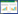中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 许少怡, 肖锐, 柴文波, 王保战, 逯慧杰. 2021
- Shaoyi Xu, Rui Xiao, Wenbo Chai, Baozhan Wang, Huijie Lu. 2021
- 全程硝化菌微生物学特性及在水处理领域的应用潜力
- Microbial characteristics and application potential of complete ammonia oxidation bacteria in water treatment system
- 微生物学报, 61(2): 315-332
- Acta Microbiologica Sinica, 61(2): 315-332
-
文章历史
- 收稿日期:2020-04-02
- 修回日期:2020-05-23
- 网络出版日期:2020-07-29
2. 南京农业大学生命科学学院, 江苏 南京 210095
2. College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, Jiangsu Province, China
传统硝化理论认为微生物自养硝化过程包括由氨氧化细菌(Ammonia-oxidizing bacteria,AOB)和氨氧化古菌(Ammonia-oxidizing archaea,AOA)介导的氨氧化过程以及亚硝酸盐氧化菌(Nitrite-oxidizing bacteria,NOB)介导的亚硝酸盐氧化过程组成[1]。2006年,Coast等通过最佳代谢路径长度的动力学预测理论上存在能独立完成全程硝化过程(Complete ammonia oxidation,comammox)的微生物,且具备生长速率低、细胞产率高的特点[2],这一预测直到2015年底才被证实。来自荷兰、奥地利、美国的科学家分别通过对不同环境样品的富集培养和宏基因组测序分析,发现了一种可以独立完成comammox过程的硝化螺菌属(Nitrospira)细菌[3–5]。此类微生物具备硝化关键功能酶(氨氧化过程为氨单加氧酶(AMO)和羟胺脱氢酶(HAO),亚硝酸盐氧化过程为亚硝酸盐氧化还原酶(NXR))的完整基因组[3–5],可在单个微生物体内完成两步硝化反应(式1和式2),将NH4+-N氧化为NO3–-N。

|
公式(1) |

|
公式(2) |
其中,ΔGo′=–275 kJ/mol。
近5年来,有关全程硝化菌在环境生态分布、分子遗传学和微生物学特性等方面的研究正在快速增加。全程硝化菌在自然环境(如水体,土壤,空气)和人工系统(如污水处理厂,饮用水处理系统,循环水产养殖系统等)中广泛存在,是一类生态功能重要的硝化微生物[6–10],以amo作为特异分子标志物,可将其划分为clade A和clade B型[3, 11]。此类微生物具备高氨亲和力、低氧适应性、低生长速率和代谢多样性等特点[11–13]。全程硝化菌在废水生物脱氮系统活性污泥群落中的相对丰度为0.02%–0.57%,在生物膜中相对丰度可高达17%[14–16],某些三级处理系统中全程硝化菌可成为主导硝化微生物(群落相对丰度:1.8%–19.4%)[17]。然而目前有关全程硝化菌与传统硝化微生物的互作及生态位分异的机制不清,原位硝化贡献不明,有关其在废水深度处理中的应用探索仍处于起步阶段。
本文概述了全程硝化菌的系统发育、生理生化特性和生境分布特征,重点总结了全程硝化菌在水处理系统中的研究进展,包括分布规律和影响因子等,分析了其在废水脱氮过程中的功能及潜在应用价值,以期为基于全程硝化菌的理论研究及相关技术研发提供参考。
1 全程硝化菌的分子生物学及生理生化特性 1.1 全程硝化菌的发现过程及分子生物学特性Van Kessel等从水产养殖系统滴滤池的厌氧段取样接种至序批式(Sequencing batch reactor,SBR)硝化反应器。在连续运行12个月后,通过对样品的高通量测序、宏基因组分析和分箱处理,得到了2个全程硝化菌基因草图,且基因组平均核苷酸相似性低于95%,分别命名为“Candidatus Nitrospira nitrosa”和“Candidatus Nitrospira nitrificans”[4, 18]。Daims等从1200 m深的石油勘探井管壁生物膜中采样,通过实验室富集和宏基因组分析技术证实了全程硝化菌的存在,并在2年后成功分离得到了第一株全程硝化菌Nitrospira inopinata[3, 12]。Pinto等也在同时期从饮用水处理系统中采样,通过宏基因组技术组装到一个全程硝化菌Nitrospira sp. Ga0074138的拼接基因组(Metagenome-assembled genomes,MAGs)[5]。这些研究均发现,全程硝化菌具有编码全部硝化过程关键功能酶的基因,可在单个微生物体内完成两步硝化反应[3–5]。
迄今通过富集和宏基因组分箱组装技术得到的全程硝化菌MAGs均属于硝化螺菌属的第二分支(Nitrospira lineage II),全程硝化菌与NOB型硝化螺菌(NOB-Nitrospira)的16S rRNA基因序列具有高度相似性而未独立成簇,是其长期未被发现的主要原因[19]。两类微生物的核心基因组部分重合,包括亚硝酸盐氧化途径、还原性三羧酸循环(Reductive tricarboxylic acid cycle,rTCA循环)和糖异生途径等[11, 20]。全程硝化菌祖先的amo和hao基因可能来自β-AOB中相关基因的水平转移,且大部分全程硝化菌基因组的连续区域编码单拷贝的氨氧化关键基因簇(amo、hao和细胞色素c合成基因等)[11, 20]。全程硝化菌的amo基因与AOA和AOB中的amo基因相似度低,而与甲烷单加氧酶基因(pmo)的相似度高于90%[3, 11]。因此,大量研究采用amo基因作为特征分子标志物,以区别环境样品中的全程硝化菌与传统氨氧化微生物[3, 7, 15–16, 21–22]。目前文献报道的基于amoA和amoB基因的引物序列多达10对以上,用于识别并定量总全程硝化菌以及不同分支和种水平的全程硝化菌(表 1)。全程硝化菌与AOB相似,基因组均编码amoCAB操纵子。AOB的AMO酶由3个多肽亚基组成,其中AmoA和AmoC亚基为膜结合蛋白,而AmoB亚基在周质空间且可能包含Cu2+催化的活性位点[23–24]。amoA基因较为保守,常用于作为研究AOB系统发育的标志物,而amoB基因的序列变异性较高,可用于分辨亲缘关系较近的AOB物种[25]。目前仅有一篇文献报道了基于amoB基因的全程硝化菌定量,且未分析全程硝化菌扩增amoA和amoB基因的覆盖度及差异性[16]。
| Objective (Gene) | Primers (5′→3′) | Annealing temperature/℃ | References |
| Total comammox bacteria (amoA) | comamoA AF: AGGNGAYTGGGAYTTCTGGcomamoA SR: CCGVACATACATRAAGCCCAT | 55 | [15] |
| Total comammox bacteria (amoA) | Ntsp-amoA 162F: GGATTTCTGGNTSGATTGGA Ntsp-amoA 359R: WAGTTNGACCACCASTACCA | 48 | [21] |
| Total comammox bacteria (amoA) | pmoA-189b-F: GGNGACTGGGACTTYTGGCom_amoA_1_R: CGAGATCATGGTGCTGTGAC | 56 | [6] |
| Total comammox bacteria (amoB) | Cmx_amoB 148F: TGGTAYGAYACNGAATGGGCmx_amoB 485R: CCCGTGATRTCCATCCA | 52 | [16] |
| Total comammox bacteria(amoA)* | First step: A189Y: GGNGACTGGGAYTTYTGGC576r: GAAGCCCATRTARTCNGCCSecond step:CA209f: GAYTGGAARGAYCGNCAC576r-barcode: XXXXXXXXXXXXGAAGCCCATRTARTCNGCC | 52 50 | [28] |
| Clade A comammox bacteria (amoA) | A378f: TGGTGGTGGTGGTCNAAYTATC616r: ATCATCCGRATGTACTCHGG | 55 | [28] |
| Clade A comammox bacteria (amoA) | ComaA 244F: TAYAAYTGGGTSAAYTAComaA 659R: ARATCATSGTGCTRTG | 52 | [7] |
| Clade B comammox bacteria (amoA) | ComaB 244F: TAYTTCTGGACRTTYTAComaB 659R: ARATCCARACDGTGTG | ||
| “Candidatus Nitrospira nitrosa” (amoA) | Nitrosa 469 F: GCGATTCTGTTTTATCCCAGCAAC Nitrosa 812R: CCGTGTGCTAACGTGGCG | 64 | [22] |
| “Candidatus Nitrospira inopinata” (amoA) | Inopinata 410F: TCACCTTGTTGCTAACTAGAAACTGG Inopinata 815R: TCCGCGTGAGCCAATGT | ||
| “Candidatus Nitrospira nitrosa” (amoA) | Nitrificans 463F: ATGTTCGCGGCACTGTTNitrificans 836R: CCAGAAAGTTTAGCTTTGTCGCCT | ||
| “Candidatus Nitrospira inopinata” (amoA) | Nino 19F: ATAATCAAAGCCGCCAAGTTGC Nino 252R: AACGGCTGACGATAATTGACC | 52 | [3] |
| * represent that primers designed for partial nested PCR. | |||
根据amoA基因序列,全程硝化菌可划分为clade A型和clade B型,其中clade B型的氨氧化相关蛋白进化速率显著快于clade A型,且两类菌在基质转运、底物代谢、能量传递和环境适应性等方面存在显著差异[11]。全程硝化菌的基因组均编码复合体Ⅰ (NADH︰泛醌氧化还原酶),利用亚硝酸盐氧化过程释放的电子进行逆向电子传输并合成NADH[11, 26]。Clade A和clade B分别具有B型和E型细胞复合体Ⅱ (琥珀酸:醌氧化还原酶),在疏水域和血红素含量上存在差异[11, 27]。NOB-Nitrospira拥有至少双拷贝的复合体Ⅲ (细胞色素bc),而全程硝化菌的基因组缺少复合体Ⅲ的第二个拷贝。由于亚硝酸盐氧化过程的逆向电子传输需要复合体Ⅲ的参与,因此当以亚硝酸盐为唯一电子供体时,全程硝化菌因无法完成逆向电子传输及CO2固定而无法生长[3, 26]。最后,全程硝化菌编码具有高氧亲和力的复合体Ⅳ (bd型末端氧化酶)基因,有利于适应溶解氧条件[11]。
Xia等通过amoA基因高通量测序进一步将clade A型细分为clade A1和clade A2两个亚支,其中clade A1分支包括“Candidatus Nitrospira nitrosa”,“Candidatus Nitrospira nitrificans”和Nitrospira inopinata,clade A2不存在典型富集物[28]。Palomo等发现根据haoA基因相似性也可将clade A型全程硝化菌划分为clade A1和A2分支,且clade A2分支与clade B型的系统发育距离更近[29]。β-AOB中hao基因为多拷贝,而大多数全程硝化菌中为单拷贝,hao基因编码的HAO是具有亚铁血红素P460活性位点的周质空间型酶,具有8个结合位点的高度保守序列,可作为系统发育和功能的生物标志物[23, 30–31]。目前clade B也缺少典型富集物和代表菌株,未来需加强不同生境中的全程硝化菌富集培养和分类工作,以丰富其菌种资源,有利于开展对其微生物学特性的深入研究。
1.2 生理生化特性对迄今为止唯一的全程硝化菌纯培养物Nitrospira inopinata的动力学研究发现,与传统硝化微生物相比,其具有高氨亲和力、高底物同化率及低生长速率等特点(表 2)[12]。陆地AOA和AOB的氨半饱和常数(Km, NH3)为Nitrospira inopinata的7.3–89.8和38.8–4081.6倍。Nitrospira inopinata的高氨亲和力有利于其在陆地生态系统的氨氮限制环境中生存。事实上,地下水、循环水产养殖厂、自来水处理系统等(氨氮浓度:4–60 μmol/L)的生物膜上均发现了高丰度的全程硝化菌[3, 5–6, 8]。Nitrospira inopinata的产率系数(Yield)高于AOM和NOB,但最大比生长速率(μmax)比其他硝化微生物低1–3个数量级[12, 19, 32–33]。相较于两步硝化反应,全程硝化过程的ATP合成产率更高,释放的能量供给单一微生物生长,细胞产率更高。而基于最佳路径长度的动力学理论推测,自养硝化过程ATP通量随代谢路径的延长而降低,因此全程硝化的长代谢路径使得ATP产生速率低,导致全程硝化菌生长缓慢[2, 34]。另外,全程硝化菌的CO2固定途径为rTCA循环,与AOB基因组编码的卡尔文(Calvin Benson Bassham cycle,CBB)循环相比,固定单位CO2所消耗的ATP更少,能源利用效率更高(式3,4)[19]。因此,全程硝化菌适宜在底物浓度低、底物或生物质混合程度小的聚集体环境(如生物膜)中缓慢生长[2]。
|
公式(3) |

|
公式(4) |
| Parameters | Half saturation constant of ammonia Km, NH3/(μmol/L) | Half saturation constant of nitrite Km, NO2–/(μmol/L) | Cell yield/ (mg Protein/mol NH3 or NO2–*) | Maximum specific growth rate (μmax/h) | Maximum specific utilization rate qmax(mmol N/[(mg protein‧h]) | References |
| Nitrospira inopinata | 0.049 | 449.2 | 394.7 | 0.0061 | 0.015, 0.017* | [12] |
| NOB-Nitrospira | NA | 9–27 | 120–213* | 0.027–0.083* | 0.147–0.694* | [19, 32] |
| NOB-Nitrobacter | NA | 49–1380 | 83–108* | 0.0115–0.1250* | 0.138–1.501* | |
| AOB | 1.9–200.0 | NA | 250 | 0.007–0.083 | 0.028–0.332 | [19, 33] |
| Non_marine AOA | 0.36–4.40 | NA | 298.4–304.3 | 0.010–0.050 | 0.011–0.024 | [12, 19] |
| Marine AOA | 0.003 | NA | NA | 0.010–0.027 | 0.012–0.052 | [36–37] |
| *: parameters (i.e. Y, μ, q) obtained based on nitrite data; NA: not available. | ||||||
当氨氮为限制性底物时,高氨亲和力的Nitrospira inopinata比生长速率可高于AOA和AOB,获得生存优势;而当氨氮充足时,AOB的最大比生长速率显著高于AOA和Nitrospira inopinata,AOB更具生存优势。值得注意的是,低氨氮环境中Nitrospira inopinata的竞争优势并不代表其原位硝化活性或对硝化过程的贡献更高。Nitrospira inopinata的最大氨氮比利用速率与AOA相似,仅为AOB的0.045–0.530倍,而以亚硝酸盐为底物的比利用速率比NOB (NOB-Nitrospira和NOB-Nitrobacter)低1–2个数量级(表 2)。因此,假设Nitrospira inopinata和AOB在环境中具有同等竞争力、氨氮利用效率相等且amoA基因丰度反映活性,那么推测Nitrospira inopinata的丰度至少为AOB的1.89倍时,其氨氧化贡献率才能高于50%。同理,当其与NOB共存时,Nitrospira inopinata的丰度至少需为NOB的8.6倍,才能对亚硝酸盐氧化过程有主要贡献[35]。由于目前全程硝化菌的动力学数据均来源于Nitrospira inopinata,具有一定局限性,未来需要更多的研究来获得全程硝化菌的动力学特征,以分析其生态位特征以及与传统硝化微生物的互作机制。
Nitrospira inopinata对亚硝酸盐的底物亲和力较低,其亚硝酸盐半饱和常数(Km, NO2–)与典型NOB-Nitrobacter类似,为NOB-Nitrospira的16.7–49.9倍(表 2)[12, 32]。Nitrospira inopinata以0.1–1.0 mmol/L NH4+-N作为唯一代谢底物时出现了NO2–-N的短暂积累,推测周质空间生成的NO2–-N未被及时代谢而排出胞外,因此亚硝酸盐氧化反应可能为全程硝化过程的限速步骤[12]。除硝化作用外,全程硝化菌的基因组编码亚硝酸盐还原酶基因(nir)而缺少一氧化氮还原酶基因(nor),理论上可在好氧/缺氧环境中产生一氧化氮(Nitric oxide,NO),但是不能通过生物途径产生一氧化二氮(Nitrous oxide,N2O)[19, 38]。Kits等发现Nitrospira inopinata的氨氧化过程伴随着NO的短暂积累,而亚硝酸盐氧化过程会形成稳定的NO积累,这可能与nirK基因的高表达相关[38]。Nitrospira inopinata代谢过程产生的微量N2O主要来自羟胺的非生物转化过程(0.07%±0.006%),显著低于AOB进行氨氧化过程时的N2O产量(0.1%–8.0%)[38–39]。
氧气对于全程硝化菌的基础代谢具有重要作用。虽然文献尚未报道Nitrospira inopinata的氧亲和力数据,但研究发现全程硝化菌适宜在微需氧环境中生存,如低溶解氧(Dissolved oxygen,DO)的SBR反应器(DO < 0.6 mg/L),水产养殖滴滤池缺氧区(DO < 3.1 μmol/L)等[4, 40–41]。全程硝化菌具有高氧亲和力的bd型末端氧化酶[11],而rTCA循环的关键酶2-氧戊二酸氧化还原酶和丙酮酸氧化还原酶也对氧气敏感,因此推测全程硝化菌在生物膜等存在氧气梯度的环境中生存是应对高浓度氧气暴露的一种自我保护机制[11, 20]。另一方面,由于全程硝化菌具备维持铜离子稳态的copCD和copAB基因,以及高铁亲和力的I型细胞色素c生物合成系统相关基因,在铜、铁缺乏的环境中具备生存优势[42]。
除无机氮源外,全程硝化菌具备代谢多样性,可以直接利用或共代谢简单无机物和有机物。全程硝化菌基因组存在脲酶操纵子(ureABCDFG)和尿素转运系统(urtABCDE)基因[11, 40],Van Kessel等研究证实了全程硝化菌的富集培养物“Cadidatus Nitrospira nitrosa”和“Cadidatus Nitrospira nitrificans”可水解尿素并进一步氧化成硝酸盐[7.8±1.1 μmol NO3–-N/(L·h)][4]。Clade B型全程硝化菌具有甲酸脱氢酶(fdh)基因,而clade A型全程硝化菌具有特异性3b[NiFe]氢化酶基因,因此两者分别具有氧化甲酸和氢气的潜力[11]。Yang等在废水处理系统的缺氧段发现clade A型全程硝化菌的存在,转录组分析证实其具有糖原和多羟基烷酸酯以及同型乙酸发酵功能[13]。另外,与传统硝化微生物类似,全程硝化菌可通过共代谢作用去除痕量有机微污染物(ng/L–μg/L),如黄草灵、环酰菌胺、米安色林和雷尼替丁等[43–44]。当以氨而非亚硝酸盐作为代谢底物时,Nitrospira inopinata独特的AMO结构使其可通过芳香环羟基化共代谢降解多菌灵(一种苯并咪唑抑菌剂)[43]。与AOA和AOB相似,Nitrospira inopinata的AMO酶介导下通过羟基化和亚硝化作用转化芳香胺,从而共代谢降解磺胺类药物,例如其对磺胺甲基嘧啶和磺胺甲恶唑的去除率高达65%[44]。
2 环境分布 2.1 总体分布特征全程硝化菌的分布特征调查主要通过以下手段:(1)利用qPCR定量功能基因确认其总体丰度(表 1);(2)根据功能基因(amo,hao)序列分析系统发育特征;(3)通过宏基因组分析获得MAGs并定量全部关键功能基因的分布特征等。本文通过挖掘NCBI数据库中全程硝化菌的amoA基因序列,结合不同研究的采样生境构建了较为完整的全程硝化菌系统发育树(图 1)。由图 1可知,目前发现的全程硝化菌以clade A型为主,广泛分布于废水处理系统(SBR、流化床生物膜和旋转生物滤池等不同工艺),饮用水处理系统(快速砂滤池和饮用水处理厂等)、自然水体(地表水和地下水)及土壤系统(农田土、森林土)等环境中[3, 8–9, 14]。Clade B型主要分布于农田及森林土、饮用水处理系统及自然水体中,未在废水处理系统中发现[3, 7, 9, 21]。Xia等发现clade A1主要分布在活性污泥、厌氧污泥、湖泊底泥等环境,而clade A2主要分布在煤矿土、旱地土和自来水样中[28]。Palomo等基于526个环境宏基因组数据发现,clade A1在污水处理厂中出现频率较高,而clade A2主要存在于饮用水处理厂[29]。Clade A型和clade B型全程硝化菌的分布差异与环境适应性有关,基因组证据显示clade A型全程硝化菌编码低氨亲和力的Rhesus型氨转运蛋白(mmol/L级别,也存在于大部分β-AOB内),而Clade B编码高亲和力的Amt型氨转运蛋白(μmol/L级别),更适宜在寡营养生境而非较高氨氮浓度的污水处理系统中生存[11, 20, 45–46]。全程硝化菌在农田土壤中普遍存在且clade A2型占主导,关键环境因子如pH、C/N是影响全程硝化菌分布的重要因素,该研究还发现未添加氮源的土壤中clade B型全程硝化菌具有生长活性[9]。Wang等通过DNA-SIP技术在无外加氮源的土壤中成功标记了clade B型,证实了其寡营养适应性[47]。Clade A和clade B的分支最初基于特征amoA基因的差异性[3],然而部分clade A型全程硝化菌(如Nitrospira sp. RCA)的amoA基因与clade B的系统发育关系更近(图 1)。因此,仅根据amoA基因进行全程硝化菌的分型具有局限性,可综合考虑其他分子标志物(hao,amoB)或基于微生物核心基因组信息得到更准确的系统发育关系[48]。
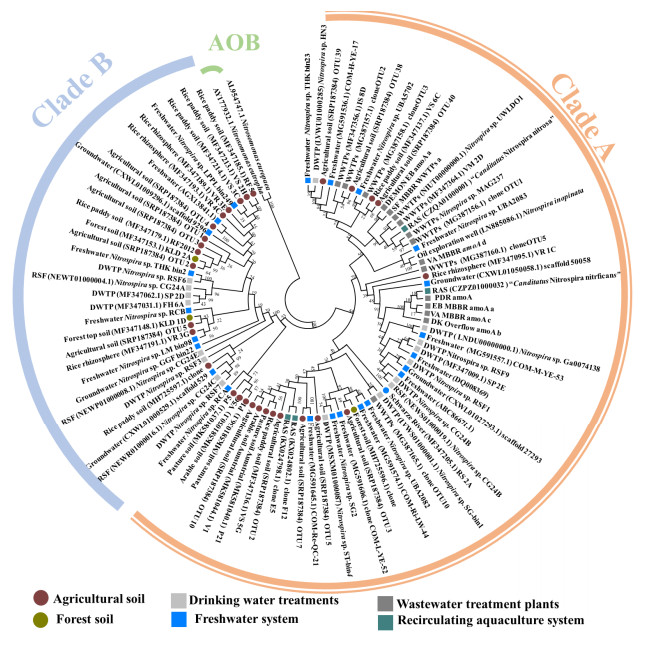
|
| 图 1 全程硝化菌的amoA基因系统发育树及其分布生境 Figure 1 amoA gene based phylogenetic tree of comammox bacteria and the associated habitats. The tree was constructed using the neighbor-joining methods in MEGA 7, based on [94] representative sequences collected from [26] articles. The evolutionary distances were computed using the Tamura 3-parameter method and the rate variation among sites was modeled with a gamma distribution. |
2.2 水处理系统的分布特征
2.2.1 总体分布规律: 饮用水处理系统(NH4+-N < 0.5 mg/L)及给水管网是全程硝化菌的适宜生境之一(表 3)[49]。Wang等调查了自来水水源、自来水处理厂及管道终端出水处的硝化细菌分布,发现某石英砂滤床中全程硝化菌分别为AOA和AOB的60.7和5.86倍,海南某处自来水龙头0.45 μm滤膜处的全程硝化菌丰度为AOB的19.7倍[8]。Fowlers等调查了丹麦的12个DWTPs,发现全程硝化菌的相对丰度(6.5%–59.8%)为AOB的2.1–12.0倍,对于氨氧化过程的贡献更高,且clade B型分布广泛,约占总全程硝化菌的75%[21]。Palomo等在丹麦某快速砂滤池样品取样,得到了3个clade B型和1个clade A型的全程硝化菌MAGs,并指出在该寡营养环境中clade B型可能更具优势[11]。循环水产养殖系统(Recirculating aquaculture system,RAS)主要通过养殖水的循环使用减少水资源消耗,需使用生物膜处理循环水中由于剩余饲料及水产排泄物等产生的含氮废物[6, 50]。Van Kessel等通过将RAS中的生物膜接种到SBR反应器内,得到了两种全程硝化菌富集物[4]。Barteleme等考察了稳定运行15年的RAS (NH4+-N:9.02±4.76 μmol/L),发现全程硝化菌约占总硝化菌群的27%,平均丰度为氨氧化菌(AOA和AOB)的1.5–2.5倍,且此类全程硝化菌的amoA基因与农田土壤中发现的amoA序列系统发育距离较近[6]。
| Habitats | Reactor/ Sample type | Environmental/Operating conditions | Abundance and diversity of comammox bacteria | References |
| Freshwater recirculating aquaculture system | SBRa | Influent ammonium: 0.1–0.5 mmol/L, anoxic: DO < 3.1 μmol/L | Relative abundance: ~15% | [4] |
| UWM fluidized sand biofilter | pH: 7.01±0.09; influent ammonium: 9.02±4.76 μmol/L; DO: 8.20±0.18. | 21% of the total nitrifying communities, and 1.5–2.5 times more abundant than AOA and AOB | [6] | |
| Drinking water system | Drinking water treatment plants (DWTPs) | Influent ammonium and nitrate: 0.01–0.53 mg N/L, 0.01–2.42 mg N/L; DO: 4.41–11.04 mg/L; Temperature: 8.6–13.2 ℃ | 0.50%–59.82%, 2.1–12.0 times more abundant than AOB; clade B accounted for about 75% of total comammox bacteria | [21] |
| Wastewater treatment plants(WWTPs) | Rapid sand filters (RSF) | 9.3±0.4 ℃; pH: 7.4±0.3; DO: 1.09±0.59; Ammonium: 0.41±0.2 mg N/L | Nitrospira: 26.08%±0.94%; 3 clade B bins and 1 clade A bins. | [11, 56–57] |
| Household tap water filters (0.2 μm or 0.45 μm) | Ammonium < 0.2 mg N/L, Nitrate: 0–4.6 mg N/L | Number of amoA sequences divided by the sequencing size (GB): AOA: 0.24–76.81; AOB: 7.02–251.19; comammox bacteria: 59.63–138.51 | [8] | |
| SBRa | Anaerobic-microaerobic (DO < 0.6 mg/L)- precipitation.~91% ammonia removal without NO and N2O production | 4.52% of the total sequencing reads; similar to “Canditatus Nitrospira nitrosa” | [40] | |
| Biological nitrogen removal system (BNR) | Reactor configurations were covered: conventional BNR, MBBR, DEMON, ClearGreen cyclic low-energy ammonium removal, and a hydrocyclone. | 0.28%–0.64% of the total sequencing reads | [14] | |
| Activated sludge in BNR system | Influent ammonia:16.1–43.6 mg N/L; COD: 161–430 mg/L; pH: 6.85–7.75; DO: 1.20–5.19; SRT: 8–30 d. | 0.02%–0.36% of the total community. dominated by “Canditatus Nitrospira nitrosa” (93.34%), with amo transcription activity | [15, 58] | |
| SBRa | 20.3±1.1 ℃; DO during intermittent aeration: 0.2–1.0 mg/L, influent ammonia: 14.3±3.8 mg N/L, SRT: 99 d | Comammox bacteria amoA gene accounted for 94% of the total ammonia-oxidizing microorganisms, the majority were “Canditatus Nitrospira nitrosa” | [41] | |
| SBRa | Influent: municipal sewage; pH: 7.3–7.8; 15.0–18.5 ℃,influent ammonia: 69.3–77.8 mg N/L; COD: 229.6–305.4 mg/L; DO: 2–4 mg/L. | amoA gene abundance:6.25×107– 4.16×108 copies/gram VSS; The addition of hydroxylamine can inhibit the nitrite oxidation activity of comammox bacteria | [55] | |
| BNR (ND, SND, PNA) | PNA influent ammonium: 468.1–1085.1 mg N/L, ND and SND influent ammonium: 12.4–52.1 mg N/L. | Abundance positively correlated with SRT, relative abundance in suspended flocs: 0.5%–3%, in biofilm: up to 17%. | [16] | |
| Rotating biological contactors (tertiary treatment) | Influent ammonium, nitrite and nitrate: 0.2–16.3 μmol/L; 0.9–66.1 μmol/L; 358.5–1777.8 μmol/L | 1.8%–19.4% of the total community, all belong to clade A | [17] | |
| a:Lab-scale reactors with inoculum from specific habitats. SBR: sequencing batch reactor; COD: chemical oxygen demand; ND: nitrification-denitrification; SND: simultaneous nitrification-denitrification; PNA: partial nitrification-Anaerobic ammonia oxidation; DMEON: deammonification; MBBR: moving bed biofilm reactor. | ||||
全程硝化菌在市政污水脱氮系统中被广泛检出,包括硝化反硝化、同步硝化反硝化和厌氧氨氧化工艺等,约占总细菌群落的0.28%–0.64% (表 3)。随着污水排放标准的提高,三级处理系统逐渐普及,全程硝化菌在三级处理系统中的丰度高于二级处理系统,如在旋转生物滤池中全程硝化菌相对丰度为1.8%–19.4%,显著高于AOB,且分箱拼接得到10个MAGs均为clade A型,其中一个MAG与“Candidatus Nitrospira nitrosa”基因组相似度高于95%[17]。Wang等调查发现全程硝化菌在WWTPs的活性污泥中相对丰度在0.02%–0.36%,是氨氧化菌(AOA和AOB)丰度的2.16–148.59倍,且主要为“Candidatus Nitrospira nitrosa”,占总全程硝化菌序列的94.34%[15]。Cotto等发现全程硝化菌的丰度与处理系统中微生物的生长方式有关,全程硝化菌在悬浮态污泥中相对丰度为0.5%–3.0%,而生物膜中最高可达17%[16]。该研究通过宏基因组得到的4个MAGs均属于“Candidatus Nitrospira nitrosa”[16]。另外,Roots和Camejo等分别在低DO的SBR反应器得到以“Candidatus Nitropsira nitrosa”为主导的硝化菌群[40–41]。与“Candidatus Nitropsira nitrosa”相比,Nitrospira inopinata的检出率和丰度均不高,这可能与其特殊的分离生境和生存条件有关[3, 29]。Palomo等调查分析了969个宏基因组样品,发现Nitrospira inopinata仅在低于1%环境样品中被检出,且在废水处理系统中的检出率很低[29]。本文通过构建系统发育树发现,与“Candidatus Nitropsira nitrosa”相似度较高的amoA基因序列均来自WWTPs,而Nitrospira inopinata与其他clade A型全程硝化菌亲缘关系较远(图 1)。由于WWTPs的废水脱氮系统中氨氮浓度较高,推测“Candidatus Nitropsira nitrosa”的氨亲和力低于Nitrospira inopinata,但目前尚未有关于其动力学实验的报道。
2.2.2 环境影响因子: (1) 氨氮浓度:循环水产养殖系统(NH4+-N: 1.4–7.0 mg/L)、饮用水处理系统(NH4+-N < 0.53 mg/L)、三级处理旋转生物接触池(NH4+-N:0.003–0.230 mg/L)等低氨氮浓度系统中全程硝化菌相对丰度最高可达59.82%,为主导硝化微生物。主流废水脱氮系统(NH4+-N < 77.8 mg/L)中全程硝化菌广泛检出(相对丰度 < 5%),而高浓度短程硝化-厌氧氨氧化工艺(进水NH4+-N > 400 mg/L)中未检测到全程硝化菌(表 3)。目前,短程硝化-厌氧氨氧化工艺主要用于处理高浓度氨氮废水,而游离氨和游离亚硝酸可协同抑制NOB生长从而实现短程硝化(抑制浓度分别为:0.08–4.00 mg/L和0.06–0.83 mg/L)[51–52]。全程硝化菌与NOB- Nitrospira在遗传学和生理生化特征上具有相似性[53–54],因此其在高浓度短程硝化-厌氧氨氧化工艺中未检出暗示游离氨/游离亚硝酸可能对全程硝化菌也具有较强的抑制作用。中低浓度废水脱氮生境中全程硝化菌也可成为主导菌群。Wang等向SBR反应器中接种某中试硝化反应器污泥(进水NH4+-N: 58.1–77.8 mg/L),稳定运行期间发现全程硝化菌丰度是AOB和Nitrobacter的5.52–5.63、4.64–9.86倍,且5 mg/L的羟胺间歇/连续投加均可抑制亚硝酸盐氧化过程,使反应器内的亚硝酸盐积累率超过90%[55]。(2) DO和污泥停留时间(Solids retention time,SRT):Cotto等研究了14个废水处理系统的全程硝化菌分布特征,发现全程硝化菌丰度与工艺的SRT成正相关。当SRT > 12.5 d时,全程硝化菌的相对丰度至少比氨氧化微生物高20%,且在集成固定膜-活性污泥系统(IFAS)样品中相对丰度可高达17%[16]。Roots和Camejo等分别通过低DO (< 1mg/L)、长SRT (80 d和99 d)的SBR反应器中也得到以“Candidatus Nitrospira nitrosa”为主导的全程硝化菌[40–41]。Annavajhala等发现虽然全程硝化菌在脱氮工艺里普遍存在,但生物膜样品上存在较多独特的全程硝化菌相关序列[14]。生物膜的污泥停留时间很长,一方面可为全程硝化菌提供附着基质,抵抗水利扰动和毒性冲击;另一方面,生物膜中存在的底物(如氧气)梯度有利于全程硝化菌在高浓度氧气暴露下实现自我保护。
3 全程硝化菌在废水脱氮中的应用潜力全程硝化菌广泛存在于废水处理系统中,可能对硝化过程有较大贡献,并与多种氮循环微生物之间存在复杂的竞争合作关系,如AOA、AOB、NOB、反硝化菌、厌氧氨氧化菌(Anaerobic ammonia oxidation bacteria,AnAOB)、硝酸盐/亚硝酸盐型厌氧甲烷氧化(Nitrate/nitrite dependent anaerobic methane oxidation,N-DAMO)微生物等(图 2)。虽然通过分子生物手段能获得全程硝化菌在废水处理中的分布规律,但无法准确评估其在污水处理系统中的原位硝化活性,其与上述微生物间的代谢互作关系也尚不明晰。基于目前已知的全程硝化菌生理生化特性,全程硝化菌在废水处理领域的应用探索可从以下三方面开展:(1)低氨氮废水在低DO条件下的全程硝化,以节省曝气能耗并减少污泥产量;(2)通过抑制全程硝化菌的亚硝酸盐氧化活性,实现低氨氮废水的短程硝化;(3)全程硝化菌与反硝化、厌氧氨氧化、厌氧甲烷氧化等微生物的耦合实现深度脱氮[19]。

|
| 图 2 全程硝化菌与其他脱氮微生物基于基础代谢物的关系网络图 Figure 2 Metabolic network of comammox bacteria and other N-cycle microorganisms. |
3.1 低氨氮废水、低DO条件下的全程硝化
全程硝化菌具有高氨亲和力、低氧适应性及生长缓慢等特点,以其作为主要硝化功能微生物时,理论上可降低污水脱氮过程的曝气能耗和污泥产量。硝化过程所需实际氧气量与硝化微生物的氧气亲和力、生物膜中的氧气梯度以及微生物在反应器内的生长形式有关。活性污泥中硝化微生物的表观氧气亲和力受单细胞聚集体(cell cluster)的结构大小影响[59]。由于聚集体内部氧气消耗梯度大,氧气易成为限制因素,且AOB聚集体比NOB略大(AOB:2–16 μm;NOB:1–8 μm),因此AOB的表观氧亲和力低于NOB[59–60]。全程硝化菌在形态上与NOB-Nitrospira相似、且氧亲和力高于AOB (表 2),控制低DO条件可能淘汰AOB,使全程硝化菌获得生存优势。实际上,通过控制硝化污泥反应器DO < 1 mg/L确实实现了全程硝化菌的富集[40–41]。另一方面,生物膜反应器中微生物的空间分层常会影响底物传质效率。利用移动床生物膜反应器(MBBR)、固定膜活性污泥系统(IFAS)可以使AOB和NOB在载体填料上以生物膜形式生长,提高固体停留时间[61–62],然而AOB与NOB在载体表面共存会加剧其对于氧气和生存空间的竞争。若以全程硝化菌取代AOB和NOB作为主要硝化微生物附着在载体上,可提高底物传质效率。Cotto等在IFAS系统中检测到全程硝化菌丰度高达17%,证实其在该系统中已成为优势硝化微生物[16]。由于全程硝化菌的氨氧化亲和力高于亚硝酸氧化亲和力,需控制生物膜反应器的DO及水力停留时间,否则可能会造成亚硝酸盐和氨氮的积累,影响全程硝化效率。
3.2 全程硝化菌在低氨氮废水短程硝化中的应用短程硝化过程指通过调节环境因子(pH、DO、NH4+-N,温度等)控制AOB为优势菌群并抑制NOB的生长,将硝化反应控制在产生NO2–-N阶段[52, 63]。短程硝化常与厌氧氨氧化、短程反硝化等过程耦合实现高效脱氮。短程硝化-厌氧氨氧化工艺理论上可节省60%的曝气能耗,减少75%的污泥产量,以及减少CO2和N2O气体的排放[64–65]。短程硝化-反硝化可以减少40%的有机物、25%的曝气量,同时分别减少硝化和反硝化过程33%和55%的污泥产量[64–66]。由于短程硝化工艺目前主要应用于处理高浓度氨氮废水,因此尚未在该工艺中发现全程硝化菌。但目前广受关注的废水主流短程硝化-厌氧氨氧化工艺中进水氨氮浓度较低,全程硝化菌有可能成为主要硝化微生物之一[16],因此需全面评估其在主流短程硝化过程中的应用潜力。由于Nitrospira inopinata的高氨亲和力和低亚硝酸盐亲和力,投加1.4–14 mg/L NH4+-N为底物时会导致亚硝酸盐短暂积累(最高约30%),最终完全转化为硝酸盐[3]。因此,理论上可通过抑制全程硝化菌的亚硝酸盐氧化活性,实现低氨氮废水的稳定短程硝化。低氨氮废水的稳定短程硝化可通过间歇曝气、控制低DO、缩短SRT、投加游离氨和游离亚硝酸等手段实现淘汰或抑制NOB[51]。Wang等近期向全程硝化菌主导的硝化反应器中投加5 mg/L的羟胺,也成功抑制了亚硝酸盐氧化过程,亚硝酸盐积累率超过90%[55]。以上方法均对抑制全程硝化微生物的亚硝酸盐氧化过程具有参考意义。
3.3 全程硝化菌与其他微生物耦合实现深度脱氮传统生物脱氮过程需要通过空间分离或曝气控制等手段分别实现硝化和反硝化功能,构筑物占地面积大、能耗高,而且是温室气体N2O的重要释放源。全程硝化菌没有N2O的生物合成途径,因此以其替代AOB作为主要硝化微生物可减少温室气体N2O的排放。另一方面,全程硝化菌的低氧适应性(DO < 0.1 mg/L)符合同步硝化反硝化(DO < 0.75 mg/L)的产生条件[4, 67–68]。在IFAS等生物膜体系中,絮体和生物膜分层形成的氧浓度梯度有利于反硝化微生物在生物膜内部缺氧的微环境中生存,而硝化微生物生长在膜外侧好氧区。同步硝化反硝化可在低曝气条件下达到较好的脱氮效果,提高空间利用率,减少内循环和曝气能耗[68]。Cotto等在同步硝化反硝化工艺中检测到全程硝化菌与AOB丰度相近,占总细菌群落的0.32%–1.48%[16]。DO的精确控制对实现同步硝化反硝化体系的高效脱氮作用至关重要,有研究表明设置实际DO浓度高于异养菌的氧气半饱和常数、低于自养硝化菌的氧气半饱和常数可实现高效脱氮[68]。未来可加强全程硝化菌在同步硝化反硝化上的应用探索。
废水主流短程硝化-厌氧氨氧化工艺可实现全程自养脱氮,具有节省曝气能耗、减少污泥产量和减少温室气体排放的优点[65, 69]。然而,该工艺存在微生物生长速率低、低温下(10–25 ℃)活性低、短程硝化不易稳定实现的问题[64]。全程硝化菌的存在可削弱低DO下氧气对AnAOB的抑制作用,但会与AnAOB产生对底物氨氮和亚硝氮的竞争。AnAOB富集物的氨亲和力低于Nitrospira inopinata (Km, NH3:5–132 μmol/L和0.049 μmol/L),而亚硝酸盐亲和力较高(Km, NO2–:2.5–48 μmol/L和449.2 μmol/L)[12, 70]。两者在低氨环境中共存时,全程硝化菌优先利用氨。此时,若通过抑制全程硝化菌的亚硝酸盐氧化过程,AnAOB可利用积累的NO2–-N实现自养脱氮(图 3-A);若发生全程硝化过程,AnAOB无法获得代谢底物而逐渐被淘汰。事实上,全程硝化菌与AnAOB在污水处理系统中的共存现象比较普遍[4, 14, 41]。Roots等发现在低DO、低氨条件下的短程硝化-厌氧氨氧化工艺中,全程硝化菌可逐渐成为主导群落[41]。因此,在主流短程硝化-厌氧氨氧化工艺中全程硝化菌具有一定的应用潜力。

|
| 图 3 生物膜反应器中AnAOB (A)以及甲烷基质膜生物反应器中N-DAMO微生物(B)和全程硝化菌的互作关系 Figure 3 Interaction between AnAOB (A), N-DAMO microorganisms (B) and comammox bacteria in the methane-based membrane biofilm reactor (MBfR). |
N-DAMO微生物可利用厌氧发酵过程产生的清洁能源甲烷作为电子供体实现自养脱氮,且不产生温室气体N2O[64],因此可与硝化微生物联合进行废水深度脱氮并实现节能减排。N-DAMO微生物由可以将硝酸盐还原为亚硝酸盐的N-DAMO古菌和将亚硝酸盐还原为氮气的N-DAMO细菌组成(图 2),二者分别对硝酸盐和亚硝酸盐具有较高亲和力[42, 71]。N-DAMO生长速率低,理论上全程硝化菌可与N-DAMO微生物在中空纤维膜MBfR反应器中实现耦合脱氮。甲烷通过气体渗透性纤维膜供给N-DAMO微生物作为电子供体,全程硝化菌在生物膜外侧消耗溶解氧,产生电子供体硝酸根和亚硝酸根供给内侧的N-DAMO古菌、N-DAMO细菌进行反硝化并减轻其氧气压力(图 3-B)。全程硝化菌的低氧适应性可减少曝气能耗,降低甲烷的溢出,提高甲烷利用效率。甲烷溶解度、DO、温度以及实际废水中可能存在的抑制物质等均可能影响上述微生物间的偶联,需要进一步深入研究其共存机制[65]。
4 展望全程硝化过程的发现对于重新认识土壤、地表水、污水处理等不同生境中的微生物氮循环具有重要意义。目前仍缺少有效方法准确评估全程硝化菌在不同生境中的原位活性,对其与传统硝化微生物的竞争和互作机制研究也有待深入。全程硝化过程具有节约曝气能耗、减少温室气体释放和提高深度脱氮效率等优点,在污水生物脱氮领域的应用前景广阔。然而全程硝化菌的富集培养缓慢、富集物活性低,成为限制其应用的最大阻碍。无论对于地球化学循环还是污水处理过程,全程硝化微生物都具有重要的研究和应用价值。因此,应继续探索全程硝化菌的生境分布特征、对陆地微生物氮循环的贡献以及与传统硝化微生物的生态位分化机制,并推进相关污水处理工艺的研发。
| [1] | Xia WW, Zhang CX, Zeng XW, Feng YZ, Weng JH, Lin XG, Zhu JG, Xiong ZQ, Xu J, Cai ZC, Jia ZJ. Autotrophic growth of nitrifying community in an agricultural soil. The ISME Journal, 2011, 5(7): 1226-1236. DOI:10.1038/ismej.2011.5 |
| [2] | Costa E, Pérez J, Kreft JU. Why is metabolic labour divided in nitrification?. Trends in Microbiology, 2006, 14(5): 213-219. DOI:10.1016/j.tim.2006.03.006 |
| [3] | Daims H, Lebedeva E V, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen P H, Wagner M. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [4] | Van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. Complete nitrification by a single microorganism. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [5] | Pinto AJ, Marcus DN, Ijaz UZ, Santos QMBDL, Dick GJ, Raskin L. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere, 2016, 1(1): e00054. |
| [6] | Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Frontiers in Microbiology, 2017, 8: 101. |
| [7] | Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, Steinberger M, Jetten MSM, Lücker S, Wagner M, Daims H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.3389/fmicb.2017.01508 |
| [8] | Wang YL, Ma LP, Mao YP, Jiang XT, Xia Y, Yu K, Li B, Zhang T. Comammox in drinking water systems. Water Research, 2017, 116: 332-341. DOI:10.1016/j.watres.2017.03.042 |
| [9] | Xu SY, Wang BZ, Li Y, Jiang DQ, Zhou YT, Ding AQ, Zong YX, Ling XT, Zhang SY, Lu HJ. Ubiquity, diversity, and activity of comammox Nitrospira in agricultural soils. Science of the Total Environment, 2020, 706: 135684. DOI:10.1016/j.scitotenv.2019.135684 |
| [10] | Gao JF, Fan XY, Pan KL, Li HY, Sun LX. Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Scientific Reports, 2016, 6(1): 38785. DOI:10.1038/srep38785 |
| [11] | Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. The ISME Journal, 2018, 12(7): 1779-1793. DOI:10.1038/s41396-018-0083-3 |
| [12] | Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [13] | Yang YC, Daims H, Liu Y, Herbold CW, Pjevac P, Lin JG, Li M, Gu JD. Activity and metabolic versatility of complete ammonia oxidizers in full-scale wastewater treatment systems. mBio, 2020, 11(2): e03175-19. |
| [14] | Annavajhala MK, Kapoor V, Santo-Domingo J, Chandran K. Comammox functionality identified in diverse engineered biological wastewater treatment systems. Environmental Science & Technology Letters, 2018, 5(2): 110-116. |
| [15] | Wang MY, Huang GH, Zhao ZR, Dang CY, Liu W, Zheng MS. Newly designed primer pair revealed dominant and diverse comammox amoA gene in full-scale wastewater treatment plants. Bioresource Technology, 2018, 270: 580-587. DOI:10.1016/j.biortech.2018.09.089 |
| [16] | Cotto I, Dai ZH, Huo LX, Anderson CL, Vilardi KJ, Ijaz U, Khunjar W, Wilson C, De Clippeleir H, Gilmore K, Bailey E, Pinto AJ. Long solids retention times and attached growth phase favor prevalence of comammox bacteria in nitrogen removal systems. Water Research, 2020, 169: 115268. DOI:10.1016/j.watres.2019.115268 |
| [17] | Spasov E, Tsuji JM, Hug LA, Doxey AC, Sauder LA, Parker WJ, Neufeld JD. High functional diversity among Nitrospira populations that dominate rotating biological contactor microbial communities in a municipal wastewater treatment plant. The ISME Journal, 2020, 14(7): 1857-1872. DOI:10.1038/s41396-020-0650-2 |
| [18] | Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou JZ, Oren A, Zhang YZ. A proposed genus boundary for the prokaryotes based on genomic insights. Journal of Bacteriology, 2014, 196(12): 2210-2215. DOI:10.1128/JB.01688-14 |
| [19] | Lawson CE, Lücker S. Complete ammonia oxidation:an important control on nitrification in engineered ecosystems?. Current Opinion in Biotechnology, 2018, 50: 158-165. DOI:10.1016/j.copbio.2018.01.015 |
| [20] | Koch H, van Kessel MAHJ, Lücker S. Complete nitrification:insights into the ecophysiology of comammox Nitrospira. Applied Microbiology and Biotechnology, 2019, 103(1): 177-189. DOI:10.1007/s00253-018-9486-3 |
| [21] | Fowler SJ, Palomo A, Dechesne A, Mines PD, Smets BF. Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environmental Microbiology, 2018, 20(3): 1002-1015. DOI:10.1111/1462-2920.14033 |
| [22] | Beach NK, Noguera DR. Design and assessment of species-level qPCR primers targeting comammox. Frontiers in Microbiology, 2019, 10: 36. DOI:10.3389/fmicb.2019.00036 |
| [23] | Beeckman F, Motte H, Beeckman T. Nitrification in agricultural soils:impact, actors and mitigation. Current Opinion in Biotechnology, 2018, 50: 166-173. DOI:10.1016/j.copbio.2018.01.014 |
| [24] | Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature, 2010, 465(7294): 115-119. DOI:10.1038/nature08992 |
| [25] | Junier P, Kim OS, Junier T, Ahn TS, Imhoff JF, Witzel KP. Community analysis of betaproteobacterial ammonia-oxidizing bacteria using the amoCAB operon. Applied Microbiology and Biotechnology, 2009, 83(1): 175-188. DOI:10.1007/s00253-009-1923-x |
| [26] | Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damsté JSS, Spieck E, Le Paslier D, Daims H. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(30): 13479-13484. DOI:10.1073/pnas.1003860107 |
| [27] | Lancaster CRD. Wolinella succinogenes quinol:fumarate reductase-2.2-Å resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochimica et Biophysica Acta (BBA)-Biomembranes, 2002, 1565(2): 215-231. DOI:10.1016/S0005-2736(02)00571-0 |
| [28] | Xia F, Wang JG, Zhu T, Zou B, Rhee SK, Quan ZX. Ubiquity and diversity of complete ammonia oxidizers (comammox). Applied and Environmental Microbiology, 2018, 84(24): e01390-18. |
| [29] | Palomo A, Dechesne A, Smets BF. Genomic profiling of Nitrospira species reveals ecological success of comammox Nitrospira. bioRxiv, 2019. |
| [30] | Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MMM, Pommerening-Roeser A, Op Den Camp HJM, Jetten MSM. Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environmental Microbiology, 2008, 10(11): 3140-3149. DOI:10.1111/j.1462-2920.2008.01732.x |
| [31] | Bergmann DJ, Hooper AB, Klotz MG. Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria:evidence for their evolutionary history. Applied and Environmental Microbiology, 2005, 71(9): 5371-5382. DOI:10.1128/AEM.71.9.5371-5382.2005 |
| [32] | Nowka B, Daims H, Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria:nitrite availability as a key factor in niche differentiation. Applied and Environmental Microbiology, 2015, 81(2): 745-753. DOI:10.1128/AEM.02734-14 |
| [33] | Jiang QQ, Bakken LR. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiology Ecology, 1999, 30(2): 171-186. DOI:10.1111/j.1574-6941.1999.tb00646.x |
| [34] |
Dong XS, Wang ZH, Huang XR, Jiang XJ. Recent discovery in nitrification:One-step nitrification and complete ammonia oxidizing microorganisms. Chinese Journal of Applied Ecology, 2017, 28(1): 345-352.
(in Chinese) 董兴水, 王智慧, 黄学茹, 蒋先军. 硝化作用研究的新发现:单步硝化作用与全程氨氧化微生物. 应用生态学报, 2017, 28(1): 345-352. |
| [35] | Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil:the quest for niche specialisation and differentiation. Trends in Microbiology, 2012, 20(11): 523-531. DOI:10.1016/j.tim.2012.08.001 |
| [36] | Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature, 2009, 461(7266): 976-979. DOI:10.1038/nature08465 |
| [37] | Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(34): 12504-12509. DOI:10.1073/pnas.1324115111 |
| [38] | Kits KD, Jung MY, Vierheilig J, Pjevac P, Sedlacek CJ, Liu SR, Herbold C, Stein LY, Richter A, Wissel H, et al. Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata. Nature Communications, 2019, 10(1): 1836. DOI:10.1038/s41467-019-09790-x |
| [39] | Prosser JI, Hink L, Gubry-Rangin C, Nicol GW. Nitrous oxide production by ammonia oxidizers:Physiological diversity, niche differentiation and potential mitigation strategies. Global Change Biology, 2020, 26(1): 103-118. DOI:10.1111/gcb.14877 |
| [40] | Camejo PY, Domingo JS, McMahon KD, Noguera DR. Genome-enabled insights into the ecophysiology of the comammox bacterium "Candidatus Nitrospira nitrosa". mSystems, 2017, 2(5): e00059-17. |
| [41] | Roots P, Wang YB, Rosenthal AF, Griffin JS, Sabba F, Petrovich M, Yang FH, Kozak JA, Zhang H, Wells GF. Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Research, 2019, 157: 396-405. DOI:10.1016/j.watres.2019.03.060 |
| [42] | Ren Y, Hao NH, Guo WS, Wang DB, Peng L, Ni BJ, Wei W, Liu YW. New perspectives on microbial communities and biological nitrogen removal processes in wastewater treatment systems. Bioresource Technology, 2020, 297: 122491. DOI:10.1016/j.biortech.2019.122491 |
| [43] | Han P, Yu YC, Zhou LJ, Tian ZY, Li Z, Hou LJ, Liu M, Wu QL, Wagner M, Men YJ. Specific micropollutant biotransformation pattern by the comammox bacterium Nitrospira inopinata. Environmental Science & Technology, 2019, 53(15): 8695-8705. |
| [44] | Zhou LJ, Han P, Yu YC, Wang BZ, Men YJ, Wagner M, Wu QL. Cometabolic biotransformation and microbial-mediated abiotic transformation of sulfonamides by three ammonia oxidizers. Water Research, 2019, 159: 444-453. DOI:10.1016/j.watres.2019.05.031 |
| [45] | Andrade SLA, Einsle O. The Amt/Mep/Rh family of ammonium transport proteins. Molecular Membrane Biology, 2007, 24(5/6): 357-365. |
| [46] | Weidinger K, Neuhäuser B, Gilch S, Ludewig U, Meyer O, Schmidt I. Functional and physiological evidence for a Rhesus-type ammonia transporter in Nitrosomonas europaea. FEMS Microbiology Letters, 2007, 273(2): 260-267. DOI:10.1111/j.1574-6968.2007.00805.x |
| [47] | Wang ZH, Cao YQ, Zhu-Barker X, Nicol GW, Wright AL, Jia ZJ, Jiang XJ. Comammox Nitrospira clade B contributes to nitrification in soil. Soil Biology and Biochemistry, 2019, 135: 392-395. DOI:10.1016/j.soilbio.2019.06.004 |
| [48] | Poghosyan L, Koch H, Lavy A, Frank J, van Kessel MAHJ, Jetten MSM, Banfield JF, Lücker S. Metagenomic recovery of two distinct comammox Nitrospira from the terrestrial subsurface. Environmental Microbiology, 2019, 21(10): 3627-3637. DOI:10.1111/1462-2920.14691 |
| [49] |
Xu JY, Mao YP. From canonical nitrite oxidizing bacteria to complete ammonia oxidizer:discovery and advances. Microbiology China, 2019, 46(4): 879-890.
(in Chinese) 徐建宇, 毛艳萍. 从典型硝化细菌到全程氨氧化微生物:发现及研究进展. 微生物学通报, 2019, 46(4): 879-890. |
| [50] | Brown MN, Briones A, Diana J, Raskin L. Ammonia-oxidizing archaea and nitrite-oxidizing Nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiology Ecology, 2013, 83(1): 17-25. DOI:10.1111/j.1574-6941.2012.01448.x |
| [51] | Duan HR, Wang QL, Erler DV, Ye L, Yuan ZG. Effects of free nitrous acid treatment conditions on the nitrite pathway performance in mainstream wastewater treatment. Science of the Total Environment, 2018, 644: 360-370. DOI:10.1016/j.scitotenv.2018.06.346 |
| [52] | Cao YS, van Loosdrecht MCM, Daigger GT. Mainstream partial nitritation-anammox in municipal wastewater treatment:status, bottlenecks, and further studies. Applied Microbiology and Biotechnology, 2017, 101(4): 1365-1383. DOI:10.1007/s00253-016-8058-7 |
| [53] | Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends in Microbiology, 2016, 24(9): 699-712. DOI:10.1016/j.tim.2016.05.004 |
| [54] | Mehrani MJ, Sobotka D, Kowal P, Ciesielski S, Makinia J. The occurrence and role of Nitrospira in nitrogen removal systems. Bioresource Technology, 2020, 303: 122936. DOI:10.1016/j.biortech.2020.122936 |
| [55] | Wang Z, Zhang L, Zhang FZ, Jiang H, Ren S, Wang W, Peng YZ. Nitrite accumulation in comammox-dominated nitrification-denitrification reactors:Effects of DO concentration and hydroxylamine addition. Journal of Hazardous Materials, 2020, 384: 121375. DOI:10.1016/j.jhazmat.2019.121375 |
| [56] | Palomo A, Fowler SJ, Gülay A, Rasmussen S, Sicheritz-Ponten T, Smets BF. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp.. The ISME Journal, 2016, 10(11): 2569-2581. DOI:10.1038/ismej.2016.63 |
| [57] | Gülay A, Musovic S, Albrechtsen HJ, Al-Soud WA, Sørensen SJ, Smets BF. Ecological patterns, diversity and core taxa of microbial communities in groundwater-fed rapid gravity filters. The ISME Journal, 2016, 10(9): 2209-2222. DOI:10.1038/ismej.2016.16 |
| [58] | Zheng MS, Wang MY, Zhao ZR, Zhou N, He SS, Liu SF, Wang JW, Wang XK. Transcriptional activity and diversity of comammox bacteria as a previously overlooked ammonia oxidizing prokaryote in full-scale wastewater treatment plants. Science of the Total Environment, 2019, 656: 717-722. DOI:10.1016/j.scitotenv.2018.11.435 |
| [59] | Picioreanu C, Pérez J, van Loosdrecht MCM. Impact of cell cluster size on apparent half-saturation coefficients for oxygen in nitrifying sludge and biofilms. Water Research, 2016, 106: 371-382. DOI:10.1016/j.watres.2016.10.017 |
| [60] | Stadler LB, Love NG. Impact of microbial physiology and microbial community structure on pharmaceutical fate driven by dissolved oxygen concentration in nitrifying bioreactors. Water Research, 2016, 104: 189-199. DOI:10.1016/j.watres.2016.08.001 |
| [61] | Rosso D, Lothman SE, Jeung MK, Pitt P, Gellner WJ, Stone AL, Howard D. Oxygen transfer and uptake, nutrient removal, and energy footprint of parallel full-scale IFAS and activated sludge processes. Water Research, 2011, 45(18): 5987-5996. DOI:10.1016/j.watres.2011.08.060 |
| [62] | Shao YX, Shi YJ, Mohammed A, Liu Y. Wastewater ammonia removal using an integrated fixed-film activated sludge-sequencing batch biofilm reactor (IFAS-SBR):Comparison of suspended flocs and attached biofilm. International Biodeterioration & Biodegradation, 2017, 116: 38-47. |
| [63] | 陈亚松.综合污水处理厂深度脱氮的优化运行与微生态机理研究.南京大学学位论文, 2019. |
| [64] | Winkler MKH, Straka L. New directions in biological nitrogen removal and recovery from wastewater. Current Opinion in Biotechnology, 2019, 57: 50-55. DOI:10.1016/j.copbio.2018.12.007 |
| [65] | Liu T, Hu SH, Guo JH. Enhancing mainstream nitrogen removal by employing nitrate/nitrite-dependent anaerobic methane oxidation processes. Critical Reviews in Biotechnology, 2019, 39(5): 732-745. DOI:10.1080/07388551.2019.1598333 |
| [66] | Chung J, Bae W, Lee YW, Rittmann BE. Shortcut biological nitrogen removal in hybrid biofilm/suspended growth reactors. Process Biochemistry, 2007, 42(3): 320-328. DOI:10.1016/j.procbio.2006.09.002 |
| [67] | Holman JB, Wareham DG. COD, ammonia and dissolved oxygen time profiles in the simultaneous nitrification/denitrification process. Biochemical Engineering Journal, 2005, 22(2): 125-133. DOI:10.1016/j.bej.2004.09.001 |
| [68] | Insel G. Effects of design and aeration control parameters on Simultaneous nitrification and denitrification (SNdN) performance for activated sludge process. Environmental Engineering Science, 2007, 24(5): 675-686. DOI:10.1089/ees.2006.0029 |
| [69] | Wang Q, Tan GYA, Azari M, Huang XW, Denecke M, Men Y, Jung JY, Okabe S, Ali M, Huang YT, Wu ZY, Lo WH, Gu JD, Lin JG, Lee PH. Insights into the roles of anammox bacteria in post-treatment of anaerobically-treated sewage. Critical Reviews in Environmental Science and Technology, 2018, 48(6): 655-684. DOI:10.1080/10643389.2018.1474679 |
| [70] | Straka LL, Meinhardt KA, Bollmann A, Stahl DA, Winkler MKH. Affinity informs environmental cooperation between ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (Anammox) bacteria. The ISME Journal, 2019, 13(8): 1997-2004. DOI:10.1038/s41396-019-0408-x |
| [71] | He ZF, Cai C, Geng S, Lou LP, Xu XY, Zheng P, Hu BL. Mdodeling a nitrite-dependent anaerobic methane oxidation process:Parameters identification and model evaluation. Bioresource Technology, 2013, 147: 315-320. DOI:10.1016/j.biortech.2013.08.001 |
 2021, Vol. 61
2021, Vol. 61