中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 黄志强, 邱景璇, 李杰, 许东坡, 刘箐. 2021
- Zhiqiang Huang, Jingxuan Qiu, Jie Li, Dongpo Xu, Qing Liu. 2021
- 基于16S rRNA基因测序分析微生物群落多样性
- Exploration of microbial diversity based on 16S rRNA gene sequence analysis
- 微生物学报, 61(5): 1044-1063
- Acta Microbiologica Sinica, 61(5): 1044-1063
-
文章历史
- 收稿日期:2020-05-25
- 修回日期:2020-07-30
- 网络出版日期:2020-09-01
微生物资源是生物技术创新的重要源泉,对生命科学基础研究和生态经济发展具有重要意义,其多样性的研究有利于微生物资源的充分挖掘与利用。微生物群落多样主要包括物种多样性、遗传多样性和功能多样性3个方面[1],在环境[2-3]、能源[4]、食品[5]与人体健康[6]等诸多领域有着广泛的研究与应用。传统的微生物群落多样性研究方法包括纯培养、理化鉴定等方式,然而自然环境中大部分微生物不可培养、难以鉴定[7-8]。随着宏基因组概念的提出与测序技术的发展,基于16S rRNA (16S ribosomal RNA)基因的高通量测序技术克服了上述困难,在微生物群落多样性研究中备受关注。
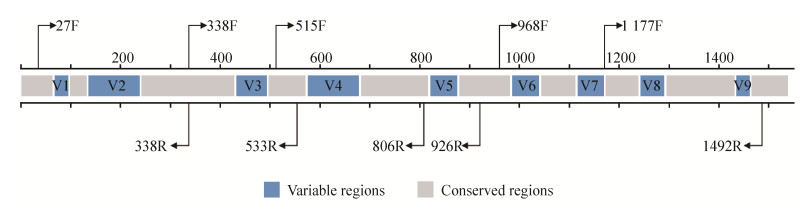
16S rRNA基因普遍存在于细菌和古细菌中,具有多个拷贝数,全长1500 bp左右,其结构由9个可变区(variable region)和10个保守区(conserved region)交替组成(图 1)。保守区有利于扩增引物的设计,可变区体现了物种间的进化差异。这些特性使16S rRNA基因成为原核生物鉴定分类、系统进化以及多样性分析等研究中常用的分子标志物。在2006年,Sogin等[9]首次对水体样本中微生物群落的16S rRNA基因进行焦磷酸测序,发现北大西洋深海水团和热液喷口的微生物群落复杂度比之前报道的任何环境高1–2个数量级。人类微生物组计划(human microbiome project,HMP)在探索人体不同部位微生物群落变化与健康疾病关系时,也将16S rRNA基因测序分析作为重要手段[10-12]。Wang等[13]通过建立小鼠酒精成瘾模型,利用16S rRNA基因测序并结合代谢组学分析,证明了肠道菌群与酒精成瘾存在关联性。目前基于16S rRNA基因测序的微生物群落多样性分析方法已被广泛应用,通过分析微生物群落中物种分布、群落特征和功能,寻找不同样本或组间的差异菌群,挖掘样本表型与微生物群落特征的关联,进而阐明微生物与环境间的相互作用关系。本文对基于16S rRNA基因测序微生物群落多样性分析流程中测序平台与扩增区的选择、测序数据预处理以及多样性分析方法四个方面进行综述,就其面临的问题与挑战进行讨论。
|
| 图 1 16S rRNA基因结构与引物示意图 Figure 1 Schematic of 16S rRNA gene and primer targets. A schematic figure of 16S rRNA gene with nine variable regions marked as V1–V9. Arrows show forward and reverse primers covering variable regions. The number indicate primer position according to the E. coli gene numbering. |
1 测序平台的选择 1.1 DNA测序技术的发展
自1953年Watson和Crick提出DNA分子双螺旋结构以来,许多研究者开始了对DNA测序技术的探索。20世纪70年代,人们先后提出加减法(plus and minus method)[14]、化学降解法(chemical cleavage method)[15]等DNA测序方法。1977年,Sanger等[16]在加减法的基础上开发了经典的双脱氧链终止法(dideoxynucleotide method),并测定了φX174噬菌体的DNA序列。这些方法及其衍生技术统称为第一代测序技术,其中又以双脱氧链终止法应用最为广泛,因此第一代测序技术也常被认为是Sanger测序。20世纪90年代初,基于双脱氧链终止法并结合荧光标记与电泳分离技术开发出了自动测序仪,它在人类基因组计划(human genome project,HGP)中发挥重要作用。第一代测序技术测序优势在于读长长,准确率高,但因其测序成本高、通量低、速度慢而未能大规模推广。2005年后,以Roche 454焦磷酸测序、Illumina Solexa聚合酶测序和ABI SOLiD连接酶测序为代表的第二代测序技术相继出现,通过对DNA片段引入碱基标签,实现了大量样本的平行测序[17-18]。第二代测序技术特点是高通量,降低了测序成本同时还提高了测序速度,并且有着较高的准确性,但序列读长比第一代测序技术要短。2008年后,Helicos (Helicos Bioscience)公司的Heliscope测序技术[19]、PacBio (Pacific Biosciences)公司的SMRT技术[20]、ONT (Oxford Nanopore Technologies)公司的纳米孔单分子技术[21],这些则被认为是第三代测序技术,与其他测序技术不同的是,纳米孔单分子技术基于电信号而不是光信号。相比于前两代技术,三代测序特点是采用单分子测序,测序读长长,不需要PCR扩增环节,但测序成本和错误率相对较高。各类测序平台技术参数见表 1。
| Sequencing types | Platforms | Reads length/bp | Yields | Run time/h | Error types | Error rate/% |
| First Generation | 3730xl (ABI)a | 1 000 | 0.08 M | 10 | Substitution | 0.001 |
| Next Generation | MiniSeq (Illumina) | 2×150 | 7.5 G | 4–24 | Substitution | 0.1–1.0 |
| Miseq Series | 2×300 | 15 G | 65 | |||
| NextSeq 550 Series | 2×150 | 120 G | 12–30 | |||
| NextSeq 2000 | 2×150 | 300 G | 24–48 | |||
| NovaSeq 6000 | 2×250 | 6 000 G | 13–38 | |||
| GS FLX+ (Roche 454)b | 1 000 | 700 M | 23 | Indels | 1 | |
| GS Junior | 700 | 35 M | 10 | |||
| SOLiD 5500 (ABI) | 35–75 | 90 G | 168 | A-T bias | 4 | |
| SOLiD 5500xl | 35–75 | 300 G | 168 | |||
| Ion PGM (Ion Torrent)c | 200–400 | 2 G | 7 | Indels | 1 | |
| Ion Proton | 200 | 10 G | 4 | |||
| Ion S5 | 200–600 | 50 G | 19 | |||
| Third Generation | HeliScope (Helicos)d | 55 | – | – | Indels | 2–7 |
| RS II (PacBio) | Mean 14 k | 1 G | 6 | Indels | 0.1–16.0 | |
| Sequel | Mean 10 k | 10 G | 20 | |||
| MinION (ONT) | > 2 M | 30 G | 72 | Indels & Substitution | 12 | |
| GridION | > 2 M | 150 G | 72 | |||
| PromethION | > 2 M | 148 G | 64 | |||
| Flongle | > 2 M | 1.8 G | 24 | |||
| a: In 2008, Life Technologies was created by the combination of Invitrogen Corporation and Applied Biosystems Inc, and Life Technologies was acquired by Thermo Fisher Scientific in 2014. b: In 2007, 454 Life Sciences was acquired by Roche Diagnostics. Roche shut down 454 in 2013, and stop supporting the platform by 2016. c: Ion Torrent was acquired by Life Technologies in 2010. d: Helicos Bioscience went bankrupt in 2012. | ||||||
1.2 不同测序平台的比较
尽管Sanger测序有着超高的准确度,但不能直接对混合样本进行16S rRNA基因的测序分析,单样本的文库构建繁琐费时且通量较低[25]。近年来,Illumina测序平台在16S rRNA基因测序应用最为广泛,主要原因是其在二代测序中有着读长相对较长、准确率较高以及成本较低的优势。Roche公司在2013年宣布停止测序业务并于2016年淘汰了454测序仪,基于其平台的相关研究日趋式微。而三代测序技术凭借超长读长的特点,基于其平台的相关研究逐渐增多。利用不同平台的16S rRNA基因测序相关研究趋势见图 2。针对诸多的测序平台,研究者们就其在16S rRNA基因测序方面进行了比较分析。Salipante等[26]利用PGM与MiSeq平台对模拟菌群的V1-V2区进行测序,结果显示PGM有着相对较高的错误率,而且测序过程由于测序方向的不同,A. odontolyticus与P. acnes正向测序测得的序列较短,致使群落结构评估有偏差。Allali等[27]使用MiSeq、PGM和GS FLX三种平台对鸡盲肠微生物群落的V1–V2区测序,发现不同测序平台在物种相对丰度与多样性上有差异,但测序结果能区分不同实验组得出相似的生物学结论。二代测序仅能对16S rRNA基因的部分可变区测序,而三代测序突破读长限制实现了16S rRNA基因的全长测序,进而能更全面、准确地解析微生物群落结构。Singer等[28]采用PacBio RS II和MiSeq两种测序平台对模拟菌群以及湖水样本进行了群落多样性分析,两者在门水平的菌群组成较为相似,但在属、种更小的阶元水平上存在差异,且差异随着菌群复杂度增加而增大,SMRT测序技术提高了物种分类学注释(taxonomic assignment) 的准确性。MinION与MiSeq相比,MinION在属水平上正确注释的序列比例相对较高,而Miseq测双可变区正确注释的序列比例高于测单可变区[25]。尽管三代测序技术通过对16S rRNA基因进行全长测序,使得一些物种能鉴定到种水平[29-30],但其结果受限于数据库中全长序列的数量。此外,三代测序技术有着较高的错误率,其对结果分析的影响也还需要进一步研究。真实样本中微生物种类复杂且未知,这使得评估测序平台对微生物多样性分析的影响变得困难,设计复杂度更高的模拟菌群作为参照十分必要。针对于16S rRNA基因测序不同平台之间比较,应关注序列质量、有效序列数、序列注释准确率等方面,做出更为全面的评估。SMRT技术与纳米孔单分子技术同属三代测序,它们在基于16S rRNA基因测序分析群落结构上的差异比较也还有待完善。总之,不同测序平台各有优势与不足,需要综合考虑实验设计与成本进行选择。

|
| 图 2 基于不同平台16S rRNA基因测序相关研究发表文章的数量统计 Figure 2 Statistics of the number of published papers about 16S rRNA sequencing based on different platforms. The data collected from ScienceDirect database with the keywords: "16S rRNA 454 pyrosequencing", "16S rRNA Illumina sequencing", "16S rRNA Ion Torrent sequencing", "16S rRNA PacBio sequencing", "16S rRNA Nanopore sequencing". |
2 扩增区的选择
提取样本中微生物总基因组后,需要选择合适的引物来扩增16S rRNA基因构建测序文库。一般认为测序片段越长,物种鉴定的准确度就越高,进而能更真实地反映样品中的微生物群落结构。但二代测序的读长不能覆盖16S rRNA基因的全长,因此常采用一个或多个V区进行测序,例如测单V区(V3、V4、V6),双V区(V3–V4、V4–V5),三V区(V1–V3、V4–V6)等。不同可变区分类学注释的准确率差异较大,对菌群多样性分析结果有影响[31],如何选择最佳扩增区显得尤为重要。这主要涉及两个问题,不同可变区能否有效区分各个物种以及引物能否扩增出绝大部分原核微生物的16S rRNA基因片段。
2.1 计算机模拟评估可变区利用计算机与数据集评估可变区的物种区分能力是一种经济、便捷的方法,而构建进化树则是其中较为简单的方式。Chakravorty等[32]通过构建进化树分析了不同可变区并鉴定110株细菌,结果显示任何单一可变区都不能完全区分所有细菌,除Enterobacteriaceae以外,单可变区V2和V3对其他细菌在属水平上能有较好区分的效果,而且V2、V3和V6联用使部分细菌能在种水平上得以区分。这种方式本质是基于序列的相似性,但其结果不能将可变区的物种区分能力量化比较,而且也不适合分析成千上万的种属。目前,依据朴素贝叶斯原理开发的RDP classifier[33]在物种分类学注释中已被广泛使用,研究者们基于此方法分析了不同可变区的物种注释准确率。Wang等[33]发现在16S rRNA基因的100 bp子序列中,V2和V4区附近的子序列在属水平上注释准确率最高。但可变区长度受其位置及所属物种的影响,采用固定长度的子序列分析并不符合实际情况,仅能间接说明单可变区中V2和V4最佳。Vilo等[34]将16S rRNA基因多序列比对后,再截取不同可变区进行分析,发现单一可变区中V2和V4在属水平注释准确率最高,而多V区中V3–V5在属水平准确率最高。此外,RDP classifier注释准确率与其参数设定有关,在较高的bootstrap值下,准确率反而降低。在提取可变区序列时,Vilo等未考虑引物结合的实际情况,但对测试序列采取了0.5%的错误率进行模拟处理,从而更接近真实测序。而Claesson等[35]根据引物结合截取6个双可变区进行分析,结果表明V3–V4在属水平注释准确率最高,然而实际的测序样本会有部分序列在属水平上未能注释。从物种注释分辨率的角度而言,V1–V3注释到种水平的序列比例仅次于全长16S rRNA基因[36]。以不同可变区在不同相似度下聚类观测到的微生物群落多样性作为衡量指标,V1–V4能提供更为准确的多样性评估结果[37],而另有研究则认为V4–V6与全长结果最接近[38]。表 2总结了16S rRNA基因不同可变区的比较研究。
| Evaluation methods | Dataset | Best single variable region | Best multiple variable regions | References | |||
| Region | Accuracya | Regions | Accuracy | ||||
| Phylogenetic analysis | 113 sequences from 110 bacterial species | V2, V3 | NA | NA | NA | [32] | |
| Taxonomic assignment | Bergey with 5014 sequences, NCBI with 23095 sequences | V2, V4 | About 80% | NA | NA | [33] | |
| Taxonomic assignment | SILVA (SSURef 102) with 274196 sequences | V2, V4 | 60% | V1–V3, V3–V5 | 80% | [34] | |
| Taxonomic assignment | SILVA (SSURef 100) with 27013 sequences | NA | NA | V3–V4 | 78% | [35] | |
| Taxonomic resolution | Greengenes (v13.8.99) HOMD (v13) | NA | NA | V1–V3 | NA | [36] | |
| Diversity estimate | RDP (Release 10) with 15825 sequences | NA | NA | V1–V4 | NA | [37] | |
| Geodesic distance | SILVA (SSURef 115) with 79096 sequences | V4 | NA | V4–V6 | NA | [38] | |
| Coverage, spectrum, POAOs | SILVA (SSURef 123), Greengenes (v13.8), RDP (Release 11.5) | V4 | NA | V1–V2, V1–V3 | NA | [40] | |
| a: Taxonomic assignment accuracy at the genus level with best single/multiple variable region(s), NA indicate the research data not show. | |||||||
通过计算机模拟分析,多数研究均表明单可变区中V4最佳[33-34, 38-40],这也是地球微生物组计划(earth microbiome project,EMP)推荐的可变区。而各项研究的最佳多可变区结论存在差异。一方面,单一研究未考虑到所有可能的多可变区组合,仅选择部分组合进行比较;另一方面,各自所用的数据集、截取可变区的引物以及分析角度也不尽相同。尽管最佳多可变区尚未达成一致意见,但总体而言,采用多可变区的结果优于单可变区,尤其是涵盖了V2或V4的多可变区。
除了计算机模拟分析之外,还有结合测序实验对不同可变区进行比较。一种是利用已知组成的模拟菌群,另一种是利用未知组成的环境样本,以鸟枪法测序分析结果作为参照。但鸟枪法测序数据库在某些环境中的可用基因组相对有限,对于某些研究较少的体系以其作为参考标准也可能会产生偏差[41]。两种模式都存在不同可变区测得物种相对丰度有差异以及某些物种可能检测不到的问题[39, 42-46]。这既因为不同可变区保守程度不同,又有来自于引物扩增的偏差。
2.2 引物覆盖率16S rRNA保守区并非绝对保守,同一引物与不同菌群模板结合的效率不同,致使某些物种的丰度评估偏高或偏低;不同引物即使扩增了相同可变区,其物种丰度结果也会有差异[25, 47]。理想的引物既能覆盖样品中所有细菌和古细菌,扩增片段又能区分不同物种之间的差异,而且扩增长度也适合测序,然而实际上单一引物并不能实现。简并引物尽管能提高对微生物物种的覆盖度,但也不能涵盖所有的菌群[47]。表 3展示了常用的16S rRNA基因扩增引物及其在不同域下的覆盖率。另外数据库中16S rRNA序列长短不一,并非都为全长,这导致可变区覆盖率的评估可能存在偏差。因此,在实验设计时应当对所用引物的覆盖能力进行评估,必要时依据样品特点、测序方法以及目标菌群设计新的引物。目前已有一些工具辅助研究者设计16S rRNA基因扩增引物,如TestPrime[48]、TestProbe、ProbeBase[49]、PrimerProspector[50]、MIPE[51]等。随着16S rRNA相关数据库的不断完善,研究者也将能设计出更多高效的引物。
| Targeting regions | Primersa | Sequences | Domain Coverageb/% | References | ||
| Bacteria | Archaea | Eukaryota | ||||
| V4 | 515F | GTGCCAGCMGCCGCGGTAA | 93.7 | 92.7 | 0.5 | [52] |
| 806R | GGACTACHVGGGTWTCTAAT | |||||
| V4 | 515Fc | GTGYCAGCMGCCGCGGTAA | 94.6 | 92.8 | 2.0 | [53-54] |
| 806Rc | GGACTACNVGGGTWTCTAAT | |||||
| V3–V4 | 341F | CCTACGGGNGGCWGCAG | 92.5 | 69.5 | 0.1 | [55] |
| 805R | GACTACHVGGGTATCTAATCC | |||||
| V3–V4 | 338F | ACTCCTACGGGAGGCAGCAG | 89.2 | 0.0 | 0.1 | [48] |
| 806R | GGACTACHVGGGTWTCTAAT | |||||
| V4–V5 | 515F | GTGNCAGCMGCCGCGGTAA | 93.4 | 91.0 | 91.4 | [56] |
| 926R | CCGYCAATTYMTTTRAGTTT | |||||
| V5–V6 | U789F | TAGATACCCSSGTAGTCC | 88.6 | 89.0 | 0.1 | [57] |
| U1068R | CTGACGRCRGCCATGC | |||||
| V1–V3 | 27F | AGAGTTTGATYMTGGCTCAG | 90.3 | 0.0 | 0.2 | [58] |
| 515R | TTACCGCGGCKGCTGGCAC | |||||
| V1–V3 | 27F | GAGTTTGATCMTGGCTCAG | 89.9 | 0.0 | 0.2 | [59] |
| 518R | WTTACCGCGGCTGCTGG | |||||
| V3–V5 | 357F | CCTACGGGAGGCAGCAG | 91.9 | 1.4 | 0.2 | [60] |
| 926R | CCGYCAATTYMTTTRAGTTT | |||||
| V4–V6 | 518F | CCAGCAGCYGCGGTAAN | 94.5 | 32.5 | 0.1 | [61] |
| 1064R | CGACRRCCATGCANCACCT | |||||
| V1–V9 | 27F | AGAGTTTGATCMTGGCTCAG | 83.3 | 0.3 | 0.1 | [62] |
| 1492R | TACCTTGTTACGACTT | |||||
| a: The number indicate primer position according to the E. coli gene numbering; b: The coverage was estimated using the SILVA TestPrime tool with the SSU r138 database RefNR sequence collection and allowing non and one mismatch during primer annealing; c: The new constructs added degeneracy (changes are shown in bold) compare to the original. | ||||||
综上所述,对于16S rRNA基因测序扩增区的选择,既要考虑不同可变区物种注释准确率又要确保引物覆盖率,以减小对菌群多样性评估带来的误差。不同研究往往采用不同的引物和可变区,相同类型的样本在扩增区选择策略上也会有差异,这也导致不同研究间数据难以比较。引物与可变区作为相互关联的两个重要因素,共同影响着多样性分析结果,但目前扩增区的最佳选择尚未达成一致。而现有的研究在分析方法与评价指标上存在不足,例如没有考虑引物结合,以注释分辨率作为评价指标而未考虑注释准确率等,加之数据库与物种注释算法的不断更新,扩增区的最佳选择仍需要进行全面系统的评估。16S rRNA基因的全长测序能解决可变区的选择之争,如何从全长中选择有效信息尽可能区分不同物种将是新的研究方向。
3 下机数据预处理高通量测序技术使得微生物群落多样性分析进入大数据时代,同时催生了各种数据处理与分析软件(表 4)。16S rRNA测序数据预处理一般包括序列质控、拼接、去除嵌合体、操作分类单元聚类、分类学注释等步骤。由于测序过程碱基判读存在一定的错误,下机数据首先要去除低质量的碱基或序列[63],构建文库时加入的接头、标签以及过短的序列等也需要去除,以免影响下游数据的分析。双端测序的数据经过质控后还需要拼接。嵌合体是PCR扩增中产生的错误序列,由来自两条或者多条模板链的序列组成。而前期质控无法将其除尽,会导致物种多样性评估偏高。常用的去除嵌合体软件有Chimera Slayer[64]、UCHIME[65],无论哪一种工具其识别结果仅为可能的嵌合体。
| Major functions | Local softwares and web service |
| Quality control | mothur[66], QIIME[67], Trimmomatic[68], AmpliconNoise[69], FASTQC[70], VSEARCH[71] |
| Merging of paired-end reads | mothur[66], QIIME[67], FLASH[72], PANDAseq[73] |
| Chimera detection | mothur[66], QIIME[67], USEARCH[74], VSEARCH[71], Chimera Slayer[64], UCHIME[65], DECIPHER[75], CATCh[76] |
| OTU clustering | mothur[66], QIIME[67], USEARCH[74], VSEARCH[71], CD-HIT[77], UCLUST[78], DADA2[79], UNOISE2[80], Deblur[81] |
| Taxonomic assignment | mothur[66], QIIME[67], RTAX[82], RDP Classifier[33] |
| Reference database | Greengenes[83], RDP[84], SILVA[85] |
| Diversity analysis | mothur[66], QIIME[67], STAMP[86], LefSe[87], PICRUSt2[88], Tax4FUN[89], FAPROTAX[90], Galaxy[91], MG-RAST[92], VAMPS[93], Calypso[94], MicrobiomeAnalyst[95] |
操作分类单元(operational taxonomic unit,OTU)早期应用在数值分类学(numerical taxonomy)中,在扩增子数据分析中是指依据某种距离度量方法将序列按照分类阈值聚类而形成的不同组别。OTU聚类作为数据处理的关键环节,对含有大量未知微生物的群落多样性评估具有重要意义。传统的OTU聚类是将序列依据相似度进行分类,既消除PCR或测序导致的序列变异又降低了后续物种注释的计算量。早期研究认为若菌株之间16S rRNA相似度小于97%则属于不同物种[96-97],因此97%也常作为与种水平对应聚类阈值,之后将其修订为98.7%[98]。Kim等建议对于16S rRNA全长序列采用98.65%有利于发现新物种[99],而Edgar认为对于全长序列最佳聚类阈值约为99%,对于V4区约100%[100]。不同聚类相似度提供不同的分辨率来观察群落组成差异,但其聚类结果不能完全等价于某个分类学水平。
能实现OTU聚类的工具算法较多,它们在代表序列的选择上存在差异。CD-HIT[77]将序列按长度排序,取最长序列作为OTU的代表序列。而Uparse[74]算法将序列按丰度排序,取丰度最高的序列作为OTU的代表序列,原因是高丰度的序列更有可能是正确序列,并且在聚类的同时完成嵌合体去除。UCLUST[78]采用贪婪算法,其聚类速度快,既可按照序列长度也可以按照序列丰度选择OTU代表序列。QIIME[67]作为比较全面的测序数据处理与分析平台,不仅嵌套了可用于聚类的工具,诸如CD-HIT[77]、UCLUST[78]、BLAST[101]等,还提供3种聚类策略,分别为de novo、closed-reference、open-reference。de novo聚类不需要参考序列库,所有序列相互比对按相似度划分OTU。运用此策略所有序列都能分配至OTU中,但由于不支持并行计算,时间开销较大。closed-reference聚类则将序列与参考序列库比对,库中序列作为聚类中心,与之相似度大于设定阈值的序列将聚类成OTU,而比对不上的序列则被舍弃。此策略支持并行计算,时间开销较小,适用于较大的数据集;而且使得不同可变区的扩增子分析数据具有可比性。需要注意的是,这要求于库中序列覆盖这些可变区,不同可变区物种区分能力不同,数据可比性有限。该策略缺点在于物种信息局限于参考库,不利于发现新物种,甚至可能舍去有效序列。open-reference聚类结合了closed-reference和de novo两种方式,先将序列进行closed-reference聚类,未比对上的序列再进行de novo聚类,最后将2个OTU表合并。此策略既节约时间又有利于发现新物种。subsampled open-reference[102]是在open-reference基础上优化的聚类策略,closed-reference聚类比对不上数据库的序列随机抽取后进行de novo聚类,其中心序列构建成新的参考数据库再次进行closed-reference聚类,仍比对不上的序列进行de novo聚类,最后设定OTU的序列阈值进行过滤。此策略采用2次closed-reference聚类,有效缩短聚类时间,适用于大型数据集。在OTU代表序列选择上,QIIME支持随机、最长、丰度最高等方式。由于存在不同物种间16S rRNA序列相似度大于99%,同一物种间16S rRNA相似度小于95%的特殊情况[103-104],基于相似度聚类有一定的局限性[105]。而近几年还开发出依据精确序列变异的OTU聚类算法,如DADA2[79]、Deblur[81],这使得分辨群落结构差异能力显著提高,同时也使得研究数据具有可比性[106]。
物种分类学注释是将每条序列或OTU代表序列与数据库中参考序列通过算法进行比对,库中比对结果最佳序列的注释信息即为该序列或OTU的注释信息,常用的工具有mothur[66]、QIIME[67]、RDP Classifier[33]。注释结果准确度除了与工具算法有关还依赖于所使用的数据库,尽管GenBank数据库中含有大量的16S rRNA序列,考虑到其序列质量问题,一般采用专门针对16S rRNA的数据库,如Greengenes[83]、RDP[84]、SILVA[85]等。由于参考库之间序列数量以及注释格式存在差异,不同研究之间进行比较时需要注意使用的注释参考库是否一致。
经过以上流程,就从原始序列中得到操作分类单元表和物种分类学表等计数型数据(count data) 特征表。这些矩阵表在用于群落多样性分析前需进行数据过滤与标准化,一般去除丰度为1的OTU即单序列(singleton),或去除丰度小于总测序数据量0.005%的OTU[63]。为了使样本间的群落组成具有可比性,需将OTU表中各样本稀释至相同序列数[107],一般按样本的最小序列数稀释。稀释曲线趋于平缓则说明测序数据量能够覆盖样本中的绝大部分物种。数据过滤与稀释会使某些低丰度的物种被丢弃,若研究关注稀有物种,则应保证每个样本有足够的测序数据量。
4 群落多样性分析 4.1 群落结构可视化微生物群落分类学数据具有高维度和稀疏性的特点,在大数据背景下,如何对其进行有效的数据挖掘与可视化分析成为挑战。微生物群落结构可用较为直观的堆叠图、饼图等方式来展示,旭日图除了表示不同物种的占比信息还体现物种间的层级关系。Circos弦图展示了每个样本中优势物种的组成比例以及各优势物种在样本中的分布比例,可用于分析样本与物种关系。但随着样本数增大以及物种组成复杂度变高,这些方式呈现信息的可读性在降低。韦恩图在分析不同样本或分组群落结构相似性与重叠情况时,只考虑了微生物是否存在而忽略了相对丰度,且同时比较的样本或分组数量有限。而热图能同时呈现群落物种组成及丰度信息,通过颜色变化直观反映不同样本或分组在群落组成上的相似性和差异性。此外,还可根据物种或样本间丰度的相似性进行聚类分析。两组间的差异比较可以用火山图、曼哈顿图进行可视化分析,而三组之间的物种组成和分布情况则可用三元相图展示。每种方式都有各自的优缺点,实际应用需根据数据情况选择合适的可视化方式。微生物群落多样性分析中常用的可视化分析图例见图 3。

|
| 图 3 微生物群落多样性分析中常用的可视化分析图例[13] Figure 3 Figures commonly used in analysis of microbial diversity[13]. A: stacked bar of community structure on the family level; B: venn diagram on the OTU level; C: heatmap of community structure on the family level; D: PCA scatter plot on the OTU level; E: circos plot; F: LefSe cladogram. |
4.2 生物多样性分析
α多样性与β多样性在微生物生态研究中被广泛应用。α多样性是描述一个特定区域或生态系统内的生物多样性,即评估某个样本的生物多样性,一般依据物种丰富度或均匀度计算多样性指数来表征。Chao1[108]和ACE[109]指数常用于估算群落物种总数。低丰度的物种数量对Chao1指数影响较大,而ACE指数通过设定稀有物种丰度阈值为10来减小这种误差。而Shannon[110]和Simpson[111]指数同时考虑物种丰富度和均匀度,能相对客观地反映群落物种多样性。由于没有考虑物种在生态系统中的功能以及相互之间的关系,以物种丰富度和均匀度反映生物多样性并不全面。PD指数[112]在丰富度和均匀度的基础上结合物种间进化关系来衡量系统发育多样性。进化距离近的物种常有类似的功能,因而PD也被用于反映群落功能多样性。由于各多样性指数考虑的因素以及加权的不同,分析结果会存在差异。
β多样性是描述不同生境群落之间物种组成的差异,即比较不同样本间的差异,常用的距离算法有Jaccard[113]、Bray-Curtis[114]、Unifrac[115]等。这些距离算法考虑因素不尽相同,Jaccard只以物种有无来计算样本间的距离,而Bray-Curtis同时考虑了物种有无和丰度大小,Unifrac则考虑了OTU间系统发育关系。得到距离矩阵后可通过层次聚类、PCoA、NMDS等进行排序分析并可视化展示,直观分析样本或分组间群落组成的差异性。仅通过散点的距离判断结果较为主观,一般结合ANOSIM或PERMANOVA统计学检验判断不同分组间群落组成是否有显著差异。
4.3 差异物种筛选与环境因子关联分析为了比较某一分类学水平下两组或多组间物种丰度是否有显著性差异,可以采用经典的统计学检验方法,如T-test/ANOVA、Mann-Whitney/ Kruskal-Wallis等。近年来,还开发出组间差异分析的R包,如MetagenomeSeq[116]、edgeR[117]、DESeq2[118]等。由于样本的物种组成十分复杂,需要进行多次假设检验,常采用FDR、Bonferroni等方法来进行多重检验校正,以减小假阳性的发生率。STAMP[86]软件既可进行组间物种差异的统计学检验又提供了丰富的作图功能。随机森林分类器与LefSe[87]分析常用于筛选对分组起重要作用的生物标记物,随机森林模型通过挖掘变量之间非线性的相互依赖关系,找到能够区分两组差异的关键物种,而LefSe通过对样本按照不同的分组条件进行线性判别分析,找出对多组间样本划分产生显著性差异影响的物种。
在探索样本、微生物群落以及环境因子三者之间的复杂关系时,可进行单变量相关分析,如皮尔森(Pearson)或斯皮尔曼(Spearman)相关分析。皮尔森相关分析表明两连续变量的线性相关程度与方向,要求两变量相互独立、均为连续变量且各自总体呈正态分布;而斯皮尔曼相关分析反映等级数据或顺序数据中各变量排列顺序的相关程度,不需要变量服从正态分布。真实的微生物生态环境复杂,往往需要同时考查多个变量之间的相关性,常采用多变量相关分析方法,如典范对应分析(canonical correspondence analysis,CCA)和冗余分析(redundancy analysis,RDA)。CCA是基于对应分析演变而来的排序方法,将对应分析与多元回归分析相结合,在对应分析的迭代过程中,每次得到的排序坐标值均与环境因子进行多元线性回归。而RDA是回归分析与主成分分析结合的排序方法,将应变量矩阵与解释变量矩阵多元回归的拟合值矩阵进行PCA降维分析。CCA基于单峰模型,而RDA基于线性模型,实际应用需根据物种分布变化情况,选择合适的分析模型。此外,检验两个矩阵相关性的Mantel test也可用于分析环境因子与微生物群落组成的相关性。若群落组成与样本表型存在高度相关性,那么在此基础上建立回归模型来预测未知样本的表型具有很好的实用价值,例如利用肠道菌群组成辅助诊断某些相关疾病等。
统计学分析方法复杂多样,原理及适用情况不同,实际实验样本数量有限,加之实验环节容易引入噪音,具有统计学意义的结果并不代表具备很好的生物学意义。而且变量之间存在相关性不能代表因果关系,也不能直接反映微生物间相互作用关系。因此,通过变量间的相关性推断潜在的相互作用需要谨慎,进一步的实验验证与机理的探究十分重要。
4.4 群落功能预测相似生态环境中的微生物群落可能组成不同而功能相似[119],揭示微生物群落功能有助于理解微生物群落与环境相互作用的机理。PICRUSt[120]通过祖先状态重建实现KEGG、COG功能预测,而Tax4FUN[89]根据构建SILVA[85]数据库与KEGG数据库中生物学分类间的线性转换也可实现KEGG功能预测。相比之下,Tax4Fun预测结果与宏基因组功能分析结果的相关性要高于PICRUSt,但Tax4Fun缺乏类似PICRUSt的NSTI (nearest sequenced taxon index)质控参数。FAPROTAX[90]是联系物种与其功能注释的数据库,其结果准确性与序列分类学注释水平有关。不同预测方法适合样本类型不同,PICRUSt和Tax4FUN适用于人体肠道微生物,而FAPROTAX更适用于环境样本。通过16S rRNA基因测序预测群落功能具有一定的局限性,一方面,不同物种16S rRNA序列高度相似不代表共有的功能基因高度相似[121],另一方面,某种微生物存在也不代表发挥其相关的生物学功能。采用预测的方式尽管不能取代宏基因组测序实验,对后续宏基因组实验设计还是具有一定的指导意义。
5 问题和挑战尽管通过16S rRNA基因测序分析微生物群落多样性的研究模式已被广泛使用,但这种方法也面临着一系列的问题与挑战。
首先,嵌合体和错误序列造成群落多样性评估偏高。PCR扩增产生的嵌合体和测序造成的错误序列若未除尽,则可能被错误地分类注释甚至被误认为是新物种。除了使用Chimera Slayer[64]、UCHIME[65]等嵌合体检测软件,也可尝试从改进实验的角度来减少嵌合体的形成[122]。
其次,16S rRNA拷贝数使得物种相对丰度难以准确评估。原核生物16S rRNA基因普遍存在多拷贝数,范围从1–15不等[123],且同一菌株内16S rRNA基因序列还可能发生变异[124-125],这使得绝对定量分析难以进行,多样性评估也会有偏差。针对拷贝数的问题,Stoddard等建立的rrnDB[126]数据库收录了各种细菌和古细菌的16S rRNA基因拷贝数,可用于校正物种丰度[127]。PICRUSt[120]、CopyRighter[128]、PAPRICA[129]能利用系统发育关系预测不同物种的16S rRNA基因拷贝数,但Louca等[130]对这三者评估后认为预测效果都不佳。
最后,物种相对丰度的变化有歧义。某一类群微生物的相对丰度比例改变不一定是本身丰度变化引起的,其他类群丰度变化也会产生影响[131-132],如何精准地定量分析微生物群落多样性也还有待解决。尽管16S rRNA是物种鉴定中常用的分子标志物,但对于某些菌群的区分效果并不佳。部分是由可变区、扩增长度和数据库造成的。另外,还存在不同物种间16S rRNA序列相似大于99%,同一物种间16S rRNA序列相似度小于95%的特殊现象[103-104],这也会影响群落多样性评估。
可用于16S rRNA基因测序数据处理的工具众多,多数都是基于Linux系统以命令行的形式运行。其中以mothur[66]和QIIME[67]平台为代表,它们涵盖了较为完整的数据处理与分析流程。目前也有一些交互式的网页分析平台,如MG-RAST[92]、MicrobiomeAnalyst[95]等,这在一定程度上降低了分析难度,但并未涵盖下机数据预处理部分。随着测序技术与工具算法的不断开发与改进,在关注新进展的同时还要注意分析比较,根据具体需要和性能选择合适的工具。不同测序平台、可变区、引物、分析流程等中间因素都会导致结果有差异,这对结果分析的可重复性,研究之间的可比性造成挑战。一方面,可通过小样本的预实验或设计已知组分的内参菌群样本减小误差;另一方面,引物与可变区仍需要全面系统性的评估以求最优。此外,测序数据处理与分析流程也有待形成标准化体系。
6 展望近年来高通量测序技术发展迅速,16S rRNA基因测序已广泛应用于各种生态环境中的微生物多样性分析。三代测序技术虽然在准确率和测序成本等方面还有待提高,但随着测序准确度、读长和通量等参数不断优化以及测序费用的下降,高质量、高通量的16S rRNA基因全长测序将会成为现实。此外,16S rRNA相关数据库的扩大以及生物信息学算法的不断开发,也将为微生物群落多样性研究提供更加准确、全面的信息。测序数据处理集成化、分析结果可视化有利于提高分析效率,交互友好、操作简便的测序数据处理与分析软件还有待开发。
监测微生物多样性对分析群落的演替规律,了解种群动态变化特征至关重要,全面理解微生物多样性有助于揭示微生物与环境相互作用的关系。16S rRNA基因测序与宏基因组测序、代谢组学等多组学联用也将成为新趋势。16S rRNA基因测序解析微生物群落结构,挖掘样本特征与群落特征的关联;宏基因组测序寻找重要编码基因或富集的代谢通路,而代谢组学进一步反映菌群生化功能在分子水平上的变化。这些研究方法相辅相成,克服了单一组学研究的局限性。微生物群落多样性的研究促进了环境科学和生态学的发展,也将在改善人体健康,提高作物产量,开发清洁能源等其他方面做出重要贡献。
| [1] | Noss RF. Indicators for monitoring biodiversity: a hierarchical approach. Conservation Biology, 1990, 4(4): 355-364. DOI:10.1111/j.1523-1739.1990.tb00309.x |
| [2] | Ferrera I, Sánchez O. Insights into microbial diversity in wastewater treatment systems: how far have we come?. Biotechnology Advances, 2016, 34(5): 790-802. DOI:10.1016/j.biotechadv.2016.04.003 |
| [3] | Nannipieri P, Ascher-Jenull J, Ceccherini MT, Pietramellara G, Renella G, Schloter M. Beyond microbial diversity for predicting soil functions: a mini review. Pedosphere, 2020, 30(1): 5-17. DOI:10.1016/S1002-0160(19)60824-6 |
| [4] | Logan BE, Rossi R, Ragab A, Saikaly PE. Electroactive microorganisms in bioelectrochemical systems. Nature Reviews Microbiology, 2019, 17(5): 307-319. DOI:10.1038/s41579-019-0173-x |
| [5] | Tamang JP, Watanabe K, Holzapfel WH. Review: diversity of microorganisms in global fermented foods and beverages. Frontiers in Microbiology, 2016, 7: 377. |
| [6] | Kriss M, Hazleton KZ, Nusbacher NM, Martin CG, Lozupone CA. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Current Opinion in Microbiology, 2018, 44: 34-40. DOI:10.1016/j.mib.2018.07.003 |
| [7] | Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 1995, 59(1): 143-169. DOI:10.1128/MR.59.1.143-169.1995 |
| [8] | Steen AD, Crits-Christoph A, Carini P, DeAngelis KM, Fierer N, Lloyd KG, Thrash JC. High proportions of bacteria and archaea across most biomes remain uncultured. The ISME Journal, 2019, 13(12): 3126-3130. DOI:10.1038/s41396-019-0484-y |
| [9] | Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl G. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(32): 12115-12120. DOI:10.1073/pnas.0605127103 |
| [10] | The human microbiome project consortium. Structure, function and diversity of the healthy human microbiome. Nature, 2012, 486(7402): 207-214. DOI:10.1038/nature11234 |
| [11] | The Integrative HMP (iHMP) research network consortium. The integrative human microbiome project. Nature, 2019, 569(7758): 641-648. DOI:10.1038/s41586-019-1238-8 |
| [12] | Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature, 2007, 449(7164): 804-810. DOI:10.1038/nature06244 |
| [13] | Wang GH, Liu Q, Guo L, Zeng HJ, Ding CC, Zhang WT, Xu DP, Wang X, Qiu JX, Dong QL, Fan ZQ, Zhang Q, Pan J. Gut microbiota and relevant metabolites analysis in alcohol dependent mice. Frontiers in Microbiology, 2018, 9: 1874. DOI:10.3389/fmicb.2018.01874 |
| [14] | Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. Journal of Molecular Biology, 1975, 94(3): 441-448. DOI:10.1016/0022-2836(75)90213-2 |
| [15] | Maxam AM, Gilbert W. A new method for sequencing DNA. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74(2): 560-564. DOI:10.1073/pnas.74.2.560 |
| [16] | Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74(12): 5463-5467. DOI:10.1073/pnas.74.12.5463 |
| [17] | Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nature Protocols, 2008, 3(2): 267-278. DOI:10.1038/nprot.2007.520 |
| [18] | Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods, 2008, 5(3): 235-237. DOI:10.1038/nmeth.1184 |
| [19] | Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single-molecule DNA sequencing of a viral genome. Science, 2008, 320(5872): 106-109. DOI:10.1126/science.1150427 |
| [20] | Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, deWinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong XX, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma CC, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen GN, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science, 2009, 323(5910): 133-138. DOI:10.1126/science.1162986 |
| [21] | Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotechnology, 2009, 4(4): 265-270. DOI:10.1038/nnano.2009.12 |
| [22] | Contreras AV, Cocom-Chan B, Hernandez-Montes G, Portillo-Bobadilla T, Resendis-Antonio O. Host-microbiome interaction and cancer: potential application in precision medicine. Frontiers in Physiology, 2016, 7: 606. |
| [23] | Netto GJ, Kaul KL, Best BPG. Genomic applications in pathology. 2nd ed. New York: Springer, 2019: 751–766. |
| [24] | Wadhwa G, Shanmughavel P, Singh AK, Bellare JR. Current trends in bioinformatics: an insight. Singapore: Springer, 2018: 27-38. |
| [25] | Winand R, Bogaerts B, Hoffman S, Lefevre L, Delvoye M, van Braekel J, Fu Q, Roosens NH, De Keersmaecker SCJ, Vanneste K. Targeting the 16S rRNA gene for bacterial identification in complex mixed samples: comparative evaluation of second (Illumina) and Third (Oxford Nanopore Technologies) generation sequencing technologies. International Journal of Molecular Sciences, 2020, 21(1): 298. |
| [26] | Salipante SJ, Kawashima T, Rosenthal C, Hoogestraat DR, Cummings LA, Sengupta DJ, Harkins TT, Cookson BT, Hoffman NG. Performance comparison of illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Applied and Environmental Microbiology, 2014, 80(24): 7583-7591. DOI:10.1128/AEM.02206-14 |
| [27] | Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, Koci M, Ballou A, Mendoza M, Ali R, Azcarate-Peril MA. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiology, 2017, 17(1): 194. DOI:10.1186/s12866-017-1101-8 |
| [28] | Singer E, Bushnell B, Coleman-Derr D, Bowman B, Bowers RM, Levy A, Gies EA, Cheng JF, Copeland A, Klenk HP, Hallam SJ, Hugenholtz P, Tringe SG, Woyke T. High-resolution phylogenetic microbial community profiling. The ISME Journal, 2016, 10(8): 2020-2032. DOI:10.1038/ismej.2015.249 |
| [29] | Benítez-Páez A, Portune KJ, Sanz Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION™ portable nanopore sequencer. Gigascience, 2016, 5(1): 4. DOI:10.1186/s13742-016-0111-z |
| [30] | Wagner J, Coupland P, Browne HP, Lawley TD, Francis SC, Parkhill J. Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiology, 2016, 16(1): 274. DOI:10.1186/s12866-016-0891-4 |
| [31] | Rintala A, Pietilä S, Munukka E, Eerola E, Pursiheimo JP, Laiho A, Pekkala S, Huovinen P. Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. Journal of Biomolecular Techniques: JBT, 2017, 28(1): 19-30. DOI:10.7171/jbt.17-2801-003 |
| [32] | Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiological Methods, 2007, 69(2): 330-339. DOI:10.1016/j.mimet.2007.02.005 |
| [33] | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 2007, 73(16): 5261-5267. DOI:10.1128/AEM.00062-07 |
| [34] | Vilo C, Dong QF. Evaluation of the RDP classifier accuracy using 16S rRNA gene variable regions. Metagenomics, 2012, 1: 235551. |
| [35] | Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Research, 2010, 38(22): e200. DOI:10.1093/nar/gkq873 |
| [36] | Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nature Communications, 2019, 10(1): 5029. DOI:10.1038/s41467-019-13036-1 |
| [37] | Kim M, Morrison M, Yu ZT. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. Journal of Microbiological Methods, 2011, 84(1): 81-87. DOI:10.1016/j.mimet.2010.10.020 |
| [38] | Yang B, Wang Y, Qian PY. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics, 2016, 17: 135. DOI:10.1186/s12859-016-0992-y |
| [39] | Barb JJ, Oler AJ, Kim HS, Chalmers N, Wallen GR, Cashion A, Munson PJ, Ames NJ. Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA using mock samples. PLoS One, 2016, 11(2): e0148047. DOI:10.1371/journal.pone.0148047 |
| [40] | Zhang JY, Ding X, Guan R, Zhu CM, Xu C, Zhu BC, Zhang H, Xiong ZP, Xue YG, Tu J, Lu ZH. Evaluation of different 16S rRNA gene V regions for exploring bacterial diversity in a eutrophic freshwater lake. Science of the Total Environment, 2018, 618: 1254-1267. DOI:10.1016/j.scitotenv.2017.09.228 |
| [41] | Tessler M, Neumann JS, Afshinnekoo E, Pineda M, Hersch R, Velho LFM, Segovia BT, Lansac-Toha FA, Lemke M, DeSalle R, Mason CE, Brugler MR. Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Scientific Reports, 2017, 7(1): 6589. DOI:10.1038/s41598-017-06665-3 |
| [42] | Kumar PS, Brooker MR, Dowd SE, Camerlengo T. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS One, 2011, 6(6): e20956. DOI:10.1371/journal.pone.0020956 |
| [43] | Thijs S, De Beeck MO, Beckers B, Truyens S, Stevens V, van Hamme JD, Weyens N, Vangronsveld J. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Frontiers in Microbiology, 2017, 8: 494. |
| [44] | Karabudak S, Ari O, Durmaz B, Dal T, Başyiğit T, Kalcioglu MT, Durmaz R. Analysis of the effect of smoking on the buccal microbiome using next-generation sequencing technology. Journal of Medical Microbiology, 2019, 68(8): 1148-1158. DOI:10.1099/jmm.0.001003 |
| [45] | Wang F, Men X, Zhang G, Liang KC, Xin YH, Wang J, Li AJ, Zhang HB, Liu HB, Wu LJ. Assessment of 16S rRNA gene primers for studying bacterial community structure and function of aging flue-cured tobaccos. AMB Express, 2018, 8(1): 182. DOI:10.1186/s13568-018-0713-1 |
| [46] | Wear EK, Wilbanks EG, Nelson CE, Carlson CA. Primer selection impacts specific population abundances but not community dynamics in a monthly time - series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environmental Microbiology, 2018, 20(8): 2709-2726. DOI:10.1111/1462-2920.14091 |
| [47] | Mori H, Maruyama F, Kato H, Toyoda A, Dozono A, Ohtsubo Y, Nagata Y, Fujiyama A, Tsuda M, Kurokawa K. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Research, 2014, 21(2): 217-227. DOI:10.1093/dnares/dst052 |
| [48] | Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 2013, 41(1): e1. DOI:10.1093/nar/gks808 |
| [49] | Greuter D, Loy A, Horn M, Rattei T. ProbeBase—an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Research, 2016, 44(D1): D586-D589. DOI:10.1093/nar/gkv1232 |
| [50] | Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics, 2011, 27(8): 1159-1161. DOI:10.1093/bioinformatics/btr087 |
| [51] | Zou B, Li JF, Zhou Q, Quan ZX. MIPE: a metagenome-based community structure explorer and SSU primer evaluation tool. PLoS One, 2017, 12(3): e0174609. DOI:10.1371/journal.pone.0174609 |
| [52] | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(S1): 4516-4522. |
| [53] | Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology, 2015, 75(2): 129-137. DOI:10.3354/ame01753 |
| [54] | Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology, 2016, 18(5): 1403-1414. DOI:10.1111/1462-2920.13023 |
| [55] | Parulekar NN, Kolekar P, Jenkins A, Kleiven S, Utkilen H, Johansen A, Sawant S, Kulkarni-Kale U, Kale M, Sæbø M. Characterization of bacterial community associated with phytoplankton bloom in a eutrophic lake in South Norway using 16S rRNA gene amplicon sequence analysis. PLoS One, 2017, 12(3): e0173408. DOI:10.1371/journal.pone.0173408 |
| [56] | Lee MD, Walworth NG, McParland EL, Fu FX, Mincer TJ, Levine NM, Hutchins DA, Webb EA. The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. The ISME Journal, 2017, 11(8): 1813-1824. DOI:10.1038/ismej.2017.49 |
| [57] | Bougouffa S, Yang JK, Lee OO, Wang Y, Batang Z, Al-Suwailem A, Qian PY. Distinctive microbial community structure in highly stratified deep-sea brine water columns. Applied and Environmental Microbiology, 2013, 79(11): 3425-3437. DOI:10.1128/AEM.00254-13 |
| [58] | Ceuppens S, De Coninck D, Bottledoorn N, van Nieuwerburgh F, Uyttendaele M. Microbial community profiling of fresh basil and pitfalls in taxonomic assignment of enterobacterial pathogenic species based upon 16S rRNA amplicon sequencing. International Journal of Food Microbiology, 2017, 257: 148-156. DOI:10.1016/j.ijfoodmicro.2017.06.016 |
| [59] | Ibarbalz FM, Figuerola ELM, Erijman L. Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Research, 2013, 47(11): 3854-3864. DOI:10.1016/j.watres.2013.04.010 |
| [60] | Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A, Rodríguez-Hilario A, González H, Bondy J, Lawson F, Folawiyo O, Michailidi C, Dziedzic A, Thangavel R, Hadar T, Noordhuis MG, Westra W, Koch W, Sidransky D. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget, 2016, 7(32): 51320-51334. DOI:10.18632/oncotarget.9710 |
| [61] | Kleindienst S, Grim S, Sogin M, Bracco A, Crespo-Medina M, Joye SB. Diverse, rare microbial taxa responded to the Deepwater horizon deep - sea hydrocarbon plume. The ISME Journal, 2016, 10(2): 400-415. DOI:10.1038/ismej.2015.121 |
| [62] | Klemetsen T, Willassen NP, Karlsen CR. Full‐length 16S rRNA gene classification of Atlantic salmon bacteria and effects of using different 16S variable regions on community structure analysis. MicrobiologyOpen, 2019, 8(10): e898. |
| [63] | Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods, 2013, 10(1): 57-59. DOI:10.1038/nmeth.2276 |
| [64] | Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Research, 2011, 21(3): 494-504. DOI:10.1101/gr.112730.110 |
| [65] | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [66] | Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 2009, 75(23): 7537-7541. DOI:10.1128/AEM.01541-09 |
| [67] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QⅡME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [68] | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014, 30(15): 2114-2120. DOI:10.1093/bioinformatics/btu170 |
| [69] | Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics, 2011, 12: 38. DOI:10.1186/1471-2105-12-38 |
| [70] | Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One, 2012, 7(2): e30619. DOI:10.1371/journal.pone.0030619 |
| [71] | Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ, 2016, 4(17): e2584. |
| [72] | Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 2011, 27(21): 2957-2963. DOI:10.1093/bioinformatics/btr507 |
| [73] | Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics, 2012, 13: 31. DOI:10.1186/1471-2105-13-31 |
| [74] | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 2013, 10(10): 996-998. DOI:10.1038/nmeth.2604 |
| [75] | Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Applied and Environmental Microbiology, 2012, 78(3): 717-725. DOI:10.1128/AEM.06516-11 |
| [76] | Mysara M, Saeys Y, Leys N, Raes J, Monsieurs P. CATCh, an ensemble classifier for chimera detection in 16S rRNA sequencing studies. Applied and Environmental Microbiology, 2015, 81(5): 1573-1584. DOI:10.1128/AEM.02896-14 |
| [77] | Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 2012, 28(23): 3150-3152. DOI:10.1093/bioinformatics/bts565 |
| [78] | Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 2010, 26(19): 2460-2461. DOI:10.1093/bioinformatics/btq461 |
| [79] | Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods, 2016, 13(7): 581-583. DOI:10.1038/nmeth.3869 |
| [80] | Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 2016. |
| [81] | Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Xu ZZ, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. Msystems, 2017, 2(2): e00191-16. |
| [82] | Soergel DAW, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. The ISME Journal, 2012, 6(7): 1440-1444. DOI:10.1038/ismej.2011.208 |
| [83] | DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 2006, 72(7): 5069-5072. DOI:10.1128/AEM.03006-05 |
| [84] | Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun YN, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Research, 2014, 42(D1): D633-D642. DOI:10.1093/nar/gkt1244 |
| [85] | Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research, 2007, 35(21): 7188-7196. DOI:10.1093/nar/gkm864 |
| [86] | Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics, 2014, 30(21): 3123-3124. DOI:10.1093/bioinformatics/btu494 |
| [87] | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biology, 2011, 12(6): R60. DOI:10.1186/gb-2011-12-6-r60 |
| [88] | Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2: an improved and extensible approach for metagenome inference. BioRxiv, 2019. |
| [89] | Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics, 2015, 31(17): 2882-2884. DOI:10.1093/bioinformatics/btv287 |
| [90] | Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science, 2016, 353(6305): 1272-1277. DOI:10.1126/science.aaf4507 |
| [91] | Afgan E, Baker D, Batut B, van Den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research, 2018, 46(W1): W537-W544. DOI:10.1093/nar/gky379 |
| [92] | Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. The metagenomics RAST server-a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics, 2008, 9: 386. DOI:10.1186/1471-2105-9-386 |
| [93] | Huse SM, Welch DBM, Voorhis A, Shipunova A, Morrison HG, Eren AM, Sogin ML. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics, 2014, 15: 41. DOI:10.1186/1471-2105-15-41 |
| [94] | Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, Krause L. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics, 2017, 33(5): 782-783. |
| [95] | Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia JG. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research, 2017, 45(W1): W180-W188. DOI:10.1093/nar/gkx295 |
| [96] | Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. International Journal of Systematic and Evolutionary Microbiology, 2010, 60(1): 249-266. DOI:10.1099/ijs.0.016949-0 |
| [97] | Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology, 1994, 44(4): 846-849. DOI:10.1099/00207713-44-4-846 |
| [98] | Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today, 2006, 33(4): 152-155. |
| [99] | Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 2014, 64(2): 346-351. |
| [100] | Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics, 2018, 34(14): 2371-2375. DOI:10.1093/bioinformatics/bty113 |
| [101] | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology, 1990, 215(3): 403-410. DOI:10.1016/S0022-2836(05)80360-2 |
| [102] | Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou HW, Knight R, Caporaso JG. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ, 2014, 2(5): e545. |
| [103] | Fox GE, Wisotzkey JD, Jurtshuk P Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. International Journal of Systematic and Evolutionary Microbiology, 1992, 42(1): 166-170. |
| [104] | Beye M, Fahsi N, Raoult D, Fournier PE. Careful use of 16S rRNA gene sequence similarity values for the identification of Mycobacterium species. New Microbes and New Infections, 2018, 22: 24-29. DOI:10.1016/j.nmni.2017.12.009 |
| [105] | Nguyen NP, Warnow T, Pop M, White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. npj Biofilms and Microbiomes, 2016, 2: 16004. DOI:10.1038/npjbiofilms.2016.4 |
| [106] | Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME Journal, 2017, 11(12): 2639-2643. DOI:10.1038/ismej.2017.119 |
| [107] | Hughes JB, Hellmann JJ. The application of rarefaction techniques to molecular inventories of microbial diversity. Methods in Enzymology, 2005, 397: 292-308. |
| [108] | Chao AN. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics, 1984, 11(4): 265-270. |
| [109] | Chao AN, Ma MC, Yang MCK. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika, 1993, 80(1): 193-201. DOI:10.1093/biomet/80.1.193 |
| [110] | Shannon CE. A mathematical theory of communication. Bell System Technical Journal, 1948, 27(3): 379-423. DOI:10.1002/j.1538-7305.1948.tb01338.x |
| [111] | Simpson EH. Measurement of diversity. Nature, 1949, 163(4148): 688. DOI:10.1038/163688a0 |
| [112] | Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation, 1992, 61(1): 1-10. DOI:10.1016/0006-3207(92)91201-3 |
| [113] | Jaccard P. Étude comparative de la distribution florale dans une portion des Alpes et du Jura. Bulletin de la Société Vaudoise des Sciences Naturelles, 1901, 37: 547-579. |
| [114] | Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs, 1957, 27(4): 325-349. DOI:10.2307/1942268 |
| [115] | Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology, 2005, 71(12): 8228-8235. DOI:10.1128/AEM.71.12.8228-8235.2005 |
| [116] | Paulson JN, Talukder H, Pop M, Bravo HC. MetagenomeSeq: statistical analysis for sparse high-throughput sequencing. Bioconductor Packages, 2013, 1: 1-20. |
| [117] | Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010, 26(1): 139-140. DOI:10.1093/bioinformatics/btp616 |
| [118] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 2014, 15(12): 550. DOI:10.1186/s13059-014-0550-8 |
| [119] | Gibbons SM. Microbial community ecology: function over phylogeny. Nature Ecology & Evolution, 2017, 1(1): 0032. |
| [120] | Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 2013, 31(9): 814-821. DOI:10.1038/nbt.2676 |
| [121] | Sevigny JL, Rothenheber D, Diaz KS, Zhang Y, Agustsson K, Bergeron RD, Thomas WK. Marker genes as predictors of shared genomic function. BMC Genomics, 2019, 20(1): 268. DOI:10.1186/s12864-019-5641-1 |
| [122] | Boers SA, Hays JP, Jansen R. Micelle PCR reduces chimera formation in 16S rRNA profiling of complex microbial DNA mixtures. Scientific Reports, 2015, 5: 14181. DOI:10.1038/srep14181 |
| [123] | Sun DL, Jiang X, Wu QL, Zhou NY. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Applied and Environmental Microbiology, 2013, 79(19): 5962-5969. DOI:10.1128/AEM.01282-13 |
| [124] | Takeda K, Chikamatsu K, Igarashi Y, Morishige Y, Murase Y, Aono A, Yamada H, Takaki A, Mitarai S. Six species of nontuberculous mycobacteria carry non-identical 16S rRNA gene copies. Journal of Microbiological Methods, 2018, 155: 34-36. DOI:10.1016/j.mimet.2018.08.012 |
| [125] | Miyazaki K, Tomariguchi N. Occurrence of randomly recombined functional 16S rRNA genes in Thermus thermophilus suggests genetic interoperability and promiscuity of bacterial 16S rRNAs. Scientific Reports, 2019, 9: 11233. DOI:10.1038/s41598-019-47807-z |
| [126] | Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Research, 2015, 43(D1): D593-D598. DOI:10.1093/nar/gku1201 |
| [127] | Wu LW, Yang YF, Chen S, Shi ZJ, Zhao MX, Zhu ZW, Yang SH, Qu YY, Ma Q, He ZL, Zhou JZ, He Q. Microbial functional trait of rRNA operon copy numbers increases with organic levels in anaerobic digesters. The ISME Journal, 2017, 11(12): 2874-2878. DOI:10.1038/ismej.2017.135 |
| [128] | Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome, 2014, 2: 11. DOI:10.1186/2049-2618-2-11 |
| [129] | Bowman JS, Ducklow HW. Microbial communities can be described by metabolic structure: a general framework and application to a seasonally variable, depth-stratified microbial community from the coastal West Antarctic Peninsula. PLoS One, 2015, 10(8): e0135868. DOI:10.1371/journal.pone.0135868 |
| [130] | Louca S, Doebeli M, Parfrey LW. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome, 2018, 6(1): 41. DOI:10.1186/s40168-018-0420-9 |
| [131] | Props R, Kerckhof FM, Rubbens P, De Vrieze J, Sanabria EH, Waegeman W, Monsieurs P, Hammes F, Boon N. Absolute quantification of microbial taxon abundances. The ISME Journal, 2017, 11(2): 584-587. DOI:10.1038/ismej.2016.117 |
| [132] | Vandeputte D, Kathagen G, D'hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. Quantitative microbiome profiling links gut community variation to microbial load. Nature, 2017, 551(7681): 507-511. DOI:10.1038/nature24460 |
 2021, Vol. 61
2021, Vol. 61