中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张宏, 李颖杰, 王文颖, 王禄山. 2021
- Zhang Hong, Li Yingjie, Wang Wenying, Wang Lushan. 2021
- 微生物硫循环网络的研究进展
- Research progress of the microbial sulfur-cycling network
- 微生物学报, 61(6): 1567-1581
- Acta Microbiologica Sinica, 61(6): 1567-1581
-
文章历史
- 收稿日期:2020-09-29
- 修回日期:2021-01-02
- 网络出版日期:2021-01-14
2. 青海师范大学生命科学学院, 青海 西宁 810000
2. School of Life Sciences, Qinghai Normal University, Xining 810000, Qinghai Province, China
硫元素是生物体必需的营养元素,是合成氨基酸(甲硫氨酸、半胱氨酸)所必需的原料。自然界中存在丰富的硫源,海洋生态系统中硫酸盐、硫化物储存量巨大[1],不同价态硫化合物之间的转化可主要由代谢功能多样的微生物等来完成,形成硫循环。主要由硫氧化和硫酸盐还原驱动的硫循环与其他重要元素循环(碳、氮、铁、锰)紧密交织,对细胞水平和生态系统水平的过程都具有深远的意义[2]。
研究表明,全球挥发性硫化合物的排放主要是人类活动和海洋微生物产生[3]。硫化氢(H2S)被称作是继CO、NO之后的第三类气体信号分子,能与活性氧相互作用,提高植物非生物胁迫的抗性,但浓度过高会抑制呼吸链致死[4-5]。二甲基硫(DMS)是海洋中含量最丰富的挥发性硫,主要由浮游植物硫代谢产物二甲巯基丙酸(DMSP)酶促氧化产生,二者在推动全球硫循环中发挥着重要作用,并可能对气候产生影响[6]。大气中的硫化合物最终氧化为硫酸盐(SO42–),有助于调节地球辐射,也是植物吸收利用的主要形式,但硫酸盐过多形成的酸雨会损坏森林湖泊生态系统[3]。零价态的单质硫(S0)是具有杀菌性的,因此硫磺可作为无机农药在农业中防治病虫害[7],然而确切的作用机制仍不清楚,有人推测硫对真菌细胞壁的亲脂性或者和巯基氧化性有关[8]。胞内的硫烷硫化学性质活泼,生理功能包括氰化物解毒、抗氧化、代谢调节、信号传递、维持氧化还原状态、参与生物合成等[9-12],但活性硫烷硫的过度累积会对生物体带来损伤。基于不同硫化合物的性质与变化,通过高效准确的硫代谢调控,生物才能趋利避害,因此深入了解并认识微生物硫代谢与高效转化机制就成为研究热点。
硫元素的化学性质活泼,化合价从–2价到+6价不等,根据基团结合形态可分为无机硫和有机硫。无机硫主要包括硫化物(–2价)、单质硫(0价)、二氧化硫(+4价)和硫酸盐(+6价)等。有机硫一般存在于生物体内,含硫化合物可以在微生物的氧化及还原作用下互相转化(图 1)。微生物参与硫循环的作用主要包括:生物氧化过程(硫化物氧化为硫烷、硫酸盐)、同化还原过程(硫酸盐还原为含硫有机物)以及异化还原过程(硫酸盐还原为硫化物)[13-14]。同时,与生物循环紧密结合的还有分解、沉淀、矿化等物理化学过程。
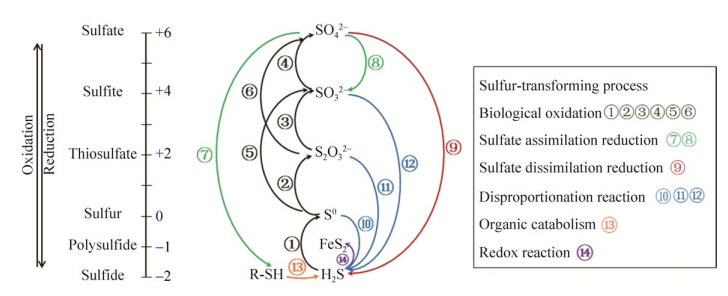
|
| 图 1 生物地球化学硫循环:过程和流动 Figure 1 Biogeochemical sulfur cycling: processes and fluxes. S(–2): H2S, FeS, DMSP; S(–1): FeS2, R-S-S–; S(0): S0, RSSH, DMSO; S(+2): S2O32–, S4O62–; S(+4): SO32–, SO2; S(+6): SO42–. |
在本综述中,我们总结了近年来国内外对微生物硫循环网络的研究进展,包括参与硫循环的主要微生物、微生物参与的硫氧化还原主要反应及其关键功能酶组分。此外,还讨论了硫转化微生物在工业应用中的现状及应用潜能。硫高效转化机制的阐明将有助于提升人们对自然生态环境中存在的硫循环过程的认识,进而为工农业生产中调控硫代谢及利用硫代谢提供理论基础与应用技术。
1 硫循环主要微生物自然界的硫循环需要依靠多种硫氧化或还原相关微生物。硫氧化细菌在好氧或厌氧光照条件下将低价态硫元素氧化为硫酸盐,硫酸盐可在厌氧条件下被硫酸盐还原细菌还原为硫化物。参与硫循环的微生物大部分属于细菌,主要属于变形菌门、厚壁菌门以及绿弯菌门等,以及少部分来自于深海热液喷口等高温环境的古菌[15]。根据不同的代谢方式可以分为光合营养型硫氧化菌、化能营养型硫氧化菌、硫还原菌、硫酸盐还原菌、有机硫降解菌等[16]。其中硫氧化细菌研究最多,根据有无光敏色素可将其大致分为四类(表 1):绿色硫细菌(green sulfur bacteria,GSB)、紫色硫细菌(purple sulfur bacteria,PSB)、紫色非硫细菌(purple non-sulfur bacteria,PNSB)以及无色硫细菌(colorless sulfur bacteria,CSB)[17]。
| Bacteria | Classification | (An)Aerobic/Lithotrophic | Electron donors | Model organisms | Distribution | References |
| Green sulfur bacteria (GSB) | Chlorobi | Anaerobic/photoautotrophy | H2S, S, S2O32– | Chlorobaculum tepidum | Deep sea hydrothermal vents rich in sulfur | [18] |
| Purple sulfur bacteria (PSB) | Chromatiaceae Ectothiorhodospiraceae | Anaerobic/photoautotrophy | S2–, S2O32– | Allochromatium vinosum | Activated sludge estuaries and swamps in freshwater ditches, ponds and lakes containing hydrogen sulfide | [19] |
| Purple non-sulfur bacteria (PNSB) | α-Proteobacteria β-Proteobacteria |
(an)aerobic/phototrophic | H2S, H2 | Rhodopseudomonas palustris | Widely distributed | [20] |
| Colorless sulfur bacteria (CSB) | γ-Proteobacteria Thermoprotei | (an)aerobic/chemolithotrophic | H2S, S2O32– | Beggiatoa leptomitoformis | Activated sludge wastewater treatment system in farmland and orchards and other natural ecological environments | [21-22] |
除硫氧化细菌外,硫酸盐还原菌也是驱动硫循环的重要角色之一。硫酸盐还原是碳循环的主要驱动因素,也是海洋沉积环境及湿地中有机碳到二氧化碳通量的重要部分[23]。大多数具有硫酸盐还原能力的原核生物是细菌,主要存在于深海热液喷口等深海极端生态位和油田环境[24]。硫酸盐还原菌(sulfate reduce bacteria,SRB)具有自养型和异养型,异养型SRB通常是严格厌氧菌。目前根据16S rRNA序列可将硫酸盐还原菌分为5个细菌类群:包括嗜中温G–-δ变形菌纲,形成芽孢的G+-厚壁菌门的Desufotomaculum、Desulfosporomusa、Desulfosporosinus,嗜热细菌SRB- Thermodesulfovibrio,热脱硫菌Thermodesulfobium和Thermodesulfobacterium;以及两个古菌类群:Archaeoglobus,热变形菌目中的Thermocladium和Caldivirga[25-27]。
2 硫转化反应相较于碳和氮代谢,硫循环代谢途径及酶类研究相对较少,但随着高通量测序技术的发展近几年相关进展迅速,已揭示出主要硫代谢细菌进行硫化合物转化的主要代谢途径和关键功能酶。目前对光能自养菌(GSB)和化能自养菌(Acidithiobacillus thiooxidans)的硫氧化关键通路相关基因/酶的模型研究较为清楚(图 2)[28-29]。其中大部分硫氧化菌氧化硫元素时会遵循从低价到高价(H2S-S0-S2O32–-SO42–)的氧化顺序[30]。

|
| 图 2 光能自养(A)和化能自养(B)硫氧化细菌的代谢途径及关键酶(改编自[28-29]) Figure 2 Overview of the key enzymes and metabolic pathways of photoautotrophic (A) and chemotrophic (B) sulfur oxidizing bacteria (adapted with permission from [28-29]). H2S could be converted to S0 by sulfide: quinone oxidoreductase (SQR, in blue) and flavocytochrome c sulfide dehydrogenase (FccAB, in pink); S2O32– could be converted to SO42– by sulfur oxidizing protein system (Sox, in purple), H2S could be converted to SO32– by dissimilatory sulfite reductase system (DSR, in orange). S4I (in cyan) catalyzed the transformation between S2O32– and S4O62–. |
2.1 H2S氧化至零价硫
硫化氢(H2S),无色刺激性气味气体,易溶于水,解离产物以HS–为主[31]。H2S来源广泛,主要来源于火山喷发、含硫的沉积物或热泉释放、工业排放,动植物等生物体也会释放少量硫化氢。微生物产生H2S的途径主要包括硫酸盐还原细菌的还原作用[32-33]、硫氧化细菌中单质硫的歧化作用[34]和含硫氨基酸降解[35-36]。硫化氢的生物氧化包括三个途径:SQR、FCSD和Sox[37-39]。
目前报道的氧化H2S最主要的酶是硫化物醌氧化还原酶(sulfide: quinone oxidoreductase,SQR) (图 3-A),是一种膜蛋白,属于二硫化物氧化还原酶家族,与过硫化物双加氧酶(PDO)广泛存在于厌氧光合细菌、化能自养细菌、无脊椎动物和哺乳动物中。该酶在光合细菌Rhodobacter capsulatus和蓝细菌Oscillatoria limnetica中的研究较为系统[40]。SQRs在结构上分为6个类型,都包含2个绝对保守的半胱氨酸(图 3-B,3-C),形成催化二硫化物氧化还原中心[41],依赖表面的黄素腺嘌呤核苷酸(FAD),催化H2S氧化为硫烷硫(图 3-C),具体催化机理如图 3-D。当有合适受体GSH时会自发形成GSSH,而脱下的2个电子转移给细胞膜上的辅酶Q或甲基奈醌,再通过呼吸链传递给氧;PDO将GSSH氧化为SO32–,SO32–与多硫化物自发反应生成S2O32–,这一途径在异养细菌中很常见[42-44]。
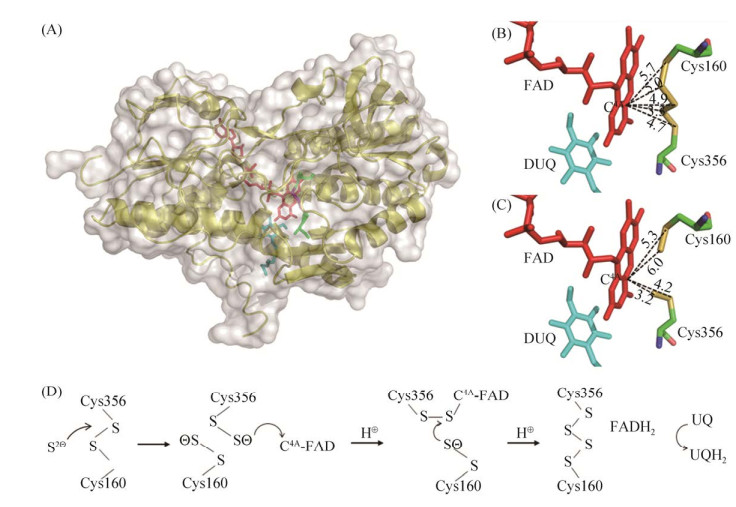
|
| 图 3 SQR的总体结构和活性位点架构(改编自[47-48]) Figure 3 The overall structure and the redox active site of the SQR. A: The overall structure of sulfide: quinone oxidoreductase (SQR) displays for the yellow cartoon, FAD shows as the red stick, DUQ (decyl ubiquinone) stick is shown as blue, green for the conservative cysteine; B: Distance change (unit Å) between the sulfur atom at the redox active site and FAD-C4A before combining with disulfide; C: Distance change (unit Å) between the sulfur atom at the redox active site and FAD-C4A after combining with disulfide; D: Mechanism of disulfide oxidation (A, B, C parts adapted from; the enzyme molecule PDB for 3T0K; part D refers to [47-48]). |
黄素细胞色素c硫化物脱氢酶(FccAB)功能与SQR相同,区别在于电子受体是细胞色素c。FccAB由小亚基细胞色素c蛋白FccA (约20 kDa)和大亚基黄素蛋白FccB (约44 kDa,可结合硫化物)组成[45],可分布于周质空间和内膜上。经KEGG数据库统计,H2S的生物氧化以SQR系统为主,FCSD主要存在于原核生物的5个门(Chlorobi,Aquificae,Deinoccoccus-Thermus,Deferribacter,Proteobacteria)[46]。
2.2 硫烷硫的氧化还原硫烷硫,化合价为0价或–1价的硫原子,一般是一个硫与另外一个或多个硫原子通过共价键形成。在生物体内可以分为低分子量的无机硫烷和结合在蛋白上的有机硫烷,如单质硫、硫代硫酸盐(S2O32–)、过硫化物(GSSH)、多硫化物(R-Sn-R)、连多硫酸盐(-SO3-Sn-SO3-)。硫烷硫在生物体内的产生途径包括SQR氧化H2S生成硫烷硫[42]、氨基酸合成酶CBS/CSE(胱硫醚β合成酶,胱硫醚β裂解酶)以半胱氨酸为底物生成半胱氨酸过硫化物[49]、3-巯基丙酮酸转移酶(MST)催化3-巯基丙酮酸产生多硫化物[50]。
在活性硫烷硫降解途径中存在最为广泛的硫烷硫氧化酶是PDO,属于金属β内酰胺酶家族,这一类酶都含有+2价的金属离子,金属结合中心高度保守,以维持金属离子配位残基和关键残基的取向[51]。在化能自养型细菌Acidithiobacillus thiooxidans外膜上,单质硫(S8)被激活,并作为硫醇结合的硫烷硫原子(R-S-SnH)进入周质空间;在周质空间内硫烷硫以半胱氨酸过硫化物(GSSH)中间体的形式存在于活性位点,在PDO的作用下氧化为SO32–和GSH[42, 52]。SO32–与GSSH可自发反应生成S2O32–,也可由硫氰酸酶或硫转移酶(ST)参与代谢,而PDO-硫氰酸酶融合蛋白在硫代谢中的作用知之甚少[53]。
此外,硫氧化还原酶(SOR)是存在于古菌和化能自养菌细胞质与细胞质膜中的单质硫氧化还原歧化酶,可以将内源或外源性零价硫氧化还原至H2S、S2O32–、SO32–[54]。该反应依赖于氧气,不需要外部辅助因子和电子供体,不伴随电子转移和底物水平磷酸化[55]。所有的SOR似乎都形成了高度热稳定的24个亚基空心球状结构。它们在活性位点携带低电势的单核非血红素铁和必不可少的半胱氨酸。但是,它们的确切反应机理尚不清楚[56]。
2.3 S2O32-氧化至SO42-硫代硫酸盐(S2O32–)的2个硫原子的价态不同,外围硫原子呈现硫烷硫性质,但目前化合价尚有争议,大部分人认为是0价。硫代硫酸盐是生物体较好的硫源,可以经过同化途径产生有机硫[57],胞外进入或S与SO32–自发形成S2O32–最主要的氧化途径是经过Sox系统氧化为SO42–排出体外[58]。
Sox系统是由7个核心蛋白SoxABCDXYZ构成的酶复合物,该系统最先在Paracoccus pantotrophus中发现,只存在于细菌的周质空间。SoxA和SoxX组成的异源二聚体SoxXA[58]与S2O32–形成复合物并释放2个电子,随后与SoxYZ复合物中SoxY的C末端保守氨基酸Cys结合,同时释放SoxXA,随后SoxB催化SoxYZ复合物上的磺酸基团水解生成SO42–,该反应未被直接证实[59]。当存在SoxCD异源四聚体复合物时,催化SoxYZ复合物上剩余的硫烷硫脱氢加氧,释放6个电子,再一次经SoxB水解生成第2个SO42–;当缺少SoxCD时,SoxYZ复合物上的硫烷硫会形成胞内硫粒,通过rDSR系统进一步氧化[40]。在这个过程中,SoxY除了能与S2O32–结合外,还可以与硫化物、零价硫和亚硫酸盐结合。体外实验将SOX系统各个元素组合在一起代谢H2S,但缺少体内验证[37]。
除此之外,连四硫酸盐代谢(S4I)途径广泛分布在β-Proteobacteria和γ-Proteobacteria中,尤其是专性化能自养菌Acidithiobacillus、Thermithiobacillus、Halothiobacillus和Tetrathiobacte等[60-61]。该途径涉及两个酶,膜结合的TQO(硫代硫酸盐醌氧化还原酶)将S2O32–氧化成S4O62–暂时储存,铁氰化物或癸基泛醌(DQ)进行电子传递;而TetH(连四硫酸盐水解酶)可以水解S4O62–生成S2O32–和其他一些产物,该酶的定位和催化机理还需进一步验证[50, 54]。
2.4 SO42-还原成H2S硫酸盐还原菌(SRB)是一类通过异化作用进行SO42–还原代谢的细菌的统称。在异化硫酸盐还原(DSR,也称为硫酸盐呼吸作用)过程中,SO42–通过硫酸盐转运体吸收,由ATP硫酰化酶(Sat)与细胞质中的ATP一起激活,形成腺苷-5’-磷酸硫酸盐(APS)(图 4)。随着腺苷酸(AMP)的释放和两个电子的消耗,APS被腺苷酸硫酸还原酶(Apr)还原为SO32–。最终SO32–还原为S2–,该过程存在两种机理,亚硫酸盐还原酶直接还原[62]和多种酶(亚硫酸盐还原酶、连三硫酸盐还原酶、硫代硫酸盐还原酶)间接还原[63]。DsrC在SO32–的还原过程中作为DsrAB的生理伴侣,根据目前的发现,DsrC和DsrAB是SRB代谢SO32–转化为S2–的核心和关键蛋白[23]。
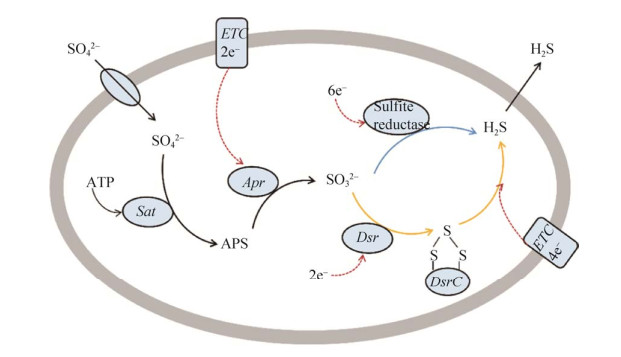
|
| 图 4 异化硫酸盐还原的代谢途径(改编自[63-64]) Figure 4 Metabolic pathway of dissimilatory sulfate reduction (adapted with permission from [63-64]). Active absorption of SO42– by membrane bound sulfate transporters, two hypotheses in the reduction pathway and the passive release of H2S. Hypothesis 1: SO32– is directly reduced to S2–(blue arrow) by sulfite reductase. Hypothesis 2: DsrAB reduces SO32– to DsrC trisulfide, and DsrMKJOP complex reduces the generated DsrC trisulfide to S2– and DsrCr (yellow arrow). |
DsrAB不仅存在于(亚)硫酸盐还原菌、硫代硫酸盐还原菌、硫歧化细菌、有机磺酸盐降解菌中[65],许多光能和化能硫氧化细菌携带一个反向操作的DsrAB (rDsrAB),负责在细胞质中将硫烷或硫化氢氧化形成亚硫酸盐,因此在硫氧化微生物中也叫作反向异化亚硫酸盐还原酶(rDSR)系统[66]。dsrAB编码异化亚硫酸还原酶(DSR)的α和β亚基,在许多以序列为基础的环境生态研究中已作为保守的分子系统发育标记。dsrEFH编码一种胞质硫转运酶,仅存在硫氧化微生物中,被认为是硫氧化剂或硫歧化作用的分子标记[67-68]。不同功能酶标记基因的检测分析,将有助于认识不同生态环境下(也可以是生物反应器中)硫代谢微生物的多样性及其硫元素的生物地球化学过程[69],表 2列举了硫循环网络中主要功能酶的扩增引物指针。
| Enzyme | Gene | Sulfur transformation | Sequence (5'→3') | References |
| Sulfur-oxidizing multienzyme complex, subunit B | soxB | [SoxYZ]-S-S-SO32–→[SoxYZ]-S-S-+SO42– | 710F-ATCGGYCAGGCYTTYCCSTA 1184R-MAVGTGCCGTTGAARTTGC |
[70] |
| Sulfide: quinone oxidoreductase | sqr | H2S→S0, HSn–+reduced quinol | 437F-CATCCTGCTTCGGCCCNGCNTAYGART 982RCCATGGATTCGATRTANCCNGTYT |
[71] |
| Dissimilatory (bi)sulfite reductase, subunits AB | dsrAB | sulfite+[DsrC protein]- dithiol→a [DsrC]-trisulfide | dsr1F-GAAGTATCCCGAGTCGAAGG dsr1R-GCGCCGGGCGGTGCATCTC |
[72] |
| Sulfite: acceptor oxidoreductase/Sulfite oxidizing enzyme | sorAB | SO32–→SO42– | 100F-ACAGCAAGGCATCCnggnttygtng 906R-CCAGAATGTGCCCtccatnayngg |
[71] |
3 微生物硫转化网络
转化硫的微生物有着惊人的多样性,每一种微生物都有不同的生理需求以获得最佳生长。由于自然界的生长条件是高度可变的,很少是最优的,因此单个微生物的硫转化必然是低效的。环境中的硫转化是由比单一微生物更有效地硫循环的微生物群落进行的。硫转化反应是由在自然和人工生态系统中形成复杂网络的微生物连接起来的,如海洋生态系统和脱硫生物反应器等。在这些不同的生态系统中,有效硫循环所需的微生物群落即使在物种组成因环境扰动而改变时,仍保持硫转化反应。
海洋生态系统是世界上最大的自然生态系统,由于广泛的硫循环,几乎达到硫平衡。海水中具有大量的硫酸盐,海洋沉积物中硫元素储存量巨大。硫酸盐在有机碳的氧化驱动下还原为硫化物(图 5-A),大多SRM属于Deltaproteobacteria,包括Desulfovibrionales和Desulfobacterales。在这样的缺氧沉积物中,Mn氧化物,特别是Fe氧化物,是硫化物的主要化学氧化剂,而一部分硫化物的氧化是由微生物通过硝酸盐介导的[63]。基于宏基因组学和16S rRNA研究,沉积物中潜在的硫化物氧化微生物种类繁多[2]。
含硫化物贫乏的深层地下水拥有一个活跃的微生物群落硫酸盐还原和硫还原细菌,共同介导了一个硫循环(图 5-B)。通过宏基因组和宏蛋白质组方法的结合揭示硫循环的主要驱动因素是微生物硫酸盐还原,Deltaproteobacteria被证明具有硫酸盐还原和可能的硫歧化的遗传能力,而Rhizobiaceae、Rhodocyclaceae、Sideroxydans和Sulfurimonas能够氧化还原硫化合物[73]。
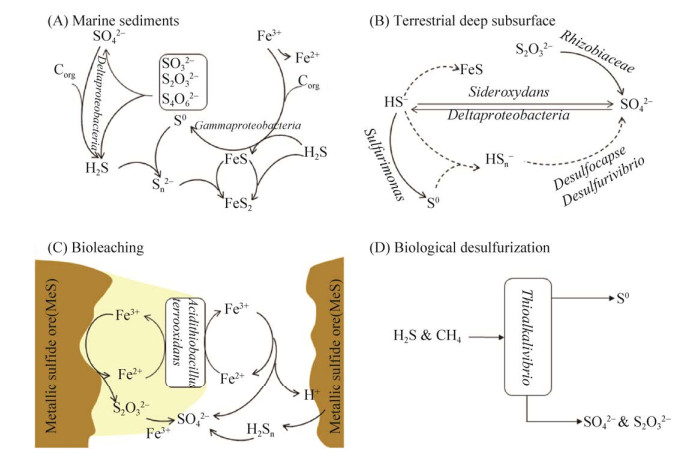
|
| 图 5 不同生境下的硫循环过程 Figure 5 Sulfur cycle in different habitats. A: marine sediments; B: Deep groundwater; C: Biological leaching; D: Biological desulfurization. |
Acidithiobacillus是一类嗜酸硫氧化菌,该菌株被广泛应用到生物浸出领域,将不溶性固体中的金属离子转化为水溶性形式,从而实现有色金属的回收。从硫化矿石中生物浸出金属的机制目前有三种解释:接触、非接触、协同作用(图 5-C)[74]。生物浸出菌主要分布在变形菌门(Acidithiobacillus,Acidiphilium,Acidiferrobacter,Ferrovum),硝化螺旋菌门(Leptospirillum),厚壁菌门(Alicyclobacillus,Sulfobacillus),放线菌门(Ferrimicrobium, Acidimicrobium,Ferrithrix),和古细菌(Sulfolobus,Acidianus,Metallosphaera,Sulfurisphaera)[75]。
生物脱硫是对工业过程、废水处理和垃圾填埋场中产生和排放的含硫化合物(例如硫化氢、二氧化硫、硫醚和硫醇)废气的一种经济有效的脱硫方法[76]。沼气中的H2S在生物脱硫过程中被硫氧化细菌(SOB)去除,形成的硫颗粒可以用作肥料和用于硫酸生产(图 5-D)[77]。Acidithiobacillus sp.、Thiobacillus sp.和Thioalkalivibrio分别是在酸性、中性和碱性pH条件下H2S生物滤池中发现的最常见的硫化物氧化细菌[77-78]。最近,一种新的硫转换相关强化生物除磷(S-EBPR)工艺已被开发用于处理富硫酸盐废水,这一过程成功地整合了硫、碳、氮和磷循环,以便同时代谢或去除碳、氮、硫和磷[79]。在检测到的SRB中,Desulfovibrio、Desulfobulbus、Desulfomicrobium和Desulfobacter是硫酸盐还原生物反应器中最常见的SRB属[80-81]。
4 展望硫是生物必需的营养元素之一,自然界中广泛存在不同形式的硫。微生物在有机硫和无机硫的转化过程中发挥了关键作用,有机硫化物依次被降解为甲硫醇(CH3SH)、含硫氨基酸(半胱氨酸、甲硫氨酸)和硫化氢,硫氧化细菌再将还原态硫化物(不)完全氧化为多种无机硫化合物[43]。基于分子生物学的方法已经揭示了参与硫循环微生物极其多样的代谢途径、功能酶及电子传递机制等信息,提高了人们对硫循环过程的认知。目前,硫代谢微生物已经被广泛应用于生物浸出、含硫废弃物脱硫等工业领域[71-73]。然而由于工业硫化物排放的减少以及生物硫化氢解毒技术的开发应用,使得硫循环平衡受到明显的影响,如土壤生态系统中硫元素的缺乏导致作物减产[82-83],如何分析研究并维持自然界的硫循环及其动态平衡成为新的研究热点。我们前期工作研究表明,在添加硫粉的生物质堆肥过程中,发生了S0→S2O32–→SO42–的物质转化,通过微生物多样性测序发现Proteobacteria含量明显增加(课题组待发表数据),该过程的发现可为农业应用中硫元素的补充提供一条安全环保的途径。随着新一代组学技术的发展与应用,将有助于人们利用更好更全面的技术分析在不同生态系统下微生物参与硫循环的动态过程及其网络协同变化,如基于宏基因组学及宏转录组学探究地下水中不同生物地球化学碳、氮、硫循环的基因功能相耦联机制[84]。整合多组学技术、大数据平台及其合成组学的思想与技术的成熟,将会使得人们全面深入了解更多生态环境中硫代谢微生物的动态转化机制与控制策略,结合现代控制理论与工艺技术就可以形成硫元素高效循环与利用的新途径,从而形成新的应用方案与工艺路线。
| [1] |
Hu X, Liu JH, Liu HW, Zhuang GC, Xun LY. Sulfur metabolism by marine heterotrophic bacteria involved in sulfur cycling in the ocean. Science China Earth Sciences, 2018, 48(12): 1540-1550.
(in Chinese) 胡欣, 刘纪化, 刘怀伟, 庄光超, 荀鲁盈. 异养细菌硫代谢及其在海洋硫循环中的作用. 中国科学: 地球科学, 2018, 48(12): 1540-1550. |
| [2] | Wasmund K, Mußmann M, Loy A. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environmental Microbiology Reports, 2017, 9(4): 323-344. DOI:10.1111/1758-2229.12538 |
| [3] | Kesselmeier J. The global sulfur cycle and China's contribution to atmospheric sulfur loads. Landbauforschung Volkenrode, 2005, 283: 67-72. |
| [4] | Wang R. Hydrogen sulfide: the third gasotransmitter in biology and medicine. Antioxidants & Redox Signaling, 2010, 12(9): 1061-1064. |
| [5] | Singh S, Kumar V, Kapoor D, Kumar S, Singh S, Dhanjal DS, Datta S, Samuel J, Dey P, Wang SQ, Prasad R, Singh J. Revealing on hydrogen sulfide and nitric oxide signals co-ordination for plant growth under stress conditions. Physiologia Plantarum, 2020, 168(2): 301-317. |
| [6] | Zhang XH, Liu J, Liu JL, Yang GP, Xue CX, Curson ARJ, Todd JD. Biogenic production of DMSP and its degradation to DMS-their roles in the global sulfur cycle. Science China Life Sciences, 2019, 62(10): 1296-1319. DOI:10.1007/s11427-018-9524-y |
| [7] | Griffith CM, Woodrow JE, Seiber JN. Environmental behavior and analysis of agricultural sulfur. Pest Management Science, 2015, 71(11): 1486-1496. DOI:10.1002/ps.4067 |
| [8] | Williams JS, Cooper RM. The oldest fungicide and newest phytoalexin-a reappraisal of the fungitoxicity of elemental sulphur. Plant Pathology, 2004, 53(3): 263-279. DOI:10.1111/j.0032-0862.2004.01010.x |
| [9] | Iciek M, Wlodek L. Biosynthesis and biological properties of compounds containing highly reactive, reduced sulfane sulfur. Polish Journal of Pharmacology, 2001, 53(3): 215-225. |
| [10] | Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nature Chemical Biology, 2006, 2(4): 185-194. DOI:10.1038/nchembio779 |
| [11] | Toohey JI. The conversion of H2S to sulfane sulfur. Nature Reviews Molecular Cell Biology, 2012, 13(12): 803. |
| [12] | Toohey JI. Sulphane sulphur in biological systems: a possible regulatory role. Biochemical Journal, 1989, 264(3): 625-632. DOI:10.1042/bj2640625 |
| [13] | Leloup J, Fossing H, Kohls K, Holmkvist L, Borowski C, Jørgensen BB. Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environmental Microbiology, 2009, 11(5): 1278-1291. DOI:10.1111/j.1462-2920.2008.01855.x |
| [14] | Thiel J, Byrne JM, Kappler A, Schink B, Pester M. Pyrite formation from FeS and H2S is mediated through microbial redox activity. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(14): 6897-6902. DOI:10.1073/pnas.1814412116 |
| [15] | Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nature Communications, 2018, 9(1): 4999. DOI:10.1038/s41467-018-07418-0 |
| [16] | Sievert SM, Kiene RP, Schulz-Vogt HN. The sulfur cycle. Oceanography, 2007, 20(2): 117-123. DOI:10.5670/oceanog.2007.55 |
| [17] | 罗剑飞. 硫氧化菌群落结构分析及其特性研究. 华南理工大学博士学位论文, 2011. |
| [18] | Sakurai H, Ogawa T, Shiga M, Inoue K. Inorganic sulfur oxidizing system in green sulfur bacteria. Photosynthesis Research, 2010, 104(2/3): 163-176. |
| [19] | Ghosh S, Bagchi A. Insight into the molecular mechanism of the sulfur oxidation process by reverse sulfite reductase (rSiR) from sulfur oxidizer Allochromatium vinosum. Journal of Molecular Modeling, 2018, 24(5): 117. DOI:10.1007/s00894-018-3652-5 |
| [20] | Baars O, Morel FMM, Zhang XN. The purple non-sulfur bacterium Rhodopseudomonas palustris produces novel petrobactin-related siderophores under aerobic and anaerobic conditions. Environmental Microbiology, 2018, 20(5): 1667-1676. DOI:10.1111/1462-2920.14078 |
| [21] | Fomenkov A, Vincze T, Grabovich MY, Dubinina G, Orlova M, Belousova E, Roberts RJ. Complete genome sequence of the freshwater colorless sulfur bacterium Beggiatoa leptomitiformis neotype strain D-402T. Genome Announcements, 2015, 3(6): e01436-15. |
| [22] | Jørgensen BB, Revsbech NP. Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp., in O2 and H2S microgradients. Applied and Environmental Microbiology, 1983, 45(4): 1261-1270. DOI:10.1128/aem.45.4.1261-1270.1983 |
| [23] | Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, Rappé MS, Pester M, Loy A, Thomas BC, Banfield JF. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. The ISME Journal, 2018, 12(7): 1715-1728. DOI:10.1038/s41396-018-0078-0 |
| [24] | Meier DV, Pjevac P, Bach W, Markert S, Schweder T, Jamieson J, Petersen S, Amann R, Meyerdierks A. Microbial metal-sulfide oxidation in inactive hydrothermal vent chimneys suggested by metagenomic and metaproteomic analyses. Environmental Microbiology, 2019, 21(2): 682-701. DOI:10.1111/1462-2920.14514 |
| [25] | Barton LL, Fauque GD. Chapter 2 biochemistry, physiology and biotechnology of sulfate-reducing bacteria. Advances in Applied Microbiology, 2009, 68: 41-98. |
| [26] | Castro HF, Williams NH, Ogram A. Phylogeny of sulfate-reducing bacteria. FEMS Microbiology Ecology, 2000, 31(1): 1-9. |
| [27] | Mori K, Kim H, Kakegawa T, Hanada S. A novel lineage of sulfate-reducing microorganisms: thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles, 2003, 7(4): 283-290. DOI:10.1007/s00792-003-0320-0 |
| [28] | Gregersen LH, Bryant DA, Frigaard NU. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Frontiers in Microbiology, 2011, 2: 116. |
| [29] | Yin HQ, Zhang X, Li XQ, He ZL, Liang YL, Guo X, Hu Q, Xiao YH, Cong J, Ma LY, Niu JJ, Liu XD. Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiology, 2014, 14(1): 179. DOI:10.1186/1471-2180-14-179 |
| [30] | 辛玉峰. 异养细菌硫化物氧化途径及产物分析. 山东大学博士学位论文, 2016. |
| [31] | Kabil O, Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxidants & Redox Signaling, 2014, 20(5): 770-782. |
| [32] | Kováč J, Vítězová M, Kushkevych I. Metabolic activity of sulfate-reducing bacteria from rodents with colitis. Open Medicine, 2018, 13(1): 344-349. DOI:10.1515/med-2018-0052 |
| [33] | Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. The ISME Journal, 2015, 9(5): 1152-1165. DOI:10.1038/ismej.2014.208 |
| [34] | Sun CW, Chen ZW, He ZG, Zhou PJ, Liu SJ. Purification and properties of the sulfur oxygenase/reductase from the acidothermophilic archaeon, Acidianus strain S5. Extremophiles, 2003, 7(2): 131-134. DOI:10.1007/s00792-002-0304-5 |
| [35] | Mironov A, Seregina T, Nagornykh M, Luhachack LG, Korolkova N, Lopes LE, Kotova V, Zavilgelsky G, Shakulov R, Shatalin K, Nudler E. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(23): 6022-6027. DOI:10.1073/pnas.1703576114 |
| [36] | Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science, 2011, 334(6058): 986-990. DOI:10.1126/science.1209855 |
| [37] | Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. Journal of Bacteriology, 2000, 182(17): 4677-4687. DOI:10.1128/JB.182.17.4677-4687.2000 |
| [38] | Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS Journal, 2008, 275(13): 3352-3361. DOI:10.1111/j.1742-4658.2008.06482.x |
| [39] | Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R. Organization of the human mitochondrial hydrogen sulfide oxidation pathway. Journal of Biological Chemistry, 2014, 289(45): 30901-30910. DOI:10.1074/jbc.M114.602664 |
| [40] | Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Advances in Microbial Physiology, 2008, 54: 103-200. |
| [41] | Shen JC, Peng H, Zhang YX, Trinidad JC, Giedroc DP. Staphylococcus aureus sqr encodes a type II sulfide: quinone oxidoreductase and impacts reactive sulfur speciation in cells. Biochemistry, 2016, 55(47): 6524-6534. DOI:10.1021/acs.biochem.6b00714 |
| [42] | Liu DX, Zhang JJ, Lü CJ, Xia YZ, Liu HW, Jiao NZ, Xun LY, Liu JH. Synechococcus sp. strain PCC7002 uses sulfide: quinone oxidoreductase to detoxify exogenous sulfide and to convert endogenous sulfide to cellular sulfane sulfur. mBio, 2020, 11(1): e03420-19. |
| [43] | Xia YZ, Lü CJ, Hou NK, Xin YF, Liu JH, Liu HL, Xun LY. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. The ISME Journal, 2017, 11(12): 2754-2766. DOI:10.1038/ismej.2017.125 |
| [44] | Xin YF, Liu HL, Cui FF, Liu HW, Xun LY. Recombinant Escherichia coli with sulfide: quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environmental Microbiology, 2016, 18(12): 5123-5136. DOI:10.1111/1462-2920.13511 |
| [45] | Barton LL, Fardeau ML, Fauque GD. Hydrogen sulfide: a toxic gas produced by dissimilatory sulfate and sulfur reduction and consumed by microbial oxidation//Kroneck P, Torres M. The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. Dordrecht: Springer, 2014: 237-277. |
| [46] | Sousa FM, Pereira JG, Marreiros BC, Pereira MM. Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2018, 1859(9): 742-753. DOI:10.1016/j.bbabio.2018.04.004 |
| [47] | Zhang YF, Weiner JH. Characterization of the kinetics and electron paramagnetic resonance spectroscopic properties of Acidithiobacillus ferrooxidans sulfide: quinone oxidoreductase (SQR). Archives of Biochemistry and Biophysics, 2014, 564: 110-119. DOI:10.1016/j.abb.2014.09.016 |
| [48] | Cherney MM, Zhang YF, Solomonson M, Weiner JH, James MNG. Crystal structure of sulfide: quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. Journal of Molecular Biology, 2010, 398(2): 292-305. DOI:10.1016/j.jmb.2010.03.018 |
| [49] | Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(21): 7606-7611. DOI:10.1073/pnas.1321232111 |
| [50] | Wang R, Lin JQ, Liu XM, Pang X, Zhang CJ, Yang CL, Gao XY, Lin CM, Li YQ, Li Y, Lin JQ, Chen LX. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Frontiers in Microbiology, 2019, 9: 3290. DOI:10.3389/fmicb.2018.03290 |
| [51] | Sattler SA, Wang X, Lewis KM, DeHan PJ, Park CM, Xin YF, Liu HL, Xian M, Xun LY, Kang CH. Characterizations of two bacterial persulfide dioxygenases of the metallo-β-lactamase superfamily. Journal of Biological Chemistry, 2015, 290(31): 18914-18923. DOI:10.1074/jbc.M115.652537 |
| [52] | Holdorf MM, Owen HA, Lieber SR, Yuan L, Adams N, Dabney-Smith C, Makaroff CA. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiology, 2012, 160(1): 226-236. DOI:10.1104/pp.112.201855 |
| [53] | Motl N, Skiba MA, Kabil O, Smith JL, Banerjee R. Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation. Journal of Biological Chemistry, 2017, 292(34): 14026-14038. DOI:10.1074/jbc.M117.790170 |
| [54] | Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiology Reviews, 2009, 33(6): 999-1043. DOI:10.1111/j.1574-6976.2009.00187.x |
| [55] | Urich T, Gomes CM, Kletzin A, Frazão C. X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science, 2006, 311(5763): 996-1000. DOI:10.1126/science.1120306 |
| [56] | Rühl P, Pöll U, Braun J, Klingl A, Kletzin A. A sulfur oxygenase from the haloalkaliphilic bacterium Thioalkalivibrio paradoxus with atypically low reductase activity. Journal of Bacteriology, 2017, 199(4): e00675-16. |
| [57] | Stoffels L, Krehenbrink M, Berks BC, Unden G. Thiosulfate reduction in Salmonella enterica is driven by the proton motive force. Journal of Bacteriology, 2012, 194(2): 475-485. DOI:10.1128/JB.06014-11 |
| [58] | Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism?. Applied and Environmental Microbiology, 2001, 67(7): 2873-2882. DOI:10.1128/AEM.67.7.2873-2882.2001 |
| [59] | Grabarczyk DB, Chappell PE, Johnson S, Stelzl LS, Lea SM, Berks BC. Structural basis for specificity and promiscuity in a carrier protein/enzyme system from the sulfur cycle. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(52): E7166-E7175. DOI:10.1073/pnas.1506386112 |
| [60] | Kanao T, Onishi M, Kajitani Y, Hashimoto Y, Toge T, Kikukawa H, Kamimura K. Characterization of tetrathionate hydrolase from the marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Bioscience, Biotechnology, and Biochemistry, 2018, 82(1): 152-160. DOI:10.1080/09168451.2017.1415128 |
| [61] | Wang L, Guo X, Li H, Qi HZ, Qian J, Yan SS, Shi JL, Niu WN. Hydrogen sulfide from cysteine desulfurase, not 3-mercaptopyruvate sulfurtransferase, contributes to sustaining cell growth and bioenergetics in E. coli under anaerobic conditions. Frontiers in Microbiology, 2019, 10: 2357. DOI:10.3389/fmicb.2019.02357 |
| [62] | Peck HD, LeGall J. Biochemistry of dissimilatory sulphate reduction. Philosophical Transactions of the Royal Society B, 1982, 298(1093): 443-466. |
| [63] | Jørgensen BB, Findlay AJ, Pellerin A. The biogeochemical sulfur cycle of marine sediments. Frontiers in Microbiology, 2019, 10: 849. DOI:10.3389/fmicb.2019.00849 |
| [64] | Zeng Q, Hao TW, Mackey HR, van Loosdrecht MCM, Chen GH. Recent advances in dissimilatory sulfate reduction: from metabolic study to application. Water Research, 2019, 150: 162-181. DOI:10.1016/j.watres.2018.11.018 |
| [65] | Löffler M, Feldhues J, Venceslau SS, Kammler L, Grein F, Pereira IAC, Dahl C. DsrL mediates electron transfer between NADH and rDsrAB in Allochromatium vinosum. Environmental Microbiology, 2020, 22(2): 783-795. DOI:10.1111/1462-2920.14899 |
| [66] | Loy A, Duller S, Baranyi C, Mußmann M, Ott J, Sharon I, Béjà O, Le Paslier D, Dahl C, Wagner M. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environmental Microbiology, 2009, 11(2): 289-299. DOI:10.1111/j.1462-2920.2008.01760.x |
| [67] | Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, Deuster O, Brune DC. Novel genes of the dsr gene cluster and evidence for close interaction of dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. Journal of Bacteriology, 2005, 187(4): 1392-1404. DOI:10.1128/JB.187.4.1392-1404.2005 |
| [68] | Thorup C, Schramm A, Findlay AJ, Finster KW, Schreiber L. Disguised as a sulfate reducer: growth of the deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. mBio, 2017, 8(4): e00671-17. |
| [69] | Luo JF, Tan XQ, Liu KX, Lin WT. Survey of sulfur-oxidizing bacterial community in the Pearl River water using soxB, sqr, and dsrA as molecular biomarkers. 3 Biotech, 2018, 8(1): 73. DOI:10.1007/s13205-017-1077-y |
| [70] | Tian HM, Gao PK, Chen ZH, Li YS, Li Y, Wang YS, Zhou JF, Li GQ, Ma T. Compositions and abundances of sulfate-reducing and sulfur-oxidizing microorganisms in water-flooded petroleum reservoirs with different temperatures in China. Frontiers in Microbiology, 2017, 8: 143. |
| [71] | Luo JF, Lin WT, Guo Y. Functional genes based analysis of sulfur-oxidizing bacteria community in sulfide removing bioreactor. Applied Microbiology and Biotechnology, 2011, 90(2): 769-778. DOI:10.1007/s00253-010-3061-x |
| [72] | Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M, Reinhardt R, Brinkhoff T, Harder J, Amann R, Mußmann M. Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. The ISME Journal, 2012, 6(12): 2178-2187. DOI:10.1038/ismej.2012.66 |
| [73] | Bell E, Lamminmäki T, Alneberg J, Andersson AF, Qian C, Xiong WL, Hettich RL, Frutschi M, Bernier-Latmani R. Active sulfur cycling in the terrestrial deep subsurface. The ISME Journal, 2020, 14(5): 1260-1272. DOI:10.1038/s41396-020-0602-x |
| [74] | Zhang S, Yan L, Xing WJ, Chen P, Zhang Y, Wang WD. Acidithiobacillus ferrooxidans and its potential application. Extremophiles, 2018, 22(4): 563-579. DOI:10.1007/s00792-018-1024-9 |
| [75] | Fonti V, Dell'Anno A, Beolchini F. Does bioleaching represent a biotechnological strategy for remediation of contaminated sediments?. Science of the Total Environment, 2016, 563-564: 302-319. DOI:10.1016/j.scitotenv.2016.04.094 |
| [76] | Li L, Zhang JY, Lin J, Liu JX. Biological technologies for the removal of sulfur containing compounds from waste streams: bioreactors and microbial characteristics. World Journal of Microbiology and Biotechnology, 2015, 31(10): 1501-1515. DOI:10.1007/s11274-015-1915-1 |
| [77] | Kiragosyan K, Klok JBM, Keesman KJ, Roman P, Janssen AJH. Development and validation of a physiologically based kinetic model for starting up and operation of the biological gas desulfurization process under haloalkaline conditions. Water Research X, 2019, 4: 100035. DOI:10.1016/j.wroa.2019.100035 |
| [78] | Montebello AM, Mora M, López LR, Bezerra T, Gamisans X, Lafuente J, Baeza M, Gabriel D. Aerobic desulfurization of biogas by acidic biotrickling filtration in a randomly packed reactor. Journal of Hazardous Materials, 2014, 280: 200-208. DOI:10.1016/j.jhazmat.2014.07.075 |
| [79] | Guo G, Ekama GA, Wang YY, Dai J, Biswal BK, Chen GH, Wu D. Advances in sulfur conversion-associated enhanced biological phosphorus removal in sulfate-rich wastewater treatment: a review. Bioresource Technology, 2019, 285: 121303. DOI:10.1016/j.biortech.2019.03.142 |
| [80] | Hao TW, Xiang PY, Mackey HR, Chi K, Lu H, Chui HK, van Loosdrecht MCM, Chen GH. A review of biological sulfate conversions in wastewater treatment. Water Research, 2014, 65: 1-21. DOI:10.1016/j.watres.2014.06.043 |
| [81] | Kushkevych I, Kováč J, Vítězová M, Vítěz T, Bartoš M. The diversity of sulfate-reducing bacteria in the seven bioreactors. Archives of Microbiology, 2018, 200(6): 945-950. DOI:10.1007/s00203-018-1510-6 |
| [82] | Bouranis DL, Malagoli M, Avice JC, Bloem E. Advances in plant sulfur research. Plants (Basel), 2020, 9(2): 256. |
| [83] | Wang SP, Wang YF, Chen ZZ, Schnug E, Haneklaus S. Sulphur concentration of soils and plants and its requirement for ruminants in the Inner Mongolia steppe of China. Grass and Forage Science, 2001, 56(4): 418-422. DOI:10.1046/j.1365-2494.2001.00285.x |
| [84] | Wegner CE, Gaspar M, Geesink P, Herrmann M, Marz M, Küsel K. Biogeochemical regimes in shallow aquifers reflect the metabolic coupling of the elements nitrogen, sulfur, and carbon. Applied Microbiology and Biotechnology, 2019, 85(5): e02346-18. |
 2021, Vol. 61
2021, Vol. 61




