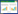中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Feiyan Lin, Yingyi Duan, Jingjie Jiang, Tingting Huang, Shuangjun Lin, Zixin Deng. 2021
- 林飞燕, 段颖异, 江晶洁, 黄婷婷, 林双君, 邓子新. 2021
- Identification and characterization of the biosynthetic pathway of naphthoquinone-oxindole alkaloid coprisidins
- 萘醌-氧吲哚生物碱coprisidins生物合成途径的研究
- Acta Microbiologica Sinica, 61(5): 1184-1199
- 微生物学报, 61(5): 1184-1199
-
文章历史
- 收稿日期:2020-05-12
- 修回日期:2020-06-03
- 网络出版日期:2020-06-10
Microbial-derived natural products often feature complex structures and a diverse array of biological activities[1]. Both skeleton and functional groups of natural products provide a source of inspiration for the development of valuable bioactive substances for the pharmaceutical industry[2]. Since re-isolation of known compounds has become one of the major obstacles for microbial natural product discovery, the exploration of potential from the bacterial symbionts emerges as one of the powerful strategies to identify novel compounds[3].
Microbial endosymbionts form ubiquitous micro-niches in nature with ecological significance[4]. Recent studies of insect symbionts clearly show their tremendous effects on the symbiotic host, ranging from the contribution to food digestion, provision of the nutrients and signaling molecules, and production of a variety of secondary metabolites to defend them by competing with other enemy microbes[5]. Insect symbiotic bacteria have been largely exploited for new natural products and become the "new frontier" for natural products chemistry. For instance, natalamycin A, a geldanamycin-like natural product with unusual post-PKS modifications, is produced by a termite-associated Streptomyces[6]. Besides, enormous microbial genome sequences obtained by advanced sequencing technologies revealed that the biosynthetic potential of microorganisms is still far from being fully exploited. Genome mining combines with disclosing the potent secondary metabolomes of insect symbiotic bacteria enable for deeper investigation of new compounds and their biosynthetic mechanisms[7].
Natural naphthoquinone derivatives are a group of functional metabolites from plants and various microorganisms including fungi and actinomycetes, etc. Certain amounts of naphthoquinone-containing compounds have been evaluated for bioactivities and pharmacological properties[8]. Among several isoforms of naphthoquinones core, a series of bioactive 1, 4-naphthoquinone derivatives such as doxorubicin, aclarubicin has been developed for medical markets[9]. In 1, 4-naphthoquinone system, the quinone core is easily susceptible to reduction, oxidation, or addition of nucleophiles, which endows naphthoquinone core with specific structure features and various biological properties. The in-depth exploration of the biosynthetic mechanisms of naphthoquinone will enable us to extend the naphthoquinone compounds family and expand their diversity via synthetic biology techniques.
Coprisidins A and B (1 and 2, Figure 1) were isolated from a gut-associated Streptomyces sp. SNU607 in the dung beetle Copris tripartitus[10]. Coprisidin A shows inhibitory effects towards Na+/K+-ATPase, and coprisidin B can induce the NQO1 [NAD(P)H: quinone oxidoreductase 1], a phase II detoxifying enzyme which has an important impact on cancer prevention. Coprisidins were the first example of natural alkaloids with naphthoquinone-oxindole skeleton[10]. Generally, 2-oxindole motif is a pharmacophore in natural products and is also incorporated into synthetic therapeutic candidates[11]. Different from the synthetic naphthoquinone oxindole compounds, oxindole moieties of coprisidins tether to the benzene ring of naphthoquinone rather than quinone ring[12]. Feeding experiments indicated that L-tryptophan and pentanoic acid can't be incorporated into the coprisidins[10]. The biosynthetic mechanism of the special naphthoquinone-oxindole structure of coprisidins remains unclear.

|
| Figure 1 Structures of coprisidin A (1) and coprisidin B (2). |
Here we sequence the genome of the dung beetle associate Streptomyces sp. SNU607 and identify a type Ⅱ PKS gene cluster governing the biosynthesis of coprisidins with a 33.3 kb DNA region containing 30 open reading frames (ORFs). Based on in vivo mutagenesis, heterologous expression experiments and bioinformatics analysis, we proposed a putative pathway for the biosynthesis of coprisidins. CopH/I/M/N/O constituted a five-enzyme cassette for the formation of butyryl-S-ACP, which served as a starter unit to install propyl moiety of coprisidins. Our proposed pathway of coprisidins biosynthesis provides opportunities to improve the fermentation level of coprisidins by reconstructing the metabolic network, explore their ecology functions in the natural habitat, and develop new naphthoquinone-oxindole analogues by modification of the biosynthetic pathway.
1 Materials and methods 1.1 Strains, plasmids, and cultures conditionsThe strains and plasmids used in this study are summarized in Table 1. The coprisidin-producing strain Streptomyces sp. SNU607 was isolated from the gut of the dung beetle Copris tripartitus from Jeju Island, Korea[10]. Streptomyces sp. SNU607 and its derivatives were cultivated at 30 ℃ in tryptic soy broth (TSB) liquid medium for growth of mycelia, on SFM medium (20 g of mannitol, 20 g of soybean meal, 20 g of agar per 1 L of tap water and adjusted to pH 7.1) for fermentation and on SFM with 10 mmol/L MgCl2 for conjugation. Luria-Bertani (LB) liquid and agar were used for culturing Escherichia coli strains. The suicide E. coli- Streptomyces shuttle vector pYH7 was used for gene deletion and the integrative E. coli-Streptomyces shuttle vector pIB139 containing the ermE* promoter for complementation[13-14]. E. coli ET12567/pUZ8002 was used for conjugal transfer of plasmids between E. coli and Streptomyces[15]. The final antibiotic concentrations used for selection of E. coli and Streptomyces were as follows: 40 µg/mL apramycin for Streptomyces sp. SNU607; 50 µg/mL apramycin, 50 µg/mL kanamycin, 50 µg/mL chloramphenicol, 80 µg/mL TMP for E. coli.
| Strains/ Plasmids | Descriptions | Sources/References |
| Strains | ||
| SNU607 | Streptomycete, wild type producer of coprisidins | [10] |
| TK24 | Streptomycete lividans, heterologous expression host | [15] |
| TK24/26D12 | Streptomycete lividans TK24, containing cosmid 26D12 | This work |
| ∆copB | SNU607 mutant with copB gene deletion | This work |
| ∆copC | SNU607 mutant with copC gene deletion | This work |
| E. coli DH10B | Host strain for general cloning | GIBCO BRL |
| E. coli ET12567/pUZ8002 | Donor strain for intergeneric conjugation | [15] |
| E. coli EPI300TM | Host strain for genomic library construction | EPICENTRE |
| Plasmids | ||
| pJTU2463 | A shuttle vector derivative of pOJ446 with SCP2 replicon replaced by int and attp from pSET152 for construction of genomic library, ApraR | [16] |
| pYH7 | A shuttle vector derivative of pHZ1358, ThioR, ApraR | [13] |
| pIB139 | Integrative vector for gene complementation, PermE*, ApraR | [14] |
| cosmid 26D12 | A sequenced cosmid, containing genes related to the biosynthesis of coprisidins | This work |
| pYH7-∆copB | pYH7 derived construct for copB inactivation | This work |
| pYH7-∆copC | pYH7 derived construct for copC inactivation | This work |
1.2 DNA isolation and general manipulations
All endonucleases and T4 DNA ligase were purchased from Thermo Scientific (Waltham, USA). The ClonExpress One Step Cloning Kit, FastPure Plasmid Mini Kit, and FastPure Gel DNA Extraction Mini Kit were purchased from Vazyme Biotech Co., Ltd (Nanjing, China). PCR amplifications were performed using 2×Taq Plus Master Mix or 2×Phanta Max Super-Fidelity DNA polymerase (Vazyme Biotech Co., Ltd, Nanjing, China). PCR primers were synthesized at BioSune Biotech Co. Ltd. (Shanghai, China) and listed in Table 2. All the constructed plasmids were confirmed by DNA sequencing (Biotech Co. Ltd, Shanghai, China).
| Primes | Sequences (5′→3′) | Usage |
| Screen-orf34-F | TCACCGCAGCAGGCGGGCGAACTGT | Primers for screening of genomic cosmid library of Streptomyces sp. SNU607 |
| Screen-orf34-R | ATGACCGACAGACCCCTCGTCCTCG | |
| Screen-orf42-F | TCAGCGCTTGCGCGCGATGGT | |
| Screen-orf42-R | ATGGAATTTCCTGCCGACCAG | |
| Screen-orf58-F | GTGAAGATCGGCATCGTCTGC | |
| Screen-orf58-R | TCAGTCCCTCGCGAGCCCGAA | |
| ∆copB-F1 | TCCACCGGGACTGATCAAGGCGAATACTTCATATGCGGCCCGGGAGCCGGCGACGGCTG | Primers used to construct knockout plasmid pYH7-∆copB |
| ∆copB-R1 | GCCATGGTGCTCACCCGGCCCGAAGGGACACGCCGCCCATCCCTAGACAAATCCCCAGT | |
| ∆copB-F2 | GGTCACCTTTCACTGGGGATTTGTCTAGGGATGGGCGGCGTGTCCCTTCGGGCCGGGTG | |
| ∆copB-R2 | CCGTCCGGGACCCGCGCGGTCGATCCCCGCATATGCGTTCCGGCTGCGTGAAGGGCGCG | |
| ∆copC-F1 | TCCACCGGGACTGATCAAGGCGAATACTTCATATGGGTGCGGGAGGGCCAGCGCGCCGC | Primers used to construct knockout plasmid pYH7-∆copC |
| ∆copC-R1 | TGGTGCAGATCCGGCCATTCCGTGGACGTGCCGAGCCGCCCCCGCACCCGCGTCCACGG | |
| ∆copC-F2 | GGTTCCGGGTGCCGTGGACGCGGGTGCGGGGGCGGCTCGGCACGTCCACGGAATGGCCG | |
| ∆copC-R2 | CCGTCCGGGACCCGCGCGGTCGATCCCCGCATATGCTGCTGGTGACGGCGGGTCCCGGA |
1.3 Genome sequencing and bioinformatic analysis
The genomic DNA of Streptomyces sp. SNU607 was sequenced using PacBio sequencing (BGI group, China). The coprisidin cluster was deposited to GenBank with the accession number CP050436.
The analysis of open reading frames (ORFs) was performed using RAST server by automatic annotation[17], followed by manual re-inspection to correct function assignments using Frame Plot[18]. For the identification of secondary metabolite gene clusters, the genome of Streptomyces sp. SNU607 was analyzed by using antiSMASH (antibiotics and Secondary Metabolite Analysis Shell)[19] and scanned for homologues to known biosynthetic genes in the databases via NCBI BLAST server. Multiple nucleotide sequence alignments, analysis and designs of PCR primers were performed using the BioEdit Sequence Alignment Editor 7.0 (available online). The modular organization of the polyketide and nonribosomal peptide megasynthases were assigned by using web tools[20]. 16S rDNA sequences were aligned using BioEdit and the phylogenetic tree was built by using MEGA 5.0 (neighbor-joining method). The ribosomal DNA sequences for all strains were obtained from the GenBank database exclude Streptomyces sp. SNU607.
1.4 Construction and screening of genomic cosmid library of Streptomyces sp. SNU607A pJTU2463-derived genomic cosmid library of Streptomyces sp. SNU607 was constructed according to a standard protocol[15, 21]. The genomic cosmid library was screened using three sets of PCR primers designed according to the 34th ORF, the 42th ORF and the 58th ORF (Table 2).
1.5 Construction of gene deletion mutantsPYH7-∆copB and pYH7-∆copC were constructed according to a method using the ClonExpress One Step Cloning Kit[13]. The plasmids were transferred to Streptomyces sp. SNU607 by E. coli-Streptomyces conjugation, respectively. The exconjugants were selected on SFM medium supplemented with apramycin (40 µg/mL), TMP (80 µg/mL)and then the in-frame deletion mutants via double-crossover homologous recombination were confirmed by PCR amplification.
1.6 Construction of complementation strainsPCR-amplified copB and copC were digested with Nde I–EcoR I and cloned downstream of the ermE* promoter into pIB139 integrative vector digested with the same restriction enzymes to generate complementary plasmids. The plasmids were introduced into respective mutants by E. coli- Streptomyces conjugation. The exconjugants were selected with apramycin (40 µg/mL), TMP (80 µg/mL) and confirmed by PCR amplification.
1.7 Heterologous expressionsThe cosmid harboring all the biosynthetic genes for coprisidin was introduced into Streptomyces lividans TK24 by E. coli-Streptomyces conjugation. The exconjugants were selected with apramycin (40 µg/mL), TMP (80 µg/mL) and confirmed by PCR amplification.
1.8 Fermentation, extraction, and isolationThe coprisidin-producing strain Streptomyces sp. SNU607 and its derivatives were cultivated in SFM medium for fermentation. The seed inoculum was prepared by inoculating 20 mL of seed medium (TSB) with 100 µL of mycelium in a 250 mL Erlenmeyer flask and incubating at 30 ℃ for 2 days on a rotary shaker (220 r/min). Then, the seed cultures (20 mL) were centrifuged and the supernatant was removed. An appropriate amount of mycelium was dipped in a sterile toothpick and transferred to an SFM medium plate (a 9 cm petri dish containing 25 mL solid medium) at 30 ℃ for 6 additional days. After cutting up two culture plates of solid fermentation of Streptomyces, the fragments were soaked with ethyl acetate. Ultrasonic extraction was performed for 15 min with one-hour waiting time. The extraction was repeated for 3 times and the extract was evaporated to dryness at 37 ℃ on a vacuum evaporator. The resulting residues were dissolved with 500 µL methanol and the resulting fermentation product was concentrated 100 times. All the organic extracts were performed in fume cupboard.
1.9 HPLC analysis and LC-MS analysisThe resulting crude extract was analyzed by HPLC on an Agilent HPLC series 1260 with an Agilent Eclipse XDB-C18 column (5 µm, 4.6 mm× 250 mm). The injection quantity is 10 µL and the column was eluted by a gradient elution system of H2O/ACN for 40 min at a flow rate of 0.6 mL/min. The elution methods were: 5% B to 40% B (linear gradient, 0–15 min), 40% B to 70% B (linear gradient, 15–22 min), 70% B to 100% B (linear gradient, 22–27 min), 100% B (constant gradient, 27–30 min), 100% B to 5% B (linear gradient, 30–33 min), 5% B (constant gradient, 33–40 min). HPLC analysis was monitored at 208 nm and 297 nm which are characteristic absorption of the coprisidins. The LC-QTOF-MS analysis was carried out on a 6545 QTOF LC/ MS spectrometer coupled to an Agilent HPLC 1290 series (Agilent Technologies) with electrospray ionization (ESI) source. All the parameters are as the same as the HPLC analysis, except the change of rate to 0.4 mL/min. The MS system was operated in negative ionization mode and the mass scan range was set between 20 and 1000 m/z.
2 Results 2.1 Sequencing and analysis of the Streptomyces sp. SNU607 genomeTo investigate the biosynthetic potential of dung beetle-associated Streptomyces sp. SNU607, we obtained a whole genome comprised of 6, 850, 319 bp (71.11% G+C content) by using PacBio sequencing strategy. As expected, the chromosome of Streptomyces sp. SNU607 has a linear topology which is a common characteristic of Streptomyces (Figure 2-A). The linear genome encodes 6, 294 predicted protein-coding open reading frames (ORF). A nucleotide BLAST analysis of the DNA sequence corresponding to the 16S rRNA gene from Streptomyces sp. SNU607 revealed the highest similarity to Streptomyces flavogriseus and Streptomyces viridochromogenes (Figure 2-B).
The complete sequence and annotation of the Streptomyces sp. SNU607 genome allowed for rational mining to identify many biosynthetic pathways of secondary metabolites. Genome analysis using antiSMASH revealed 23 gene clusters potentially involved in secondary metabolism (Table 3).
| ID | Types | Size/kb | Most similar know clusters |
| 1 | Blactam | 23.5 | Clavulanic acid |
| 2 | Terpene | 26.4 | Hopene |
| 3 | T1 PKS | 94 | Sceliphrolactam |
| 4 | Bacteriocin | 10.3 | – |
| 5 | NRPS | 57.4 | Cadaside A/cadaside B |
| 6 | Siderophpre | 13.1 | Ficellomycin |
| 7 | Terpene | 19.7 | – |
| 8 | Bacteriocin | 9.5 | – |
| 9 | Butyrolactone | 8.3 | Lactonamycin |
| 10 | NRPS, T1 PKS | 56.2 | Istamycin |
| 11 | Siderophore | 11.8 | Desferrioxamin B/ desferrioxamin E |
| 12 | Lanthipeptide | 23.1 | Niphimycin C-E |
| 13 | Terpene | 19.8 | – |
| 14 | Ectoine | 8.6 | Ectoine |
| 15 | T2 PKS, PKS-like | 71.6 | Cinerubin B |
| 16 | Terpene | 20.6 | Steffimycin D |
| 17 | Terpene, Ectoine | 20.9 | Ectoine |
| 18 | Bacteriocin | 10.2 | – |
| 19 | T3 PKS | 41.1 | Tetronasin |
| 20 | Melanin | 10.5 | Melanin |
| 21 | T2 PKS, terpene | 72.5 | Spore pigment |
| 22 | NRPS | 53.5 | Rimosamide |
| 23 | Butyrolactone | 10.9 | – |
The most represented type of secondary metabolite biosynthetic genes within the Streptomyces sp. SNU607 genome is terpenoid (6 out of 23). Among six NRPS and PKS biosynthetic machineries, four of them were related to PKS: a type Ⅰ PKS, a type Ⅱ PKS, a type Ⅲ PKS and a hybrid PKS-NRPS. Type Ⅰ PKSs are module-organized enzyme complexes and each module is responsible for the catalysis of one cycle of polyketide chain elongation[22]. Type Ⅱ PKSs consist of discrete enzymes which catalyze a single set of iteratively acting activities and typically produce polycyclic aromatic compounds[23]. Type Ⅲ PKSs, also known as chalcone synthase-like PKSs, consist of a homodimer ketosynthase that iteratively condenses malonyl-CoA to form small benzol or naphthol rings. The Cluster 3 is a type Ⅰ PKS gene cluster, sharing 88% similarity with sceliphrolactam, a 26 membered polyene macrolactam isolated from a Streptomyces strain in tropical mangrove sediments[24]. Cluster 10 contains a type Ⅰ PKS and NRPS combined biosynthetic pathway. Cluster 15 harbors KSα-KSβ-ACP, the "minimal PKS" core of Type Ⅱ PKS normally for aromatic polyketides. The Cluster 19 consists of a gene encoding single module type Ⅲ PKS domain. The core region, including the genes encoding type Ⅲ PKS, a methyltransferase and an oxidoreductase is responsible for the assembly of the alkylresorcinol in Streptomyces species[25].
In addition, several other types of secondary metabolites including the ectoine, siderophores, lanthiopeptide, and butyrolactone could be possibly produced by S. sp. SNU67 (Table 3). Accordingly, the gene clusters from Streptomyces sp. SNU607 possess a relatively high level of structural diversity of secondary metabolites and most of these gene clusters show no homology to any other sequenced microbial genomes.
2.2 Identification of the coprisidin biosynthetic gene clusterThe chemical structure of coprisidins together with previous feeding experiments indicated the aromatic core is PKS origin[10]. Among the polyketide gene clusters identified from the genome, the Cluster 19 (type Ⅲ PKS) was not expected to govern the biosynthesis of coprisidins because it is syntenic with the characterized alkylresorcinol[25]. We then concluded that Cluster 15, the type Ⅱ PKS gene cluster located within the "arm region" (5515974–5587573 nt) of the linear chromosome, may contribute to the biosynthesis of coprisidins.
To identify the coprisidins biosynthetic gene cluster, we intended to knock out the conserved type Ⅱ PKS core in Cluster 15. The genes encoding KSα and KSβ were inactivated by in frame deletion to afford mutants ∆copB and ∆copC, both of which no longer produced coprisidins. The coprisidins production was partially restored by introducing a full-length copB and copC into corresponding mutants for complementation (Figure 3-B). These results demonstrated that the type Ⅱ PKS in Cluster 15 was responsible for the biosynthesis of coprisidin A.

|
| Figure 3 Identification of the coprisidin biosynthetic gene cluster. A: Schematic representation of the fragment from the Streptomyces sp. SNU607 in cosmid26D12, range from the 31st ORF to the 60th ORF; B: HPLC analysis of fermentation products of the mutants ∆copB, ∆copC and complementation strains ∆copB: : copB, ∆copC: : copC; C: LC/MS analysis of heterologous expression strain S. lividans TK24/26D12 during 5 days fermentation. WT: wild-type producer Streptomyces sp. SNU607; TK24/26D12: S. lividans TK24 containing cosmid26D12; ★: coprisidin A; coprisidin A: m/z [M-H]– 408.1089. |
We further carried out heterologous expression to confirm the essential region for coprisidins biosynthesis. A genomic cosmid library of Streptomyces sp. SNU607 was constructed in the integrating pJTU2463 (an E. coli-Streptomyces shuttle vector)[16], and three sets of PCR primers were designed according to the 34th ORF, 42th ORF and 58th ORF for library screening (Table 2). Cosmid 26D12 containing all three ORFs has been identified and contained a 33.3-kb fragment, ranging from the 31st ORF to the 60th ORF (Figure 3-A).Cosmid 26D12 was introduced into a heterogeneous host, S. lividans TK24 by E. coli-Streptomyces conjugation to afford recombinant strain TK24/26D12. The formation of coprisidin A was confirmed by LC-MS analysis (m/z: [M-H]– 408.1089, identical to that of the coprisidin A; Figure 3-C). All these results demonstrated that the aromatic core of coprisidins is type Ⅱ PKS origin and the DNA region from the 31st gene to the 60th gene in cosmid 26D12 is adequate for coprisidins biosynthesis. The identified coprisidin biosynthetic genes along with their proposed functions are listed in Table 4.
| Deduced proteins | Size/aa | Proposed function | Homologous protein origin | Similarity/ Identity/%* |
| Orf31 | 218 | Membrane protein | KcsA with V76ester mutation, [Streptomyces lividans] | 48/38 |
| Orf32 | 190 | AP-4-A phosphorylase | Rv2613c, [Mycobacterium tuberculosis (strain ATCC 25618/H37Rv)] | 69/56 |
| Orf33 | 77 | – | – | – |
| CopQ | 300 | NAD-dependent ketoductse/epimerase | Nucleotide-diphosphate-sugar epimerase [Mycolicibacterium monacense] BBZ62295.1 | 51/34 |
| CopA | 81 | ACP | ACP, [Streptomyces violaceoruber] P12885.2 | 72/56 |
| CopB | 422 | Chain length factor, KSβ | Tetracenomycin C KS, [Streptomyces glaucescens] P16539.2 | 71/57 |
| CopC | 423 | KSα | Tetracenomycin C KS, [Streptomyces glaucescens] P16539.2 | 68/57 |
| CopR1 | 167 | Resistance protein | MarR family transcriptional regulator, [Nocardia sp. ET3-3] MVU82195.1 | 50/34 |
| CopD | 151 | Dehydratase/cyclase | SsfY4, [Streptomyces sp. SF2575] ADE34486.1 | 65/53 |
| CopR2 | 268 | Transcriptional regulator | SARP family regulator, [Streptomyces lavendulae] BAG74714.1 | 63/49 |
| CopS | 518 | Cholesterol oxidase | Oxidase, [Streptomyces griseoruber] AQW35067.1 | 79/64 |
| CopT | 224 | O-methyltransferase | MdmC, [Streptomyces mycarofaciens] Q00719.1 | 60/44 |
| CopE | 267 | KR | AknA, [Streptomyces galilaeus] BAB72043.1 | 82/71 |
| CopF | 347 | Cyclase/dehydrase | SsfY1, [Streptomyces sp. SF2575] ADE34490.1 | 61/47 |
| CopG | 615 | Acyl CoA ligase (ATP dependent) | SsfL2, [Streptomyces sp. SF2575] ADE34493.1 | 57/45 |
| CopH | 246 | 3-oxoacyl-ACP reductase | FabG, [Aquifex aeolicus (strain VF5)] AAC07575.1 | 45/28 |
| CopI | 263 | Enoyl-ACP reductase Ⅲ | FabL, [Bacillus subtilis (strain 168)] QGU25416.1 | 52/33 |
| CopJ | 423 | FAD-binding monooxygenase | TjhO2, [Streptomyces aureus] AYU66242.1 | 66/52 |
| CopK | 257 | Cyclase | TjhC1, [Streptomyces aureus] AYU66231.1 | 76/64 |
| CopL | 328 | Aldo/ketoreductase | Aldo/keto reductase, [Acidobacteria bacterium] PYX56021.1 | 74/58 |
| CopM | 343 | KSⅢ | CerJ, [Streptomyces tendae] AEI91069.1 | 46/35 |
| CopN | 333 | AT | AknF, [Streptomyces galilaeus ATCC 25435] BAB72049.1 | 70/62 |
| CopO | 558 | Acyl-CoA carboxyl transferase | SsfE, [Streptomyces sp. SF2575] ADE34513.1 | 84/78 |
| CopP | 560 | Membrane protein | Membrane protein, [Streptomyces venezuelae ATCC 10712] CCA54417.1 | 78/65 |
| Orf55 | 741 | Translation elongation factor G-related protein |
[Streptomyces sp. Tue6314] QED90636.1 | 99/99 |
| Orf56 | 222 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase | [Streptomyces griseus subsp. rhodochrous] KOG79528.1 | 97/96 |
| Orf57 | 307 | Phosphatidylinositol mannoside acyltransferase | [Streptomyces sp. SID6648] NED01444.1 | 93/91 |
| Orf58 | 386 | GDP-mannose-dependent alpha-(1-2) phosphatidylin-ositol mannosyltransferase | [Streptomyces sp. WM6378] KOU54515.1 | 96/94 |
| Orf59 | 181 | Unknown | FHW51_10450, [Streptomyces argenteolus] TWF62658.1 | 96/95 |
| Orf60 | 306 | Pyridoxal 5′-phosphate synthase lyase subunit | PdxS, [Streptomyces sp. SID6648] NED01441.1 | 99/96 |
| %*: indicates protein similarity/identity in comparison to homologues of coprisidin cluster. | ||||
2.3 Genes putatively involved in the biosynthesis of starter unit
Most type Ⅱ PKS pathways use acetyl-CoA as a starter unit, while several examples using non-acetate starter units thereby enrich the structure diversity of type Ⅱ polyketides[26]. For example, the benzoyl-CoA starter unit of enterocin is derived from L-phenylalanine, the unusual 4-methylvaleryl moiety of R1128C is synthesized from L-valine, and a propionyl starter unit is involved in the biosynthesis of doxorubicin and daunorubicin[27].
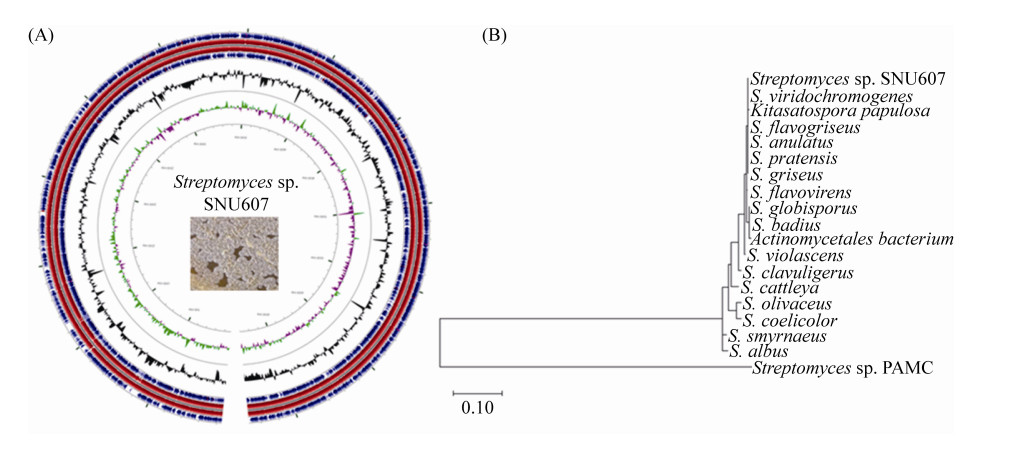
Butyryl starter unit is reasonable for coprisidins biosynthesis according to the chemical structure and the type Ⅱ PKS catalytic logic. The natural butyryl-CoA is formed by the sequential conversion of two acetyl-CoA catalyzed by a series of enzymes: thiolase, 3-hydroxybutyryl-CoA dehydrogenase, crotonase, and butyryl CoA dehydrogenase[28]. However, no homologues were found in the coprisidins cluster. A set of genes encoding enzymes involve in the formation of the ACP tethered short acyl chain (Table 4, Figure 3-A). These genes include copH [encoding 3-oxoacyl-(ACP) reductase], copI (encoding enoyl-ACP reductases), copM (ketoacylsynthase for starter unit, KSⅢ), copO (acetyl-CoA carboxylase), and copN (acyltransferase, AT).
The proposed pathway for butyryl-CoA formation may start with acetyl-CoA and malonyl-CoA directly from primary metabolism. Carboxylase CopO is possible to synthesize malonyl-CoA from acetyl-CoA. Next, CopM, functions as a priming KSⅢ, catalyzes the Claisen condensation of acetyl-CoA and malonyl-CoA to form acetoacetyl-CoA, and transferred onto a standalone ACP by CopN. CopH and CopJ catalyzed ketoreduction and enol reduction of β-ketoacyl-ACP thioester intermediates to form the butyryl-S-ACP as the starter unit for chain elongation (Figure 4-A). These genes resemble those found in fatty acid biosynthesis leads to a functional cross talk between primary (fatty acid) and secondary (polyketide) metabolism. Alternatively, instead of using a discrete ACP, CopM may condense acetyl-CoA and malonyl-CoA directly to form acetoacetyl-CoA, further CopH and CopI catalyzed the butyryl-CoA formation and primed the PKS.
|
| Figure 4 Proposed biosynthetic pathway for coprisidins. Two hypothetic pathways for coprisidins have been proposed according to the biosynthetic genes (A) and the chemical structure (B). The biosynthetic pathway for butyryl-S-ACP precursor is highlighted in dash. |
2.4 Prediction of the coprisidins biosynthetic pathway
Several genes encode type Ⅱ PKS enzymes for the naphthoquinone core of coprisidins: KSα (CopC), KSβ (also referred as the chain-length factor, CopB), ACP (CopA), an acyltransferase (CopN), a ketoreductases (CopE), and three cyclases (CopF, CopK, CopG) (Figure 3-A, Table 4).
CopC and CopB show high sequence homology to the KSα (57% identities) and KSβ (57% identities) involved in the biosynthesis of tetracenomycin, respectively. The single-domain protein CopC and CopB form a dimeric complex (KSα-KSβ or KS-CLF) that acts in an iterative fashion to build a nascent polyketide chain[23].
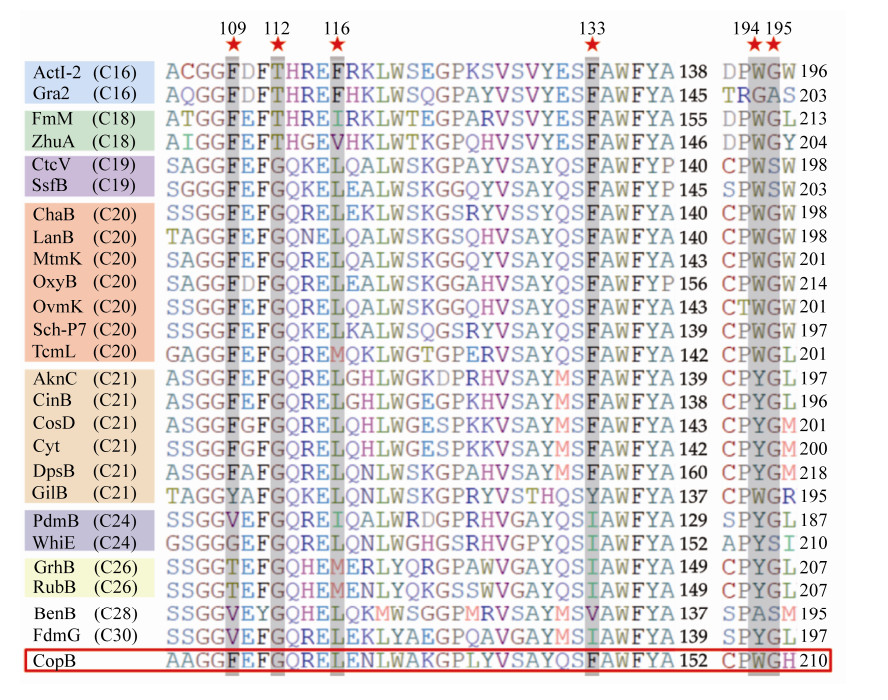
Crystal structure of actinorhodin KS-CLF reveals an amphipathic cavity spanning across the KS-CLF dimer interface for the growing polyketide chain, which is the key factor for chain length controlling[29]. The amino acid residues at six positions (109, 112, 116, 133, 194, 195, numbered according to actinorhodin CLF) have been defined as determinants contributing to the size of the cavity, and further accommodated products of different length[29-30].
To study the chain length of nascent polyketide of coprisidins, we aligned CopB with selected CLFs from structurally characterized aromatic polyketides. Sequence alignments elucidated that the six key residues in CopB are identical to the CLF of C20 polyketides (Figure 5). Thus, CopB-CopC-CopA is proposed to form a polyketide intermediate for coprisidins scaffold with a chain length of at least C20.
|
| Figure 5 Multiple sequence alignment of CopB and its CLF homologs with increasing chain length specificity. ★ represents the six residues as "gatekeeper". |
Ketoreductase and cyclases function together to afford aromatic polyketide core. CopE is 71% identical to the ketoreductase from Streptomyces galilaeus, and reduces the keto group at C9 of nascent polyketide intermediate to hydroxyl, resulting in carbonyl-reduced poly-β-keto chain. CopF, a didomain cyclase similar to SsfY1 (47% sequence identity), is predicted to catalyze the reducing C7-C12 cyclization to afford the first ring (D)[31] (Figure 4-A). CopK has sequence homology (64% identity) to TjhC1, the second ring cyclase from a cryptic aromatic polyketide gene cluster[32]. CopK is probably a C5-C14 cyclase catalyzing the second ring (C) and the third ring (B) is spontaneously formed of 5. CopG is an ATP-dependent ligase and displays high homology to SsfL2 (57% similarity and 45% identity), which catalyzes the C1-C18 Claisen cyclization in the presence of ATP to form the fourth ring (A) in tetracycline biosynthesis. According to the type Ⅱ PKS related genes, we therefore proposed that during coprisidins biosynthesis, a four-ring polyketide system 5 was formed first, underwent oxidative cleavage, and rearranged to form naphthoquinone-oxindole skeleton.
Minimal PKS forms the aromatic core ring which is afterward modified by different types of accessory enzymes such as halogenases, oxygenases, methyltransferases, prenyltransferases, etc [33]. Different tailoring enzymes of coprisidins pathway enable to diversify the chain length, oxidation states and the final structures.
CopJ is a probable FAD binding oxidoreductase, which is similar with OxyL in the oxytetracycline and SsfO2 in the tetracycline SF2575. CopL is a NADPH-dependent ketoreductase with specificity for aromatic substrate. Presumably, the A ring of 5 was doubly hydroxylated by CopJ, and CopL has been assigned as a candidate accounting for the hydroxylation to form 6.
More steps are needed to complete the biosynthesis of coprisidins from 6. CopQ located upstream of type Ⅱ PKS genes, which encodes an NAD-dependent ketoductase or epimeriase. CopD is predicted to be a dehydratase or cyclase which is similar with SsfY4 in tetracycline SF2575. CopS shows homology (50% similarity and 35% identity) to cholesterol oxidase. The roles of these enzymes in coprisidins biosynthesis remain unclear. Only one methyltransferase (CopT) is present in the coprisidins pathway based on sequence homology. CopT is similar with a SAM-dependent O-methyltransferase from Acidimicrobiia bacterium (MSO79939, 53% identity) or caffeoyl-CoA O-methyltransferase from Asanoa hainanensis (SNS73218, 61% identity). CopT is supposed to be responsible for the O-methylation of coprisidins.
A proposed biosynthetic pathway is shown in Figure 4-A. The biosynthesis of coprisidins starts with the assembly of the ketide backbone by the minimal PKS, which consists of the KS-CLF (CopC and CopB) heterodimer, an ACP (CopA). The formation of butyryl-CoA precursor shared with fatty acid synthase from primary metabolism. The butyryl precursor served as starter unit is subsequently transferred to the active site of KS, undergoes elongation using malonyl-CoA as extender unit to afford a nascent poly-β-keto chain, and subsequently the aromatic four ring intermediate is formed. At the later stage of the biosynthesis, enzymatic oxidation and rearrangement or cleavage possibly take place to form the final naphthoquinone-oxindole. The nitrogen of indole ring may be incorporated at the post-PKS step or during the oxidative arrangement. The hydroxyl of coprisidin B could be directly stereospecific installed by an oxygenase. Experimental evidence will be needed to determine these steps (Figure 4-A).
3 DiscussionCoprisidins are structurally unique naphthoquinone-oxindole alkaloids from a dung beetle-associated Streptomyces sp. SNU607 with unique bioactivities. In this work, we have demonstrated that the rare naphthoquinone-oxindole framework of coprisidins was biosynthesized by a type Ⅱ PKS system employing a non-acetate starter unit.
Aromatic ring of naphthoquinones is generally synthesized by fungi iterative type Ⅰ PKSs, or type Ⅲ PKSs through an 1, 3, 6, 8-tetrahydroxynaphthalene (T4HN) intermediate[34]. A few examples of naphthoquinone-derived compounds were biosynthesized by type Ⅱ PKSs in Streptomyces[35]. However, the coupling of naphthoquinone and oxindole moiety at the benzene ring is rarely reported in type Ⅱ PKS products. According to the chemical structure and biosynthetic logic, we initially proposed a unified pathway for coprisidin A and B biosynthesis (Figure 4-B). The proposed pathway suggested that the type Ⅱ PKS genes are dedicated to forming the naphthoquinone ring independently from the oxindole moiety. A group of enzymes catalyze the oxidative tailoring and coupling with the indole derivate to complete the biosynthesis of coprisidin A. A possible oxidase may catalyze stereospecific hydroxylation at C3 of the indole ring to afford coprisidin B.
The chain length of type Ⅱ polyketides is largely controlled by chain length factor (KSβ). Coprisidins biosynthetic gene cluster shows a 25% similarity with cinerubin B biosynthetic gene cluster (accession no. GG770539) and 32% similarity with aclacinomycin cluster (accession no. AB008466), while both of them possess the anthracycline four ring system. Sequence analysis of CopB implies that the type Ⅱ PKS of coprisidins cluster is more likely to generate decaketide intermediate. The presence of ketoreductases (CopE) and cyclases (CopF/K/G) are strongly implicated in the formation of the anthracycline-like ring system, and then transformed to a naphthoquinone-oxindole through the proposed retro-aldol cleavage to open the C ring, oxidative catalysis, and rearrangement. Since coprisidin A and B show different bioactivity, further characterization of the hydroxyl group incorporation in coprisidin B will pave the way toward structure-activity relationship studies.
Genetic elements of the type Ⅱ PKSs and the post-PKS tailoring enzymes have been shown to be interchangeable and combinable to produce diverse structures showing the potential of type Ⅱ PKS system[36]. The initial priming of the PKS with a starter unit is an important point for introducing structural diversity in type Ⅱ PKS. Butyryl starter unit generation in coprisidins biosynthesis provides a feasible gene cassette. Exploration of the novel catalytic mechanisms during coprisidins biosynthesis would contribute to the generation of "non-natural" coprisidins derivatives with better biological activities. Future studies are needed to clarify the roles of enzymes in the pathway. Another challenge is how to make coprisidins at scale. The low yield of coprisidins in Streptomyces sp. SNU607 may be due to the inability to simulate the symbiotic environment with insects in the laboratory culture. Understanding the biosynthetic pathway enables us to enhance the precursor supply for coprisidins, manipulate the transcription level of rate-limit genes in the pathway, or engineer and express biosynthetic genes in heterologous host for coprisidins production.
On the other hand, the gene cluster of coprisidins is of special interest to us because when we searched against the microbial genome database, gene clusters with high identity have been found in several Streptomyces. The phylogram clearly showed that these strains are not located in the same branch (Figure 2-B). Since Streptomyces sp. SNU607 is the only case known to produce naphthoquinone- oxindole alkaloids so far, our results also highlight the importance of discovering the pathways directly from the genome sequencing data. Additionally, being the products of beetle gut-associate bacteria, it is suggested that coprisidins may play a role in niche competition and host protection. More studies are needed to provide insight into their biological roles.
Specific insect symbiont-derived natural products present reservoirs of useful new natural products[3]. The genome sequence of Streptomyces sp. SNU607 unveiled the biosynthetic potential of enormous secondary metabolites. These unknown gene clusters in coprisidins producer suggest a promise source for new compounds discovery through traditional metabolic engineering in the wild type producer, functionally expression in the optimized chassis strains, and construction of efficient biological machineries by synthetic biology strategies.
Acknowledgments
We thank Prof. Dong-Chan Oh at Natural Products Research Institute, College of Pharmacy, Seoul National University, Republic of Korea for providing the strain Streptomyces sp. SNU607.
| [1] | Shen B. A new golden age of natural products drug discovery. Cell, 2015, 163(6): 1297-1300. DOI:10.1016/j.cell.2015.11.031 |
| [2] | Rodrigues T, Reker D, Schneider P, Schneider G. Counting on natural products for drug design. Nature Chemistry, 2016, 8(6): 531-541. DOI:10.1038/nchem.2479 |
| [3] | Beemelmanns C, Guo HJ, Rischer M, Poulsen M. Natural products from microbes associated with insects. Beilstein Journal of Organic Chemistry, 2016, 12: 314-327. DOI:10.3762/bjoc.12.34 |
| [4] | Dossey AT. Insects and their chemical weaponry: new potential for drug discovery. Natural Product Reports, 2010, 27(12): 1737-1757. DOI:10.1039/c005319h |
| [5] | Douglas AE. Lessons from studying insect symbioses. Cell Host & Microbe, 2011, 10(4): 359-367. |
| [6] | Kim KH, Ramadhar TR, Beemelmanns C, Cao SG, Poulsen M, Currie CR, Clardy J. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp.. Chemical Science, 2014, 5(11): 4333-4338. DOI:10.1039/C4SC01136H |
| [7] | Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes - a review. Natural Product Reports, 2016, 33(8): 988-1005. |
| [8] | Qiu HY, Wang PF, Lin HY, Tang CY, Zhu HL, Yang YH. Naphthoquinones: a continuing source for discovery of therapeutic antineoplastic agents. Chemical Biology & Drug Design, 2018, 91(3): 681-690. |
| [9] | Aminin D, Polonik S. 1, 4-Naphthoquinones: some biological properties and application. Chemical and Pharmaceutical Bulletin, 2020, 68(1): 46-57. DOI:10.1248/cpb.c19-00911 |
| [10] | Um S, Bach DH, Shin B, Ahn CH, Kim SH, Bang HS, Oh KB, Lee SK, Shin J, Oh DC. Naphthoquinone-oxindole alkaloids, coprisidins A and B, from a gut-associated bacterium in the dung beetle, Copris tripartitus. Organic Letters, 2016, 18(22): 5792-5795. DOI:10.1021/acs.orglett.6b02555 |
| [11] | Leoni A, Locatelli A, Morigi R, Rambaldi M. 2-Indolinone a versatile scaffold for treatment of cancer: a patent review (2008-2014). Expert Opinion on Therapeutic Patents, 2016, 26(2): 149-173. DOI:10.1517/13543776.2016.1118059 |
| [12] | Yu JS, Zhou F, Liu YL, Zhou J. Organocatalytic asymmetric Michael addition of unprotected 3-substituted oxindoles to 1, 4-naphthoquinone. Beilstein Journal of Organic Chemistry, 2012, 8: 1360-1365. DOI:10.3762/bjoc.8.157 |
| [13] | Demydchuk Y, Sun YH, Hong H, Staunton J, Spencer JB, Leadlay PF. Analysis of the tetronomycin gene cluster: insights into the biosynthesis of a polyether tetronate antibiotic. ChemBioChem, 2008, 9(7): 1136-1145. DOI:10.1002/cbic.200700715 |
| [14] | Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Böhm I, Mironenko T, Deacon M, Wheatcroft M, Wirtz G, Staunton J, Leadlay PF. Increasing the efficiency of heterologous promoters in actinomycetes. Journal of Molecular Microbiology and Biotechnology, 2002, 4(4): 417-426. |
| [15] | Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood D. Practical streptomyces genetics: a laboratory manual. Norwich: John Innes Foundation, 2000. |
| [16] | Zou Y, Yin HX, Kong DK, Deng ZZ, Lin SJ. A trans-acting ketoreductase in biosynthesis of a symmetric polyketide dimer SIA7248. ChemBioChem, 2013, 14(6): 679-683. DOI:10.1002/cbic.201300068 |
| [17] | Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics, 2008, 9: 75-90. DOI:10.1186/1471-2164-9-75 |
| [18] | Ishikawa J, Hotta K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiology Letters, 1999, 174(2): 251-253. DOI:10.1111/j.1574-6968.1999.tb13576.x |
| [19] | Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research, 2019, 47(W1): W81-W87. DOI:10.1093/nar/gkz310 |
| [20] | Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Research, 2004, 32(Web Server issue): W405-W413. |
| [21] | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. |
| [22] | Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan ARH, Håkansson K, Sherman DH, Smith JL, Skiniotis G. Structure of a modular polyketide synthase. Nature, 2014, 510(7506): 512-517. DOI:10.1038/nature13423 |
| [23] | Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type Ⅱ polyketide synthases: gaining a deeper insight into enzymatic teamwork. Natural Product Reports, 2007, 24(1): 162-190. DOI:10.1039/B507395M |
| [24] | Low ZJ, Pang LM, Ding YC, Cheang QW, Le Mai Hoang K, Thi Tran H, Li JM, Liu XW, Kanagasundaram Y, Yang L, Liang ZX. Identification of a biosynthetic gene cluster for the polyene macrolactam sceliphrolactam in a Streptomyces strain isolated from mangrove sediment. Scientific Reports, 2018, 8: 1594-1607. DOI:10.1038/s41598-018-20018-8 |
| [25] | Wang CM, Cane DE. Biochemistry and molecular genetics of the biosynthesis of the earthy odorant methylisoborneol in Streptomyces coelicolor. Journal of the American Chemical Society, 2008, 130(28): 8908-8909. DOI:10.1021/ja803639g |
| [26] | Tang Y, Lee TS, Kobayashi S, Khosla C. Ketosynthases in the initiation and elongation modules of aromatic polyketide synthases have orthogonal acyl carrier protein specificity. Biochemistry, 2003, 42(21): 6588-6595. DOI:10.1021/bi0341962 |
| [27] | Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Natural Product Reports, 2002, 19(1): 70-99. DOI:10.1039/b003939j |
| [28] | Okamura E, Tomita T, Sawa R, Nishiyama M, Kuzuyama T. Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(25): 11265-11270. DOI:10.1073/pnas.1000532107 |
| [29] | Tang Y, Tsai SC, Khosla C. Polyketide chain length control by chain length factor. Journal of the American Chemical Society, 2003, 125(42): 12708-12709. DOI:10.1021/ja0378759 |
| [30] | Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nature Structural & Molecular Biology, 2004, 11(9): 888-893. |
| [31] | Pickens LB, Kim W, Wang P, Zhou H, Watanabe K, Gomi S, Tang Y. Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. Journal of the American Chemical Society, 2009, 131(48): 17677-17689. DOI:10.1021/ja907852c |
| [32] | Ji ZY, Nie QY, Yin Y, Zhang M, Pan HX, Hou XF, Tang GL. Activation and characterization of cryptic gene cluster: two series of aromatic polyketides biosynthesized by divergent pathways. Angewandte Chemie International Edition, 2019, 58(50): 18046-18054. DOI:10.1002/anie.201910882 |
| [33] | Olano C, Méndez C, Salas JA. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Natural Product Reports, 2010, 27(4): 571-616. DOI:10.1039/b911956f |
| [34] | Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S. A new pathway for polyketide synthesis in microorganisms. Nature, 1999, 400(6747): 897-899. DOI:10.1038/23748 |
| [35] | Katsuyama Y, Sone K, Satou R, Izumikawa M, Takagi M, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y. Involvement of the Baeyer-Villiger monooxygenase IfnQ in the biosynthesis of isofuranonaphthoquinone scaffold of JBIR-76 and -77. ChemBioChem, 2016, 17(11): 1021-1028. DOI:10.1002/cbic.201600095 |
| [36] | Poust S, Hagen A, Katz L, Keasling JD. Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Current Opinion in Biotechnology, 2014, 30: 32-39. DOI:10.1016/j.copbio.2014.04.011 |
 2021, Vol. 61
2021, Vol. 61