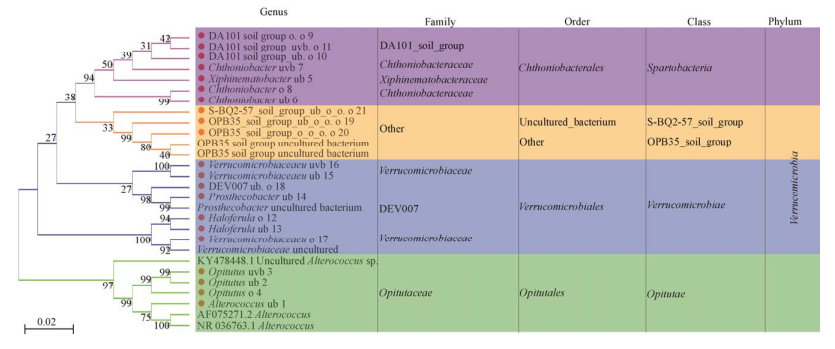
|
| 图 2 锡林河景观21个疣微菌属的分类学谱系 Figure 2 Taxonomic lineage of 21 Verrucomicrobial genera from Xilin River landscape. o: other; u: uncultured; ub: uncultured_bacterium; uvb: uncultured_Verrucomicrobia_bacterium. The number represented the serial number of each taxonomic group at genus level. |
2.2 锡林河景观疣微菌群的空间分布特征
斯巴达杆菌纲的Chthoniobacter ub 6、Chthoniobacter uvb 7、Chthoniobacter o 8成员和疣微菌纲的Prosthecobacter ub 14、Verrucomicrobiaceae_u. o17、Verrucomicrobiales_ DEV007_ub. o 18成员主要分布在RC,其最高相对丰度依次为4.430%、2.168%、0.487%和0.227%、0.702%、0.046%;Verrucomicrobiaceae_u. uvb 16和OPB35_soil_group_ub_o_o. o 19的最高相对丰度出现在RE,其最高相对丰度依次为0.080%和0.615%;Opitutus ub 2、Haloferula ub 13和Verrucomicrobiaceae_u. ub 15成员的最高相对丰度出现在RO,其最高相对丰度依次为0.631%、0.192%和1.578%;Alterococcus ub 1、Opitutus o 4、Haloferula o 12和Verrucomicrobiaceae_u. uvb 16成员主要分布在LF和HF,其最高相对丰度依次为0.095%、0.194%、0.084%和0.080%;Opitutus uvb 3、Xiphinematobacter ub 5、Chthoniobacterales_ DA101_o. o 9、Chthoniobacterales_DA101_ub. o 10、Chthoniobacterales_DA101_uvb. o 11、OPB35_soil_group_o_o_o. o 20和S-BQ2-57_soil_group_ ub_o_o. o 21成员主要分布在LT和HT,其最高相对丰度依次为:0.975%、0.207%、0.454%、10.313%、1.264%、0.148%和0.043% (图 3)。
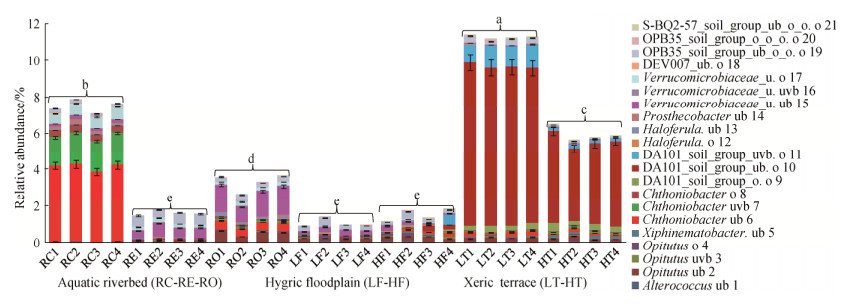
|
| 图 3 锡林河不同景观要素间疣微菌群的空间异质性 Figure 3 Spatial heterogeneity of Verrucomicrobial populations from the different landscape components of Xilin River basin. The same and different lowercase letters respectively indicated insignificant and significant differences of samples (P < 0.05) which were determined by one-way analysis of variance (ANOVA) followed by Dunnett's test. |
2.3 锡林河景观要素内及景观要素间疣微菌群的分布趋势
河床中心RC、河床边缘RE、牛轭湖床RO、低河漫滩LF、高河漫滩HF、低阶地LT、高阶地HT七个景观要素内土壤疣微菌群大致呈现趋同的分布特征,景观要素间呈现此消彼涨的趋异分布特征(图 4-A)。不同景观要素间疣微菌群差异较大,RC自成一类,RE、RO、LF与HF (HF4除外)聚成一类,LT与HT及HF4聚成一类(图 4-B)。
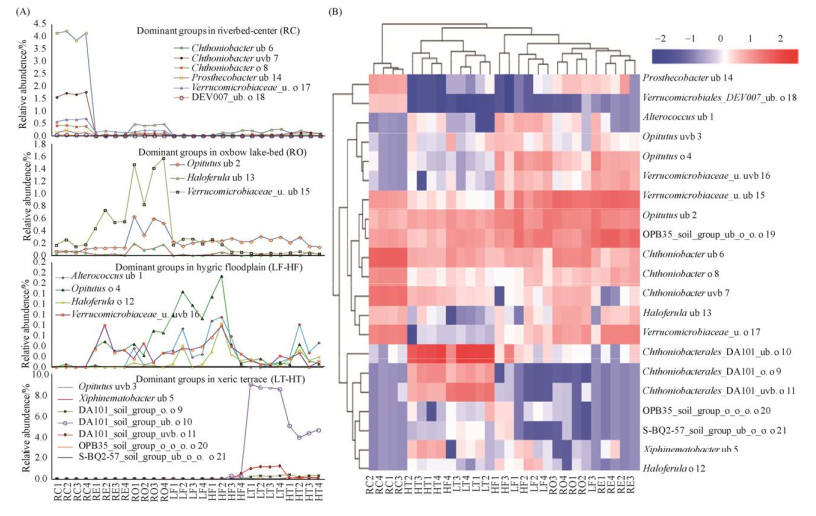
|
| 图 4 锡林河流域景观要素内及景观要素间疣微菌群的空间分布规律(A)及景观要素间疣微菌群的相似性及差异性(B) Figure 4 Spatial distribution dynamics (A) as well as similarity and dissimilarity analyses (B) of Verrucomicrobial populations within and between the landscape components in Xilin River basin. |
2.4 锡林河景观疣微菌群空间异质性的环境影响因素 2.4.1 单个疣微菌群与单个环境变量的相关性分析: 表 1中皮尔逊相关性分析结果显示,Chthoniobacter ub 6、Chthoniobacter uvb 7、Chthoniobacter o 8、Prosthecobacter ub 14、Verrucomicrobiaceae_u. o 17、DEV007_ub. o 18与pH和盐度等存在极显著负相关关系(P < 0.01),与砂粒含量存在显著正相关关系(P < 0.05)或极显著正相关关系(P < 0.01);Xiphinematobacter ub 5、DA101_soil_ group_o. o 9、DA101_soil_group_ub. o 10、DA101_ soil_group_uvb. o 11与湿度和砂粒含量存在极显著负相关关系(P < 0.01),与盐度、全氮、全磷、硝酸盐、黏粒和粉粒含量存在极显著正相关关系(P < 0.01);OPB35_soil_group_o_o_o. o 20和S−BQ2−57_soil_ group_ub_o_o. o 21与全氮、全磷和硝酸盐含量存在显著正相关关系(P < 0.05),与砂粒含量存在极显著负相关(P < 0.01),Opitutus o 4和Verrucomicrobiaceae_u. ub 15与湿度含量呈显著正相关(P < 0.01)。
| Populations | Moisture | pH | Salinity | TOC | TN | TP | Ammonia | Nitrate | Clay | Silt | Sand |
| Alterococcus. ub 1 | 0.076 | 0.505** | 0.245 | 0.136 | 0.104 | 0.235 | −0.230 | −0.014 | −0.087 | 0.247 | −0.145 |
| Opitutus ub 2 | 0.386* | 0.649** | 0.329 | 0.890** | 0.464* | 0.283 | 0.229 | 0.044 | 0.143 | 0.109 | −0.124 |
| Opitutus uvb 3 | −0.161 | 0.143 | 0.409* | 0.128 | 0.321 | 0.37 | −0.274 | 0.266 | 0.416* | 0.523** | −0.504** |
| Opitutus o 4 | 0.570** | 0.664** | −0.008 | 0.052 | −0.209 | −0.178 | 0.056 | −0.369 | −0.370 | −0.218 | 0.275 |
| Xiphinematobacter ub 5 | −0.695** | −0.036 | 0.577** | 0.174 | 0.609** | 0.760** | −0.28 | 0.603** | 0.430* | 0.652** | −0.599** |
| Chthoniobacter ub 6 | −0.143 | −0.702** | −0.824** | −0.506** | −0.550** | −0.567** | −0.405* | −0.441* | −0.323 | −0.444* | 0.418* |
| Chthoniobacter uvb 7 | −0.185 | −0.725** | −0.811** | −0.539** | −0.545** | −0.545** | −0.426* | −0.420* | −0.316 | −0.425* | 0.403* |
| Chthoniobacter o 8 | −0.165 | −0.714** | −0.807** | −0.519** | −0.541** | −0.555** | −0.393* | −0.407* | −0.304 | −0.423* | 0.397* |
| DA101_soil_group_o. o 9 | −0.833** | −0.196 | 0.699** | 0.229 | 0.815** | 0.872** | −0.340 | 0.782** | 0.805** | 0.812** | −0.835** |
| DA101_soil_group_ub. o 10 | −0.760** | −0.251 | 0.688** | 0.235 | 0.831** | 0.800** | −0.325 | 0.793** | 0.970** | 0.808** | −0.887** |
| DA101_soil_group_uvb. o 11 | −0.530** | −0.214 | 0.548** | 0.188 | 0.664** | 0.565** | −0.275 | 0.630** | 0.916** | 0.683** | −0.782** |
| Haloferula o 12 | −0.049 | 0.360 | 0.262 | 0.052 | 0.128 | 0.241 | −0.269 | 0.119 | 0.054 | 0.362 | −0.272 |
| Haloferula ub 13 | 0.418* | 0.299 | −0.249 | 0.574** | −0.019 | −0.179 | 0.263 | −0.282 | −0.304 | −0.339 | 0.338 |
| Prosthecobacter ub 14 | 0.016 | −0.572** | −0.794** | −0.333 | −0.514** | −0.570** | −0.213 | −0.496** | −0.357 | −0.531** | 0.490** |
| Verrucomicrobiaceae_u. ub 15 | 0.657** | 0.464* | −0.143 | 0.596** | −0.066 | −0.246 | 0.639** | −0.293 | −0.370 | −0.466* | 0.449* |
| Verrucomicrobiaceae_u. uvb 16 | 0.482** | 0.489** | 0.127 | 0.021 | −0.186 | −0.143 | 0.335 | −0.183 | −0.261 | −0.065 | 0.132 |
| Verrucomicrobiaceae_u. o 17 | 0.056 | −0.625** | −0.873** | −0.433* | −0.641** | −0.694** | −0.107 | −0.502** | −0.466* | −0.616** | 0.586** |
| Verrucomicrobiales _DEV007_ub. o 18 | −0.145 | −0.718** | −0.833** | −0.564** | −0.584** | −0.584** | −0.401* | −0.443* | −0.342 | −0.457* | 0.434* |
| OPB35_soil_group_ub_o_o. o 19 | 0.415* | 0.116 | −0.002 | −0.076 | −0.239 | −0.339 | 0.765** | 0.056 | −0.146 | −0.316 | 0.270 |
| OPB35_soil_group_o_o_o. o 20 | −0.335 | −0.148 | 0.364 | 0.118 | 0.416* | 0.402* | −0.230 | 0.411* | 0.581** | 0.459* | −0.514** |
| S−BQ2−57_soil_group_ub_o_o. o 21 | −0.389* | −0.039 | 0.495** | 0.149 | 0.475* | 0.464* | −0.290 | 0.405* | 0.678** | 0.699** | −0.714** |
| *: Correlation was significant at the 0.05 level (2-tailed); **: Correlation was significant at the 0.01 level(2-tailed). TOC: total organic carbon; TN: total nitrogen; TP: total phosphorus. | |||||||||||
2.4.2 不同景观要素间疣微菌群与多个环境变量的多重相关性及单个环境变量的解释度: 图 5-A中RDA显示,水生河床中心RC中斯巴达杆菌纲的Chthoniobacter ub 6、Chthoniobacter uvb 7、Chthoniobacter o 8成员和疣微菌纲的Prosthecobacter ub 14、Verrucomicrobiaceae_u. o 17、Verrucomicrobiales_DEV007_ub. o 18成员与砂粒含量相关性大;水生河床边缘RE中OPB35_soil_group_ub_o_o. o 19成员及水生牛轭湖床RO中Opitutus ub 2、Haloferula. ub 13和Verrucomicrobiaceae_u. ub 15成员与砂粒、湿度及氨态氮含量相关性大;低河漫滩LF与高河漫滩HF中Alterococcus ub 1、Opitutus o 4、Haloferula o 12和Verruco-microbiaceae_u. uvb 16成员与pH相关性大;旱生阶地LT与HT中Opitutus uvb 3、Xiphinematobacter ub 5、Chthoniobacterales_ DA101_o. o 9、Chthoniobacterales_DA101_ub. o 10、Chthoniobacterales_DA101_uvb. o 11、OPB35_ soil_group_o_o_o. o 20和S-BQ2-57_soil _group_ ub_o_o. o 21成员与盐度及养分含量相关性大。

|
| 图 5 锡林河景观疣微菌群与环境变量的冗余分析(A)及单个环境变量的变异权重分析(B) Figure 5 Redundancy analysis (RDA) between Verrucomicrobial populations and environmental variables (A) as well as variation partitioning analysis (VPA) of single environmental variables (B) in Xilin River landscape. AlterUb1- S-BQ2O21 represented Alterococcus. ub 1- S-BQ2-57_soil_group _ub_o_o. o 21, total names in Table 1. |
单个环境变量的变异权重分析(图 5-B)显示,砂粒的解释度最高达到53.3%,盐度的解释度为16.7%,全磷、氨态氮、pH、全氮和湿度的解释度分别为7.6%、5.1%、4.2%、2.2%和1.5%,Monte carlo检验显示这些环境变量的解释度均达到统计学显著性水平(P < 0.05)。已知环境变量对疣微菌群的空间分异的解释度为90.6%,其中砂粒和盐度两者的解释度为70%。
2.4.3 复合环境变量对单个疣微菌群空间变异的影响: 为进一步验证砂粒及盐度等环境变量是否为影响疣微菌群空间变异的核心因素,我们基于单个环境变量的VPA (图 5-B)结果抽选了解释度排名前六位的砂粒、盐度、全磷、氨态氮、pH、全氮进行RFA (图 6),发现盐度、氨态氮和全氮组合与疣微菌群中Chthoniobacter ub 6、Chthoniobacter uvb 7、Chthoniobacter o 8和Verrucomicrobiaceae_u. o 17的相关系数高达0.988−0.951;全磷、氨态氮、pH、全氮四者组合与Opitutus ub 2、Haloferula ub 13和Verrucomicrobiaceae_u. ub 15的相关性较大,相关性系数为0.840−0.929;全磷、氨态氮、全氮三者组合与Chthoniobacterales_DA101_ub. o 10、Haloferula. o 12、OPB35_soil_group_ub _o_o. o 19的相关性系数为0.679−0.923;全磷、全氮三者组合与Alterococcus. ub 1、Chthoniobacterales_ DA101_uvb. o 11的相关性系数为0.921−0.697。

|
| 图 6 锡林河景观单个疣微菌群与复合环境变量的随机森林分析 Figure 6 Random forest analysis (RFA) between single Verrucomicrobial populations and combined environmental variables in Xilin River landscape. |
2.4.4 复合环境变量对整个疣微菌群空间变异的影响: 基于随机森林分析的环境变量组合结果和基于单个环境变量的VPA结果发现组合变量中砂粒的相关性及解释度低于单个环境变量的解释度,推测砂粒并非环境因子中的核心变量。进一步对环境变量划分为两组和三组进行VPA,同时计算组间环境变量解释度差值(Δ)。两组间环境变量解释度差值(Δ)计算结果显示,湿度、盐度、pH、氨态氮、硝酸盐的解释度分别为24.7%、24.3%、23.1%、11.6%、1.0%;三组间环境变量解释度差值(Δ)计算结果显示,湿度、盐度、pH、氨态氮、硝酸盐的解释度分别为21.4%、22.7%、20.8%、8.8%、4.4% (图 7)。

|
| 图 7 锡林河景观疣微菌群与两组或三组复合环境变量的变异权重分析 Figure 7 Variation partitioning analysis (VPA) between Verrucomicrobial populations and environmental variables divided into 2 groups (A) and 3 groups (B) in Xilin River landscape. +: group A plus a certain environmental factor; -: group A minus a certain environmental factor. |
3 讨论
斯巴达杆菌纲的Chthoniobacter成员和疣微菌纲的突柄杆菌属等成员主要分布在水生的河床中心,这与文献报道的专性好氧性疣微菌属及突柄杆菌属主要分布在淡水生境的研究结果相一致[27],原因之一可能是锡林河流域水生的河床中心为盐度相对较低的砂质土壤[25],更适合于疣微菌属及突柄杆菌属的生存;原因之二是Chthoniobacter成员具有降解复杂有机化合物的能力,是低碳水域环境[25]的指示生物[28]。丰祐菌纲的丰祐菌属及交替球菌属成员主要分布在湿生的河漫滩,与文献报道的丰祐菌属分布在稻田土壤的研究结果具有相似性[27],推测丰祐菌属成员更适合生存在周期性淹水的河漫滩湿地土壤。Xiphinematobacter和DA101_soil_group主要分布在旱生的阶地典型草原[25],与文献报道的Xiphinematobacter和DA101_soil_group主要分布在天然草地土壤中的研究结果具有一致性[29],说明Xiphinematobacter和DA101_soil_group成员是旱生草地土壤的优势菌群。不可培养的疣微菌科成员主要分布在锡林河流域相对富营养的牛轭湖[25]和山东境内微山湖富营养化的养殖区,推测不可培养的疣微菌是富营养化水体的指示生物[30]。本研究发现一些OPB35_soil_group成员广泛分布在水生河床、湿生河漫滩、旱生阶地等景观要素中,且主要分布在河床边缘湿地土壤中,这与文献报道的OPB35主要分布在青藏高原三类湿地土壤的研究结果具有一致性[31];另一些OPB35_soil_group成员主要分布在湿偏旱生河漫滩及旱生阶地土壤中,这与文献报道的OPB35主要分布在湿地土壤中的研究结果稍有不同,但与OPB35_soil_group分布在黑麦草和白三叶草放牧土壤中的研究结果相似,原因可能是部分OPB35_soil_group成员的定殖与放牧利用的草原土壤有关[32-33]。简言之,OPB35_soil_group成员适宜的生境较广。S-BQ2-57_soil_group成员主要分布在锡林河阶地,这与S-BQ2-57_soil_group分布在密歇根内陆湖的研究结果相似,原因可能与蒙古高原内陆型锡林河与密歇根内陆湖同属于温带大陆性气候有关[18],推测S-BQ2-57_soil_group适应于降雨量较少的温带大陆性气候河湖生态系统。
锡林河流域河床中心、牛轭湖、河漫滩和河流阶地等景观要素间土壤疣微菌群存在此消彼长的分布趋势,这和北温带不同类型淡水湖泊疣微菌群的趋异特征具有一致性[9]。同一景观要素内优势疣微菌群的分布趋势相同,这和水生环境微生物的趋同性[34]具有一致性。
湿度通常是驱动锡林河流域动植物及厌氧绳菌群落空间分布异质性的限制性因子[26, 35-36]。本研究发现湿度是影响疣微菌群景观空间异质性的关键因子,最高解释度为24.7%。单个环境因子变异权重分析显示砂粒的解释度为53.3%,这和Hemkemeyer等[37]发现疣微菌更适合在土壤砂粒中生存的研究结果具有一致性。有趣的是,砂粒与其他环境变量复合时的解释度低至负值(-1.5%),这和本文锡林河流域大多数疣微菌群与砂粒存在极显著负相关关系(P < 0.01,见表 1)的研究结果具有一致性,说明在水生、湿生、旱生复合生境下砂粒主要发挥负向驱动作用。本研究发现21类疣微菌中有11类和9类疣微菌分别与砂粒和硝酸盐存在正相关关系,这可能与砂质土壤中疣微菌群进行的硝酸盐呼吸作用有关[37]。大多数陆地和水域生态系统中pH通常被认为是影响细菌群落空间变异的主要因子[4-5, 38-40],本研究发现pH的最高解释度为23.1%,说明pH也是影响锡林河流域疣微菌群景观异质性的重要因子,原因可能是疣微菌等细菌对pH值的微小变化响应敏感[39, 41]。本研究发现盐度对锡林河流域疣微菌群景观异质性的最高解释度为24.3% (P=0.002),这和斯巴达杆菌纲类群沿着波罗的海盐度梯度呈现空间异质性的研究结果相吻合[14, 42]。Mohapatra等在2020年研究咸水近岸泻湖浮游细菌群落的时空动态时发现,相比于其他细菌类群疣微菌群中的斯巴达杆菌纲类群更适合于低盐淡水环境,是寡盐-中盐体系的最佳指示生物。这为锡林河流域疣微菌群既可适应于寡盐[25]的河床中心,又可适应于中盐[25]的河床边缘、河漫滩、阶地的研究结果提供了科学证据[43]。同时发现疣微菌中的全部丰祐菌目成员、半数Chthoniobacterales成员、部分OPB35_soil_group和S-BQ2-57_soil_group成员分布在相对高盐[25]的湿地及旱地土壤中,这与疣微菌适应于高盐土壤的研究报道[44-45]一致,原因可能是疣微菌与耐盐植物相互作用在缓解调控盐胁迫方面发挥潜在作用。可见疣微菌群适应的盐度范围较为广泛,既可以适应于低盐环境,也可以适应于高盐环境[41, 46-48]。有研究显示,疣微菌群的相对丰度受无机营养物浓度的影响[49],部分疣微菌适应寡营养的生境[2, 50],随土壤深度的增加,疣微菌群的相对丰度增加,变得更优势[51],这与本研究有机碳、全氮、全磷等是影响疣微菌群景观异质性的结果相一致。其原因可能与疣微菌多样化的代谢方式[2-3]及固氮能力[6, 8-9]有关。
4 结论疣微菌群是锡林河景观异质性的指示生物。斯巴达杆菌纲的Chthoniobacter成员,疣微菌纲的突柄杆菌属、Verrucomicrobiaceae_u. o 17和Verrucomicrobiales _DEV007_ub. o 18等成员是水生河床中心景观要素的指示生物;疣微菌纲的Verrucomicrobiaceae_u. uvb 16、OPB35_soil_ group_ub_o_o. o 19是水生河床边缘景观要素的指示生物;丰祐菌纲的Opitutus ub 2成员,疣微菌纲的Haloferula ub 13和Verrucomicrobiaceae_u. ub 15成员是水生牛轭湖床景观要素的指示生物;丰祐菌纲的Alterococcus ub 1和Opitutus o 4,疣微菌纲的Haloferula o 12和Verrucomicrobiaceae_u. uvb 16等成员是河漫滩景观要素的指示生物;丰祐菌纲的Opitutus uvb 3,斯巴达杆菌纲的Xiphinematobacter和Chthoniobacterales_DA101,OPB35_soil_group_o_o_o. o 20,S-BQ2-57_soil_ group_ub_o_o. o 21成员是旱生阶地景观要素的指示生物。湿度、盐度、pH均是影响疣微菌群景观空间异质性的重要因子。湿度、盐度、pH与其他因子复合的解释度高于单一因素时各自的解释度,说明湿度、盐度、pH是协同驱动锡林河流景观异质性的核心驱动力。单个环境变量与复合环境变量在解释砂粒权重方面存在较大差异,具体原因有待后续研究证实。
| [1] | Hedlund BP, Gosink JJ, Staley JT. a new division of the bacteria containing three new species of Prosthecobacter.. Antonie Van Leeuwenhoek, 1997, 72(1): 29-38. DOI:10.1023/A:1000348616863 |
| [2] | Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biology and Biochemistry, 2011, 43(7): 1450-1455. DOI:10.1016/j.soilbio.2011.03.012 |
| [3] | Tran P, Ramachandran A, Khawasik O, Beisner BE, Rautio M, Huot Y, Walsh DA. Microbial life under ice: Metagenome diversity and in situ activity of Verrucomicrobia in seasonally ice-covered Lakes. Environmental Microbiology, 2018, 20(7): 2568-2584. DOI:10.1111/1462-2920.14283 |
| [4] | Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, den Camp HJMO. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature, 2007, 450(7171): 874-878. DOI:10.1038/nature06222 |
| [5] | Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland NK. Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia Phylum. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(1): 300-304. DOI:10.1073/pnas.0704162105 |
| [6] | Nixon SL, Daly RA, Borton MA, Solden LM, Welch SA, Cole DR, Mouser PJ, Wilkins MJ, Wrighton KC. Genome-resolved metagenomics extends the environmental distribution of the Verrucomicrobia Phylum to the deep terrestrial subsurface. mSphere, 2019, 4(6): 00613-00619. DOI:10.1128/msphere.00613-19 |
| [7] | Wertz JT, Kim E, Breznak JA, Schmidt TM, Rodrigues JLM. Genomic and physiological characterization of the Verrucomicrobia isolate Diplosphaera colitermitum gen. nov., sp. nov., reveals microaerophily and nitrogen fixation genes. Applied and Environmental Microbiology, 2012, 78(5): 1544-1555. DOI:10.1128/AEM.06466-11 |
| [8] | Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou SB, Ly B, Saw JH, Zhou ZM, Ren Y, Wang JM, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. Methane oxidation by an extremely acidophilic bacterium of the Phylum Verrucomicrobia. Nature, 2007, 450(7171): 879-882. DOI:10.1038/nature06411 |
| [9] | Chiang E, Schmidt ML, Berry MA, Biddanda BA, Burtner A, Johengen TH, Palladino D, Denef VJ. Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS One, 2018, 13(3): e0195112. DOI:10.1371/journal.pone.0195112 |
| [10] | Ajar Nath Y. Beneficial plant-microbe interactions for agricultural sustainability. Journal of Applied Biology & Biotechnology, 2021, 9(1): 1-4. DOI:10.7324/jabb.2021.91ed |
| [11] | Chin KJ, Hahn D, Hengstmann U, Liesack W, Janssen PH. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Applied and Environmental Microbiology, 1999, 65(11): 5042-5049. DOI:10.1128/AEM.65.11.5042-5049.1999 |
| [12] | Scheuermayer M, Gulder TAM, Bringmann G, Hentschel U. Rubritalea marina gen. nov., sp. nov., a marine representative of the Phylum 'Verrucomicrobia', isolated from a sponge (Porifera). International Journal of Systematic and Evolutionary Microbiology, 2006, 56(9): 2119-2124. DOI:10.1099/ijs.0.64360-0 |
| [13] | Zhang CY, Dong BL, Wang RJ, Su Y, Han SB, Yu XY, Zhao Z, Fu GY, Sun C, Wu M. Luteolibacter flavescens sp. nov., isolated from deep seawater. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(3): 729-735. DOI:10.1099/ijsem.0.001713 |
| [14] | Bergen B, Herlemann DPR, Labrenz M, Jürgens K. Distribution of the verrucomicrobial clade Spartobacteria along a salinity gradient in the Baltic Sea. Environmental Microbiology Reports, 2014, 6(6): 625-630. DOI:10.1111/1758-2229.12178 |
| [15] | Wang MJ, Sun HY, Xu L, Xu ZM. Bacterial diversity in tea plant (Camellia sinensis) rhizosphere soil from Qinling Mountains and its relationship with environmental elements. Plant and Soil, 2021, 460(1/2): 403-415. DOI:10.1007/s11104-020-04822-8 |
| [16] | Carlos FS, Schaffer N, Marcolin E, Fernandes RS, Mariot R, Mazzurana M, Roesch LFW, Levandoski B, Oliveira Camargo FA. A long-term no-tillage system can increase enzymatic activity and maintain bacterial richness in paddy fields. Land Degradation & Development, 2021, 32(6): 2257-2268. |
| [17] | Zhang JX, Zhang XL, Liu Y, Xie SG, Liu YG. Bacterioplankton communities in a high-altitude freshwater wetland. Annals of Microbiology, 2014, 64(3): 1405-1411. DOI:10.1007/s13213-013-0785-8 |
| [18] | Lindström ES, Vrede K, Leskinen E. Response of a member of the Verrucomicrobia, among the dominating bacteria in a hypolimnion, to increased phosphorus availability. Journal of Plankton Research, 2004, 26(2): 241-246. DOI:10.1093/plankt/fbh010 |
| [19] | Haukka K, Kolmonen E, Hyder R, Hietala J, Vakkilainen K, Kairesalo T, Haario H, Sivonen K. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microbial Ecology, 2006, 51(2): 137-146. DOI:10.1007/s00248-005-0049-7 |
| [20] | Arnds J, Knittel K, Buck U, Winkel M, Amann R. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Systematic and Applied Microbiology, 2010, 33(3): 139-148. DOI:10.1016/j.syapm.2009.12.005 |
| [21] | Freitas S, Hatosy S, Fuhrman JA, Huse SM, Mark Welch DB, Sogin ML, Martiny AC. Global distribution and diversity of marine Verrucomicrobia. The ISME Journal, 2012, 6(8): 1499-1505. DOI:10.1038/ismej.2012.3 |
| [22] |
Zhou YJ, Liu TX, Duan LM, Wang YX, Li X, Li MY. Driving force analysis and landscape pattern evolution in the up stream valley of Xilin River Basin. Arid Zone Research, 2020, 37(3): 580-590.
(in Chinese) 周亚军, 刘廷玺, 段利民, 王怡璇, 李霞, 黎明扬. 锡林河流域上游河谷湿地景观格局演变及其驱动力. 干旱区研究, 2020, 37(3): 580-590. |
| [23] | Li XB, Tian MR, Wang H, Wang H, Yu JJ. Development of an ecological security evaluation method based on the ecological footprint and application to a typical steppe region in China. Ecological Indicators, 2014, 39: 153-159. DOI:10.1016/j.ecolind.2013.12.014 |
| [24] | Matulich KL, Martiny JBH. Microbial composition alters the response of litter decomposition to environmental change. Ecology, 2015, 96(1): 154-163. DOI:10.1890/14-0357.1 |
| [25] |
Ma QL, Yu JL, Wei YR, Wang YJ, Li QX, Terigele, Xia JJ, Lu Y, Xininigen. Spatial differentiation of Cyanobacterial communities and their relationship with environmental factors in Xilin River basin. Acta Scientiae Circumstantiae, 2020, 40(12): 4338-4348.
(in Chinese) 马巧丽, 于景丽, 魏亚茹, 王怡静, 李千雪, 特日格乐, 夏晶晶, 芦燕, 希尼尼根. 锡林河流域Cyanobacteria群落空间分化及其与环境因子的关系. 环境科学学报, 2020, 40(12): 4338-4348. |
| [26] |
Wang YJ, Xia JJ, Yu JL, Xininigen, Li CH, Li X, Lu Y. Moisture determined spatial heterogeneity of river sediment/soil Anaerolineaceae communities in semiarid region. Microbiology China, 2020, 47(9): 2807-2821.
(in Chinese) 王怡静, 夏晶晶, 于景丽, 希尼尼根, 李传虹, 李新, 芦燕. 水分驱动半干旱区河流沉积物/土壤厌氧绳菌群落的空间异质性. 微生物学通报, 2020, 47(9): 2807-2821. |
| [27] | Schlesner H, Jenkins C, Staley JT. The Phylum Verrucomicrobia: A phylogenetically heterogeneous bacterial group. The Prokaryotes. New York, NY: Springer New York, 2006: 881-896. |
| [28] | Bill M, Chidamba L, Gokul JK, Labuschagne N, Korsten L. Bacterial community dynamics and functional profiling of soils from conventional and organic cropping systems. Applied Soil Ecology, 2021, 157: 103734. DOI:10.1016/j.apsoil.2020.103734 |
| [29] | Armbruster M, Goodall T, Hirsch PR, Ostle N, Puissant J, Fagan KC, Pywell RF, Griffiths RI. Bacterial and archaeal taxa are reliable indicators of soil restoration across distributed calcareous grasslands. European Journal of Soil Science, 2020: 12977. |
| [30] |
Fan H, Yao SM, Shu FY, Song WX. The study and comparison of the bacterial diversity from cultured area and uncultured area in Weishan lake. Journal of Qufu Normal University: Natural Science, 2014, 40(3): 81-88.
(in Chinese) 范慧, 姚淑敏, 舒风月, 宋文霞. 微山湖养殖区与非养殖区细菌多样性的研究与比较. 曲阜师范大学学报: 自然科学版, 2014, 40(3): 81-88. |
| [31] | Sangwan P, Kovac S, Davis KER, Sait M, Janssen PH. Detection and cultivation of soil Verrucomicrobia. Applied and Environmental Microbiology, 2005, 71(12): 8402-8410. DOI:10.1128/AEM.71.12.8402-8410.2005 |
| [32] | Anders H, Power JF, MacKenzie AD, Lagutin K, Vyssotski M, Hanssen E, Moreau JW, Stott MB. Limisphaera ngatamarikiensis gen. nov., sp. nov., a thermophilic, pink-pigmented Coccus isolated from subaqueous mud of a geothermal hotspring. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(Pt_4): 1114-1121. DOI:10.1099/ijs.0.000063 |
| [33] | Deng YC, Cui XY, Hernández M, Dumont MG. Microbial diversity in hummock and hollow soils of three wetlands on the Qinghai-Tibetan Plateau revealed by 16S rRNA pyrosequencing. PLoS One, 2014, 9(7): e103115. DOI:10.1371/journal.pone.0103115 |
| [34] | Brown JM, Felice NR, Scalfone NB, Hewson I. Influence of habitat confluence on aquatic microbial assemblages in experimental mesocosms. Aquatic Microbial Ecology, 2012, 66(1): 33-40. DOI:10.3354/ame01550 |
| [35] | Rocarpin P, Gachet S, Metzner K, Saatkamp A. Moisture and soil parameters drive plant community assembly in Mediterranean temporary pools. Hydrobiologia, 2016, 781(1): 55-66. DOI:10.1007/s10750-015-2604-7 |
| [36] | Borchhardt N, Baum C, Mikhailyuk T, Karsten U. Biological soil crusts of arctic svalbard-water availability as potential controlling factor for microalgal biodiversity. Frontiers in Microbiology, 2017, 8: 1485. DOI:10.3389/fmicb.2017.01485 |
| [37] | Hemkemeyer M, Dohrmann AB, Christensen BT, Tebbe CC. Bacterial preferences for specific soil particle size fractions revealed by community analyses. Frontiers in Microbiology, 2018, 9: 149. DOI:10.3389/fmicb.2018.00149 |
| [38] | Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 2009, 75(15): 5111-5120. DOI:10.1128/AEM.00335-09 |
| [39] | Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Gregory Caporaso J, Knight R, Fierer N. Soil bacterial and fungal communities across a pH gradient in an arable soil. The ISME Journal, 2010, 4(10): 1340-1351. DOI:10.1038/ismej.2010.58 |
| [40] | Cho H, Tripathi BM, Moroenyane I, Takahashi K, Kerfahi D, Dong K, Adams JM. Soil pH rather than elevation determines bacterial phylogenetic community assembly on Mt. Norikura. FEMS Microbiology Ecology, 2019, 95(3): fiy216. |
| [41] | Martiny JBH, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: a phylogenetic perspective. Science, 2015, 350(6261): aac9323. DOI:10.1126/science.aac9323 |
| [42] | Herlemann DPR, Lundin D, Andersson AF, Labrenz M, Jürgens K. Phylogenetic signals of salinity and season in bacterial community composition across the salinity gradient of the Baltic sea. Frontiers in Microbiology, 2016, 7(517): 1883. |
| [43] | Mohapatra M, Behera P, Kim JY, Rastogi G. Seasonal and spatial dynamics of bacterioplankton communities in a brackish water coastal lagoon. Science of the Total Environment, 2020, 705: 134729. DOI:10.1016/j.scitotenv.2019.134729 |
| [44] | Szymańska S, Borruso L, Brusetti L, Hulisz P, Furtado B, Hrynkiewicz K. Bacterial microbiome of root-associated endophytes of Salicornia europaea in correspondence to different levels of salinity. Environmental Science and Pollution Research, 2018, 25(25): 25420-25431. DOI:10.1007/s11356-018-2530-0 |
| [45] | Mukhtar S, Mirza BS, Mehnaz S, Mirza MS, McLean J, Malik KA. Impact of soil salinity on the microbial structure of halophyte rhizosphere microbiome. World Journal of Microbiology and Biotechnology, 2018, 34(9): 1-17. DOI:10.1007/s11274-018-2509-5 |
| [46] | Fernández AB, Ghai R, Martin-Cuadrado AB, Sánchez-Porro C, Rodriguez-Valera F, Ventosa A. Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics. FEMS Microbiology Ecology, 2014, 88(3): 623-635. DOI:10.1111/1574-6941.12329 |
| [47] | Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal, 2011, 5(10): 1571-1579. DOI:10.1038/ismej.2011.41 |
| [48] | Iwaoka C, Imada S, Taniguchi T, Du S, Yamanaka N, Tateno R. The impacts of soil fertility and salinity on soil nitrogen dynamics mediated by the soil microbial community beneath the halophytic shrub tamarisk. Microbial Ecology, 2018, 75(4): 985-996. DOI:10.1007/s00248-017-1090-z |
| [49] | Philippot L, Bru D, Saby NPA, Čuhel J, Arrouays D, Šimek M, Hallin S. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environmental Microbiology, 2009, 11(12): 3096-3104. DOI:10.1111/j.1462-2920.2009.02014.x |
| [50] | Rocha UN, Andreote FD, Azevedo JL, Elsas JD, Overbeek LS. Cultivation of hitherto-uncultured bacteria belonging to the Verrucomicrobia subdivision 1 from the potato (Solanum tuberosum L.) rhizosphere. Journal of Soils and Sediments, 2010, 10(2): 326-339. DOI:10.1007/s11368-009-0160-3 |
| [51] | Helgason BL, Konschuh HJ, Bedard-Haughn A, VandenBygaart AJ. Microbial distribution in an eroded landscape: Buried A horizons support abundant and unique communities. Agriculture, Ecosystems & Environment, 2014, 196: 94-102. |
 2021, Vol. 61
2021, Vol. 61




