中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 刘忠林, 刘一凡, 林丹丹, 周蕾, 牟伯中. 2021
- Zhonglin Liu, Yifan Liu, Dandan Lin, Lei Zhou, Bo-Zhong Mu. 2021
- 暗黑菌(Atribacteria)的环境分布和功能特征
- Environmental distribution and functions of the Atribacteria
- 微生物学报, 61(6): 1416-1427
- Acta Microbiologica Sinica, 61(6): 1416-1427
-
文章历史
- 收稿日期:2021-03-23
- 修回日期:2021-04-18
- 网络出版日期:2021-04-30
微生物是地球上最丰富的细胞生命形式,广泛参与到地球化学循环过程中。然而,绝大部分微生物难以通过现有的培养手段分离得到纯菌株[1],目前对于此类微生物的研究,仍然主要借助于宏基因组(metagenomics)[2]和单细胞测序(single cell sequencing,SCS)[3]等不依赖于单菌分离的技术手段。针对环境中含量相对丰富的微生物,可以通过宏基因组测序以及拼接(assembly) 和分箱(binning)操作来获得其完整或接近完整基因组;单细胞测序可将环境中低丰度微生物通过荧光激活细胞分选(fluorescence activated cell sorting,FACS)或微操技术(包括微流分选、光学捕获和微移液)分离得到单个细胞,并测序获得其基因组(metagenome assembly genomes,MAGs;single-cell amplified genomes,SAGs)。基于这两种测序技术手段,目前已经从多个环境中发现了许多此前未曾认识的细菌门类。这些微生物普遍缺乏分离培养的代表菌株,因此通常被称为候选门(candidate phyla)[4-5]。
暗黑菌属于目前新发现的细菌门之一,被称为暗黑菌门(意指生长在黑暗厌氧环境中的一类未获得纯培养细菌;the dark bacterial phylum[6]),它包含OP9和JS1两大分支[7]。OP9和JS1在自然界广泛分布,在一些特殊环境的微生物群落中甚至占主要地位,如海洋沉积物[8-9]、地热环境[10]、厌氧反应器[11]以及油藏环境[12]。根据目前已获得的环境样品基因组信息推断,暗黑菌门的成员具有至少3种碳氢化合物代谢途径,推测它是地球元素循环中重要的一环。然而,目前仅有一例纯培养菌株Atribacter laminatus RT761的报道,对其生态功能认知仍局限于基因组信息推测[13]。本文将对暗黑菌的分类、代谢及生态功能做一个系统性梳理,并对未来的研究方向进行展望。
1 暗黑菌门的确立暗黑菌门(Atribacteria)包含OP9和JS1两个谱系。其中OP9最早是由Hugenholtz等[14]在1998年通过16S rRNA基因克隆建库方法,在美国黄石国家公园的黑曜石池(Obsidian Pool)沉积物中发现的12种新的细菌谱系(OP1–OP12)中的一种。然而,由于长期以来缺乏纯培养菌株和相关基因组信息,对此类微生物功能的认识仍然有限。Dodsworth等[6]首次借助宏组学和单细胞测序技术,从热泉沉积物和原位富集的纤维素分解嗜热性群落种获得了2个接近完整的OP9基因组,分别命名为OP9-cSCG和OP9-77CS。根据系统生物学分析将这2个基因组所代表的菌种命名为Candidatus Caldatribacterium californiense和Ca. Caldatribacterium saccharofermentans,并建议将其归类到新的细菌门类——暗黑菌门Atribacteria。另一方面,JS1成员最早于1994年在日本海样品中发现(Japan Sea,–78 m),随后Webster等对多个环境样品(包括海底沉积物、花园土壤、海水及河水等)进行检测,在样品中均检测到与OP9相似同源片段,并提出将其命名为JS1。由于早期缺乏足够的参考基因序列,当时的进化发育学分析错误地将它归纳为一个新的细菌门水平分支[15]。后来,根据Yarza等[16]在2014年建立的基于16S rRNA基因序列同一性的微生物分类标准,Nobu等[7]重新分析后指出OP9和JS1成员的16S rRNA基因序列同一性的中值(80.8%)和最低值(74.2%)均低于细菌门类划分参考值范围(参考中间值83.68%,最低值77.43%),因此建议将这2个谱系重新归纳到暗黑菌门下。Chakraborty等[17]根据所获得的近全长(> 1000 bp) 16S rRNA基因序列,使用4种不同的计算方法计算,成功构建出一个高分辨水平系统发育进化树,根据基因序列同一性的最低值分类标准(minimum sequence identity,MSI),指出暗黑菌门类中应该包含OP9和JS1两个纲水平分支(MSI值为80.4%),3个目水平的进化枝(clade) (MSI值为83.6%),至少包含11个科水平的进化枝(MSI值为87.7%),并且随着研究的不断进行,属水平的进化枝数量在不断扩充(MSI值为94.8%),对暗黑菌的研究也成为了目前的研究焦点。
2 暗黑菌的环境分布和多样性 2.1 环境分布暗黑菌(OP9/JS1)在自然环境中分布广泛,其中OP9广泛存在于一些高温环境,如陆地热泉、高温厌氧反应器等;而JS1则多见于中低温环境,如深海沉积物、可燃冰富集区(低温、高压),在富含有机物或含天然气水合物沉积环境中往往占主导地位(丰度占比≥50%)[18]。近年来,随着宏组学的发展和多环境样品的广泛检测,在咸水湖[19]、甲烷形成区[17, 20]、地下水[21]等环境样品的宏基因组中也频繁检测到暗黑菌的同源片段存在。环境分布可以参见图 1。

|
| 图 1 暗黑菌(OP9/JS1)的环境分布 Figure 1 Environmental distribution of Atribacteria (OP9/JS1). |
目前已经在美国火山附近热溪(约80 ℃)[22]、热泉(> 73 ℃)[23]以及加拿大高温地热泉(81 ℃)[24]沉积物中,通过16S rRNA基因文库构建与分析发现了OP9细菌的存在;在多个处理生活废水的高温厌氧反应器中也发现少量OP9 (细菌中丰度占比分别为≤5%、≤6%和≤1%)[25-27]。此外,在丹麦油田产出水Dan60S样品中以及鄂霍次克海海底沉积物中都检测到了OP9 (≤3%)的16S rRNA基因序列[28-29],进一步扩大了对其环境分布的认知。
自JS1被发现以来,研究者们已在多个深海沉积物样品中检测到JS1细菌的存在。研究发现,随着海洋环境深度增加,其在细菌群落中的丰度占比呈上升趋势[30-31]。例如,在南海北部、罗斯海(Ross sea)、秘鲁海洋钻井项目(Peru margin ODP)等海洋沉积物中发现大量的JS1,在细菌中丰度占比分别为0.07%–8.78%、9.8%–40.8%、6%–7% (随深度变化)[31-33];在日本Niibori油田和Medicine Hat Glauconitic C (MHGC)油田产出水及胜利油田含油污泥富集培养物中[34-36],均检测到JS1基因片段,细菌中丰度占比分别为60%、9%及30%。此外,在厌氧反应器中也发现有JS1菌的分布(< 1%)[26]。
2.2 暗黑菌的多样性在暗黑菌的环境分布以及多样性的调查中,研究者设计并开发了一系列特异性的引物和探针应用于暗黑菌的快速鉴定(表 1)。2004年,Webster等首次设计针对JS1 16S rRNA基因的特异性引物63F/665R,并在随后的实验中使用该引物对多个环境样品进行了检测分析[8, 15, 37]。然而,结果显示,引物63F/665R的特异性不高,不能很好地区分暗黑菌和环境样品中其他微生物。为此,Blazejak等[32]在引物63F/665R基础上改进并设计出新的16S rRNA基因特异性引物519F/665R,并用q-PCR实验来定量检测深海样品中的暗黑菌。目前针对16S rRNA基因的特异性引物还较少,随着越来越多的暗黑菌相关序列的发现,相信在这些扩充的16S rRNA基因信息基础上,未来有望设计出兼顾通用性和特异性的相关引物。
| Primer | Target | Sequence (5′→3′) | References |
| 8F | Universal primers | AGAGTTTGATCCTGGCTCAG | [38] |
| 1390R | GACGGGCGGTGTGTACAA | [39] | |
| 27F | Universal primers | AGRGTTTGATCCTGGCTCAG | [40] |
| 1492R | CGGCTACCTTGTTACGACTT | ||
| 926F | (SSU) rRNA | cct atc ccc tgt gtg cct tgg cag tct cag AAA CTY AAA KGA ATT GRC GG | [41-42] |
| 1392R | cca tct cat ccc tgc gtg tct ccg act cag-XXXXX-ACG GGC GGT GTG TRC | ||
| 63F | JS1 | CAGGCCTAACACATGCAAGTC | [43] |
| 519F | CAGCAGCCGCGGTAAYAC | [43][32][15][9] | |
| 665R | ACCGGGAATTCCACYTYCCT | [15] | |
| Atri578 | ACTTTTAAGACCGCCTACGA | [9] |
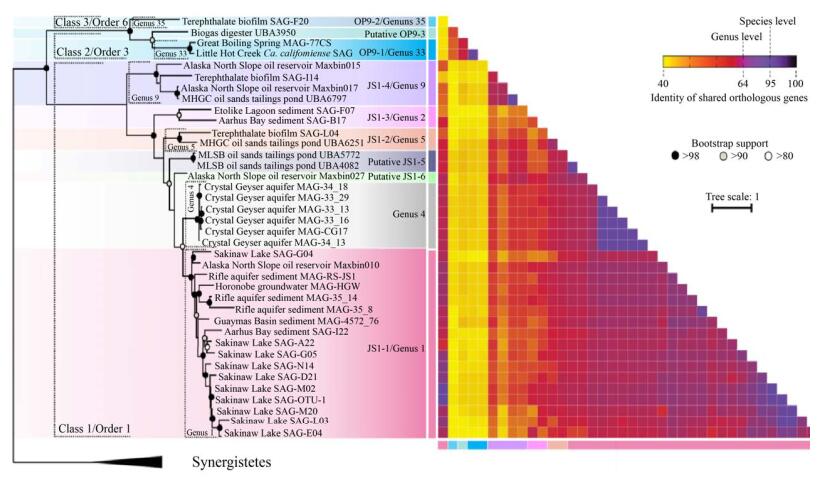
目前,基于16S rRNA基因的系统发育进化树得到的暗黑菌门的属多达35个[16],但是其中只有10个属有相关的基因组信息(图 2)。Dodsworth等[6]对获得的SAGs和MAGs进行系统进化发育分析,将Ca. Caldatribacterium californiense和Ca. Caldatribacterium saccharofermentans归类到OP9-1属下。Lloyd等[44]借助于单细胞测序技术,针对Aarhus Bay沉积物样品中获得的SAGs归类为JS1-1属。Rinke等[19]收集了9个样品点(包括海水、沉积污泥、湖泊、生物反应器) 的大量样品进行了单细胞测序、全基因组测序和SSU rRNA筛选,共获得了201个SAGs,并随后进行了系统性分析,在部分SAGs中发现暗黑菌存在,进一步证实了暗黑菌在环境中分布多样性。Nobu等[7]对Lloyd和Rinke等先前报道的部分SAGs重新组装(co-assembly)和分箱操作,将从降解对苯二甲酸酯的生物反应器中获得的SAG 232、SAG 231、SAG 167归类为OP9-2、JS1-2和JS1-4属;此外,在Sakinaw Lake (SAK) SAGs 124、130、136、217、219中也发现了JS1-1[45],这些结果拓展了我们对其环境分布了解;分析Aarhus Bay SAG B17、Etoliko Lagoon SAG 227发现了一个新的亚群JS1-3,扩充了我们对暗黑菌分支的认知。最近,我们对阿拉斯加北坡油藏样品SB1和SB2,以及先前报道的油藏、石脑油降解体系(NAPDC)、短链烷烃(C6–C10)的降解菌系(SCADC)以及厌氧反应器、地下水及沉积污泥等样品的多个宏基因组,重新拼接获得了17个高质量的MAGs,并从中分析归类发现了4个新的属水平分支,分别命名为JS1-5、JS1-6、JS1-7和OP9-3 (图 2)[21]。
3 暗黑菌的生理生化特征
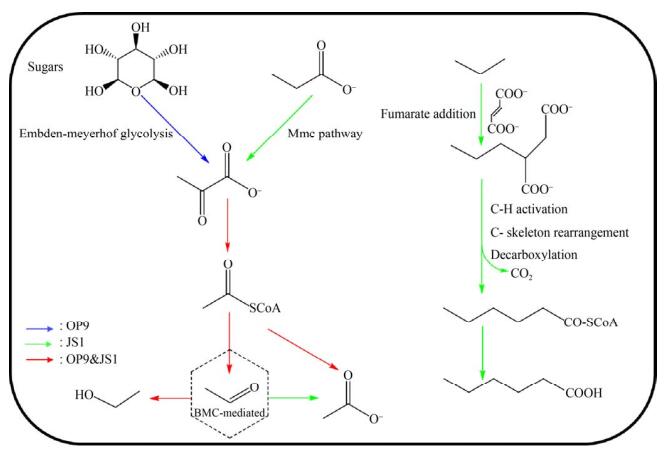
暗黑菌(OP9/JS1) 在自然界的广泛分布,暗示该类微生物可能具有多样的代谢功能,从而能适应不同的环境。同时,暗黑菌在部分环境下是细菌群落中的优势菌,因此推测该菌在环境中的地球化学循环中扮演着重要的角色。长期以来,由于缺乏纯培养物,使得对于暗黑菌的生理和生化作用的认识受到限制。近年来,分子生物信息学的发展给了我们一个认识的新窗口,通过对已获得的基因组信息分析可以推测它们的代谢潜能。目前已知的暗黑菌的代谢功能有3种:糖酵解、有机酸代谢和厌氧烃降解(图 3)。
|
| 图 3 暗黑菌的代谢模式 Figure 3 Metabolic capacities of Atribacteria. Putative metabolic reactions in OP9 are denoted as blue arrows, putative metabolic reactions in JS1 are denoted as green arrows and putative metabolic reactions shared in both OP9 and JS1 are denoted as red arrows. |
3.1 糖酵解
Webster等[36]分别用13C-乙酸盐和13C-葡萄糖作为添加底物,海滩污泥菌群作为接种源进行富集培养,将稳定同位素标记技术(SIP)[46-47]和变性梯度凝胶电泳技术(DGGE)[48]相结合,在两个培养体系中均发现属于JS1的13C-DNA片段,并首次推测得到该类菌的代谢模式,即在硫酸盐还原条件下,能够利用乙酸盐和葡萄糖或葡萄糖代谢产物。同样的,Dodsworth等[6]对OP9-cSCG和OP9-77CS进行代谢功能重构,发现这两个基因组包含编码多种糖降解途径关键酶的基因,但是几乎完全缺失氨基酸和脂肪酸的分解代谢途径,因此推测该类微生物在含糖底物中(如木聚糖、纤维素糖)主要通过糖酵解途径(embden- meyerhof pathway,EMP)或戊糖磷酸途径(pentose phosphate pathway,PPP)来降解和利用己糖和戊糖,同时产生乙酸和少量乙醇。Carr等[30]针对来自南极洲海洋环境样品基因组分析,也发现其具有乙酸氧化或糖酵解(如氨基糖)的代谢能力。以上结果表明在海洋环境和甲烷富集区,暗黑菌扮演了推动地球碳循环的重要角色。
3.2 有机酸代谢Nobu等[7, 45]研究表明,JS1-1和JS1-2缺乏糖酵解途径,但具有通过甲基丙二酰辅酶A (methylmalonyl-CoA,Mmc)途径降解丙酸的功能[49],对其相关性网络分析发现,暗黑菌常常与产甲烷菌(利用H2和甲酸)以及丙酸营养型的Cloacimonetes同时大量存在,暗示其参与对丙酸盐的降解过程[42, 50]。暗黑菌基因组中广泛存在的电子歧化甲酸脱氢酶(electron-bifurcating formate dehydrogenase,EBFdh)以及膜结合氢化酶(membrane-bound hydrogenase),能够以“反向电子传递”的方式推动丙酸氧化和氢气、甲酸的生成。通过以13C-乙酸盐为底物,Webster等[37]在海岸污泥的富集培养体系、Aoyagi等[51]在处理有机固废的生物膜反应器中均观察到底物中的13C结合到JS1 DNA分子的碳骨架上,推测其能够直接利用或者参与乙酸代谢转化过程。Lee等[31]分析罗斯海样品基因组RS JS1-Csag时发现其具有乙酸氧化代谢途径中相关功能基因,也表明其乙酸氧化潜能。此外,Nobu等[7]研究发现,除了基因完整度较低的JS1-4,在JS1-1、JS1-2、JS1-3以及OP9-1、OP9-2基因组中都含有编码细菌微区室外壳蛋白(bacterial microcompartments; BMCs被普遍认为是介导分解代谢和合成代谢的细胞器,广泛存在于细菌中[52-53])的同源基因簇,其中包含了编码醛类氧化途径中的部分关键酶如辅酶A-酰化丙醛脱氢酶、磷酸转乙酰酶以及醌氧化还原酶样NADH脱氢酶的同源基因pduP、pduL和pduS,以及连接醛氧化途径的2-脱氧-D-核糖5-磷酸醛缩酶(DERA)和戊糖单磷酸异构酶的编码基因,推测这些暗黑菌的成员具有将丙酸或糖代谢中产生的乙酰辅酶A转化成乙醛的能力,而BMC可以螯合代谢过程中产生的有毒醛类,将糖和醛类代谢相关联。在Katayama等和Lee等研究体系中也发现BMCs同源基因簇的存在[13, 31],但是在Liu等[21]烃降解体系中未发现BMCs同源基因,因此推测BMCs包裹的醛类物质转化酶类介导的醛糖转化可能不是暗黑菌固有的一种代谢方式,而是通过外源基因转移获得的一种全新代谢方式。
3.3 厌氧烃降解最近,我们在JS1-2、JS1-4、JS1-5和JS1-6的相关基因组中发现了完整的编码富马酸加成酶(FAE)的操纵子,通过针对编码基因assABC的蛋白建模分析发现了富马酸加成功能的保守催化位点,推测其具有将烷烃通过富马酸加成的方式厌氧活化的功能,但是在这些基因组中未能检测到β-氧化途径中全部的功能基因,无法在自身细胞内完成烷烃衍生的脂肪酸类物质的降解,故而推断其将短链烷烃通过富马酸加成后转化成较长链的脂肪酸后排出胞外,再被周围其他微生物逐级降解[21]。这个发现与以往报道的碳氢化合物发酵降解有很大不同,暗示着暗黑菌可能具有未知的厌氧烃降解代谢途径。此外,目前已经报道的具有富马酸加成功能的微生物均属于厚壁菌门(Firmicutes)和变形菌门(Proteobacteria)[54-55],该发现也表明暗黑菌门成为第3个检测到含有富马酸加成功能微生物的细菌门类。Garcia等[56]在未注水油井BA-1微生物群落基因组中检测到编码芳香烃富马酸加成酶的功能基因bssA,且与注水油井相比,暗黑菌的丰度有明显上升,推测其在原始未注水油井中发挥了烃降解功能。在表 2中列出目前报道的不同亚群的代谢模式。
| Subgroup | Substrate | Pathway | References |
| JS1-1 | Acetate/propionate | Methylmalonyl-CoA | [7] |
| JS1-2 | Acetate/propionate | Methylmalonyl-CoA | [7] |
| Short-chain n-alkanes | Fumarate addition | [21] | |
| JS1-3 | / | ||
| JS1-4 | Propionate | Methylmalonyl-CoA | [19] |
| Short-chain n-alkanes | Fumarate addition | [21] | |
| JS1-5 | Short-chain n-alkanes | Fumarate addition | [21] |
| JS1-6 | Propionate | Methylmalonyl-CoA | [21] |
| Short-chain n-alkanes | Fumarate addition | ||
| JS1-7 | Propionate | Methylmalonyl-CoA | [21] |
| Short-chain n-alkanes | Fumarate addition | ||
| OP9-1 | (hemi) Cellulose | Embden-Meyerh of pathway | [6] |
| OP9-2 | (hemi) Cellulose | Embden-Meyerh of pathway | [6] |
| OP9-3 | (hemi) Cellulose | Embden-Meyerh of pathway | [6] |
除了对MAGs的生物信息学分析,传统的富集分离策略也取得一定进展。最近,Katayama等[13]成功分离出了第一株OP9的代表性菌株,命名为RT761,代谢研究表明其能发酵葡萄糖,产生H2、乙酸、CO2和微量的乙醇,但不能利用外源电子受体进行厌氧呼吸(硝酸盐、铁和硫酸盐等),推测RT761主要通过产生H2来维持细胞内氧化还原电位平衡,因此,RT761的培养需要如氢营养型产甲烷菌及时地移除代谢产生的H2。这也预示着和专性氢营养型菌共培养可能是今后富集分离难培养微生物的一个方向。
4 总结与展望暗黑菌门的研究发现已有22年历史,借助于分子生物学手段,如分子探针、宏组学技术,在暗黑菌的环境分布、多样性、生态功能等方面的研究取得了长足的进展,暗黑菌的多样性也进一步扩充了生物系统发育进化树,丰富了我们对未培养环境微生物的认知。
虽然目前对暗黑菌门微生物已经开展了一系列研究,但是对其各个亚群环境分布规律、生理生化特征及生态学作用的认识仍然十分有限,需要进一步探索。近年来,生物信息学的发展无疑推动了难培养难分离微生物的研究,有助于系统认识微生物的多样性。随着测序工作的不断进行,数据库中不同环境来源的宏基因组数量激增,这些基因组信息将对后续有针对性地分离此类微生物菌株提供了基础。此外,在调研中发现,目前对暗黑菌相关研究多为国外研究报道,其研究对象主要集中于深海、热泉、火山、油藏等环境样品,然而最新报道中,在畜牧场、酒厂等地污水沟渠中,甚至人体排泄物中也发现暗黑菌存在,故而针对不同环境样品的分子探针检测能在一定程度上揭示其更多的生态分布,同时结合组学技术分析增进我们对其生态功能的认知。
暗黑菌的命名由来一定程度上是因为其难培养未分离的特性,RT761的成功分离具有重要意义,不仅有助于研究暗黑菌(OP9/JS1)的形态结构和生理功能,也为探索其难培养原因提供了一个可能;同时,分离策略无疑会给人们更多的启发去获得更多的未培养微生物的纯培养菌株,扩充对难培养微生物生态功能及其介导的地球元素循环的认知。
| [1] | Steen AD, Crits-Christoph A, Carini P, DeAngelis KM, Fierer N, Lloyd KG, Cameron Thrash J. High proportions of bacteria and archaea across most biomes remain uncultured. The ISME Journal, 2019, 13(12): 3126-3130. DOI:10.1038/s41396-019-0484-y |
| [2] | Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology, 1998, 5(10): R245-R249. |
| [3] | Kalisky T, Blainey P, Quake SR. Genomic analysis at the single-cell level. Annual Review of Genetics, 2011, 45: 431-445. DOI:10.1146/annurev-genet-102209-163607 |
| [4] | Youssef NH, Blainey PC, Quake SR, Elshahed MS. Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma). Applied and Environmental Microbiology, 2011, 77(21): 7804-7814. DOI:10.1128/AEM.06059-11 |
| [5] | Kamke J, Sczyrba A, Ivanova N, Schwientek P, Rinke C, Mavromatis K, Woyke T, Hentschel U. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. The ISME Journal, 2013, 7(12): 2287-2300. DOI:10.1038/ismej.2013.111 |
| [6] | Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PSG, Scholz MB, Lo CC, Raymond J. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nature Communications, 2013, 4(1): 1854-1854. DOI:10.1038/ncomms2884 |
| [7] | Nobu MK, Dodsworth JA, Murugapiran SK, Rinke C, Gies EA, Webster G, Schwientek P, Kille P, Parkes RJ, Sass H. Phylogeny and physiology of candidate phylum 'Atribacteria' (OP9/JS1) inferred from cultivation-independent genomics. The ISME Journal, 2016, 10(2): 273-286. DOI:10.1038/ismej.2015.97 |
| [8] | Webster G, Yarram L, Freese E, Köster J, Sass H, Parkes RJ, Weightman AJ. Distribution of candidate division JS1 and other Bacteria in tidal sediments of the German Wadden Sea using targeted 16S rRNA gene PCR-DGGE. FEMS Microbiology Ecology, 2007, 62(1): 78-89. DOI:10.1111/j.1574-6941.2007.00372.x |
| [9] | Hoshino T, Toki T, Ijiri A, Morono Y, Machiyama H, Ashi J, Okamura K, Inagaki F. Atribacteria from the subseafloor sedimentary biosphere disperse to the hydrosphere through submarine mud volcanoes. Frontiers in Microbiology, 2017, 8: 1135. DOI:10.3389/fmicb.2017.01135 |
| [10] | Lau MCY, Aitchison JC, Pointing SB. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles, 2009, 13(1): 139-149. DOI:10.1007/s00792-008-0205-3 |
| [11] | Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Andersen GL, Coates JD. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. The ISME Journal, 2008, 2(11): 1146-1156. DOI:10.1038/ismej.2008.48 |
| [12] | Pham VD, Hnatow LL, Zhang SP, Fallon RD, Jackson SC, Tomb JF, DeLong EF, Keeler SJ. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environmental Microbiology, 2009, 11(1): 176-187. DOI:10.1111/j.1462-2920.2008.01751.x |
| [13] | Katayama T, Nobu MK, Kusada H, Meng XY, Hosogi N, Uematsu K, Yoshioka H, Kamagata Y, Tamaki H. Isolation of a member of the candidate phylum 'Atribacteria' reveals a unique cell membrane structure. Nature Communications, 2020, 11(1): 1-9. DOI:10.1038/s41467-019-13993-7 |
| [14] | Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. Journal of Bacteriology, 1998, 180(2): 366-376. DOI:10.1128/JB.180.2.366-376.1998 |
| [15] | Webster G, Parkes RJ, Fry JC, Weightman AJ. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Applied and Environmental Microbiology, 2004, 70(9): 5708-5713. DOI:10.1128/AEM.70.9.5708-5713.2004 |
| [16] | Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology, 2014, 12(9): 635-645. DOI:10.1038/nrmicro3330 |
| [17] | Chakraborty A, Ruff SE, Dong XY, Ellefson ED, Li C, Brooks JM, McBee J, Bernard BB, Hubert CRJ. Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(20): 11029-11037. DOI:10.1073/pnas.2002289117 |
| [18] | Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(8): 2815-2820. DOI:10.1073/pnas.0511033103 |
| [19] | Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature, 2013, 499(7459): 431-437. DOI:10.1038/nature12352 |
| [20] | Glass JB, Ranjan P, Kretz CB, Nunn BL, Johnson AM, McManus J, Stewart FJ. Adaptations of Atribacteria to life in methane hydrates: hot traits for cold life. bioRxiv, 2019. |
| [21] | Liu YF, Qi ZZ, Shou LB, Liu JF, Yang SZ, Gu JD, Mu BZ. Anaerobic hydrocarbon degradation in candidate phylum 'Atribacteria' (JS1) inferred from genomics. The ISME Journal, 2019, 13(9): 2377-2390. DOI:10.1038/s41396-019-0448-2 |
| [22] | Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP. Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology, 2010, 8(2): 140-154. DOI:10.1111/j.1472-4669.2009.00228.x |
| [23] | Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles, 2009, 13(3): 447-459. DOI:10.1007/s00792-009-0230-x |
| [24] | Wemheuer B, Taube R, Akyol P, Wemheuer F, Daniel R. Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka peninsula. Archaea, 2013: 1-13. |
| [25] | Levén L, Eriksson ARB, Schnürer A. Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiology Ecology, 2007, 59(3): 683-693. DOI:10.1111/j.1574-6941.2006.00263.x |
| [26] | Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li TL, Camacho P, Sghir A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. The ISME Journal, 2009, 3(6): 700-714. DOI:10.1038/ismej.2009.2 |
| [27] | Tang YQ, Ji P, Hayashi J, Koike Y, Wu XL, Kida K. Characteristic microbial community of a dry thermophilic methanogenic digester: its long-term stability and change with feeding. Applied Microbiology and Biotechnology, 2011, 91(5): 1447-1461. DOI:10.1007/s00253-011-3479-9 |
| [28] | Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiology Ecology, 2008, 66(2): 181-196. DOI:10.1111/j.1574-6941.2008.00566.x |
| [29] | Gittel A, Sørensen KB, Skovhus TL, Ingvorsen K, Schramm A. Prokaryotic community structure and sulfate reducer activity in water from high-temperature oil reservoirs with and without nitrate treatment. Applied and Environmental Microbiology, 2009, 75(22): 7086-7096. DOI:10.1128/AEM.01123-09 |
| [30] | Carr SA, Orcutt BN, Mandernack KW, Spear JR. Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Frontiers in Microbiology, 2015, 6: 872. |
| [31] | Lee YM, Hwang K, Lee JI, Kim M, Hwang CY, Noh HJ, Choi H, Lee HK, Chun J, Hong SG, Shin SC. Genomic insight into the predominance of candidate phylum atribacteria JS1 lineage in marine sediments. Frontiers in Microbiology, 2018, 9: 2909. DOI:10.3389/fmicb.2018.02909 |
| [32] | Blazejak A, Schippers A. High abundance of JS-1-and Chloroflexi-related Bacteria in deeply buried marine sediments revealed by quantitative, real-time PCR. FEMS Microbiology Ecology, 2010, 72(2): 198-207. DOI:10.1111/j.1574-6941.2010.00838.x |
| [33] | Zhang Y, Su X, Chen F, Jiao L, Jiang HC, Dong HL, Ding GC. Abundance and diversity of candidate division JS1-and Chloroflexi-related bacteria in cold seep sediments of the northern South China Sea. Frontiers of Earth Science, 2012, 6(4): 373-382. DOI:10.1007/s11707-012-0324-0 |
| [34] | Kobayashi H, Endo K, Sakata S, Mayumi D, Kawaguchi H, Ikarashi M, Miyagawa Y, Maeda H, Sato K. Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. Journal of Bioscience and Bioengineering, 2012, 113(2): 204-210. DOI:10.1016/j.jbiosc.2011.09.015 |
| [35] | Cheng L, Shi SB, Li Q, Chen JF, Zhang H, Lu YH. Progressive degradation of crude oil n-alkanes coupled to methane production under mesophilic and thermophilic conditions. PLoS One, 2014, 9(11): R249. |
| [36] | Toth CRA, Gieg LM. Time course-dependent methanogenic crude oil biodegradation: dynamics of fumarate addition metabolites, biodegradative genes, and microbial community composition. Frontiers in Microbiology, 2017, 8: 2610. |
| [37] | Webster G, Watt LC, Rinna J, Fry JC, Evershed RP, Parkes RJ, Weightman AJ. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environmental Microbiology, 2006, 8(9): 1575-1589. DOI:10.1111/j.1462-2920.2006.01048.x |
| [38] | Hicks RE, Amann RI, Stahl DA. Dual staining of natural bacterioplankton with 4',6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Applied and Environmental Microbiology, 1992, 58(7): 2158-2163. DOI:10.1128/aem.58.7.2158-2163.1992 |
| [39] | Zheng D, Alm EW, Stahl DA, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Applied and Environmental Microbiology, 1996, 62(12): 4504-4513. DOI:10.1128/aem.62.12.4504-4513.1996 |
| [40] | Teske A, Hinrichs KU, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP, Sogin ML, Jannasch HW. Microbial diversity of hydrothermal sediments in the Guaymas basin: evidence for anaerobic methanotrophic communities. Applied and Environmental Microbiology, 2002, 68(4): 1994-2007. DOI:10.1128/AEM.68.4.1994-2007.2002 |
| [41] | Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. The ISME Journal, 2010, 4(5): 642-647. DOI:10.1038/ismej.2009.153 |
| [42] | Gies EA, Konwar KM, Beatty JT, Hallam SJ. Illuminating microbial dark matter in meromictic sakinaw lake. Applied and Environmental Microbiology, 2014, 80(21): 6807-6818. DOI:10.1128/AEM.01774-14 |
| [43] | Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Applied and Environmental Microbiology, 1998, 64(2): 795-799. DOI:10.1128/AEM.64.2.795-799.1998 |
| [44] | Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB. Predominant archaea in marine sediments degrade detrital proteins. Nature, 2013, 496(7444): 215-218. DOI:10.1038/nature12033 |
| [45] | Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu WT. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. The ISME Journal, 2015, 9(8): 1710-1722. DOI:10.1038/ismej.2014.256 |
| [46] | Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes RJ, Cappenberg TE. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature, 1998, 392(6678): 801-805. DOI:10.1038/33900 |
| [47] | Radajewski S, Ineson P, Parekh NR, Colin Murrell J. Stable-isotope probing as a tool in microbial ecology. Nature, 2000, 403(6770): 646-649. DOI:10.1038/35001054 |
| [48] | Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology, 1993, 59(3): 695-700. DOI:10.1128/aem.59.3.695-700.1993 |
| [49] | Webster G, Sass H, Cragg BA, Gorra R, Knab NJ, Green CJ, Mathes F, Fry JC, Weightman AJ, Parkes RJ. Enrichment and cultivation of prokaryotes associated with the sulphate-methane transition zone of diffusion-controlled sediments of Aarhus Bay, Denmark, under heterotrophic conditions. FEMS Microbiology Ecology, 2011, 77(2): 248-263. DOI:10.1111/j.1574-6941.2011.01109.x |
| [50] | Müller B, Sun L, Schnürer A. First insights into the syntrophic acetate-oxidizing bacteria-a genetic study. MicrobiologyOpen, 2013, 2(1): 35-53. DOI:10.1002/mbo3.50 |
| [51] | Aoyagi T, Inaba T, Aizawa H, Mayumi D, Sakata S, Charfi A, Suh C, Lee JH, Sato Y, Ogata A, Habe H, Hori T. Unexpected diversity of acetate degraders in anaerobic membrane bioreactor treating organic solid waste revealed by high-sensitivity stable isotope probing. Water Research, 2020, 176: 115750. DOI:10.1016/j.watres.2020.115750 |
| [52] | Axen SD, Erbilgin O, Kerfeld CA. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Computational Biology, 2014, 10(10): e1003898. DOI:10.1371/journal.pcbi.1003898 |
| [53] | Kirst H, Kerfeld CA. Bacterial microcompartments: catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biology, 2019, 17(1): 79. DOI:10.1186/s12915-019-0691-z |
| [54] | Tan B, Jane Fowler S, Laban NA, Dong XL, Sensen CW, Foght J, Gieg LM. Comparative analysis of metagenomes from three methanogenic hydrocarbon-degrading enrichment cultures with 41 environmental samples. The ISME Journal, 2015, 9(9): 2028-2045. DOI:10.1038/ismej.2015.22 |
| [55] | Hu P, Tom L, Singh A, Thomas BC, Baker BJ, Piceno YM, Andersen GL, Banfield JF. Genome-resolved metagenomic analysis reveals roles for candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. mBio, 2016, 7(1): e01669-15. |
| [56] | Sierra-Garcia IN, Belgini DRB, Torres-Ballesteros A, Paez-Espino D, Capilla R, Santos Neto EV, Gray N, de Oliveira VM. In depth metagenomic analysis in contrasting oil wells reveals syntrophic bacterial and archaeal associations for oil biodegradation in petroleum reservoirs. Science of the Total Environment, 2020, 715: 136646. DOI:10.1016/j.scitotenv.2020.136646 |
 2021, Vol. 61
2021, Vol. 61