中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 李慧, 谢建平. 2021
- Hui Li, Jianping Xie. 2021
- CRISPR/Cas系统与结核病防控新措施相关性研究
- CRISPR/Cas system and implications for novel measures against tuberculosis
- 微生物学报, 61(2): 300-314
- Acta Microbiologica Sinica, 61(2): 300-314
-
文章历史
- 收稿日期:2020-03-31
- 修回日期:2020-07-16
- 网络出版日期:2020-09-27
2. 西南大学生命科学学院, 现代生物医药研究所, 重庆 400715
2. Institute of Modern Biopharmaceuticals, School of Life Sciences, Southwest University, Chongqing 400715, China
结核分枝杆菌(Mycobacterium tuberculosis, 以下简称结核杆菌)感染引起的结核病(Tuberculosis)仍然是全球重大传染病[1]。耐药结核杆菌的出现以及与HIV共感染使结核病的防控形势越来越严峻[2]。因此,结核病新控制措施的开发极其迫切。深入研究结核杆菌生物学是开发新控制措施的基础[3]。
成簇规律性间隔短回文重复CRISPR/Cas系统是细菌和古菌中广泛存在的适应性免疫系统[4]。CRISPR系统由前导区(Leader)、重复序列(Repeat)、间隔区(Spacer)以及CRISPR相关的Cas基因组成。前导区是CRISPR阵列上游富含TA的一段序列,新获得的间隔区通常插入到第一个重复序列之后[5]。Cas蛋白多种多样,不同的Cas蛋白的功能也不尽相同[6]。
CRISPR/Cas系统是重要的基因组编辑工具。靶向切割抗性质粒或细菌基因组的CRISPR/Cas能特异性清除耐药基因或病原菌。细菌的crRNA (CRISPR RNA)引导Cas核酸酶定向切割自身DNA对细菌具有致死效应[7]。与抗生素相比,CRISPR/Cas系统靶向特定的DNA序列,具有窄谱、特异性高和对正常菌群的选择压力小的优势[8]。此外CRISPR系统单独或者与噬菌体联合使用也可能用于控制耐药结核杆菌。
CRISPR/Cas系统还可以降解入侵的遗传元件,保护细菌染色体不被外源遗传物质破坏,有利于维持细菌基因组稳定。但就具体物种而言,其利弊还需要视情况而定。比如某些外源抗生素抗性基因、毒力相关基因可能有利于细菌进化,提高其环境适应能力[9]。
1 CRISPR概述及分类 1.1 CRISPR概述1987年Ishino等在大肠杆菌中发现了一段特殊的重复序列[10]。2002年,Jansen等将该重复序列和随机间隔区(Spacer)的结构命名为CRISPR。此后,微生物基因组中发现了越来越多的重复序列[11]。2007年Barrangou等证实CRISPR/Cas系统可以发挥细菌免疫系统的功能[6]。2013年,Zhang等利用CRISPR/Cas9系统对小鼠细胞进行多位点基因编辑,证明CRISPR/Cas9具有广泛适用性[12]。
CRISPR/Cas9系统包含3个功能组分:crRNA、tracrRNA (trans-activating RNA,tracr RNA)及Cas9蛋白。三者形成复合物,寻找特定的前间隔区序列邻近基序(Protospacer Adjacent Motif,PAM) NGG (N代表ATGC中任一种碱基),引导crRNA与附近的间隔序列进行碱基互补配对,Cas9蛋白切割靶DNA。真核细胞中,断裂的DNA通过同源重组修复(Homologous-directed repair,HDR)或非同源末端连接(Non-homologous end joining,NHEJ)进行修复[13-14]。结核杆菌NHEJ效率不高,忠实性低,通常不能有效修复Cas9产生的DNA双链断裂,细菌因此死亡[15]。CRISPR/Cas系统靶向切割DNA,可以用来特异性清除耐药结核杆菌。
1.2 CRISPR/Cas系统的分类近年CRISPR-Cas系统发展迅速,分类不断更新,根据Cas蛋白、Cas基因操纵子结构、CRISPR阵列中重复序列的结构,2015年CRISPR/Cas系统分为2类、5型(Type I–V)和16个亚型(Subtype)[16]。随着新型Cas蛋白的发现,进一步分为2类(Class)、6型和33亚型[17]。
1类CRISPR/Cas系统,根据Cas蛋白结构又分为Ⅰ、Ⅲ、Ⅳ和12个亚型,常见是Ⅰ、Ⅲ型,Ⅰ型中常研究的是I-E型,广泛存在于大肠杆菌和放线菌中;Ⅳ型存在古菌中,细菌中不常见[18]。这一系统的特点是由多蛋白效应物复合体,如Cascade (The CRISPR associated complex for antiviral defense)进行有序的靶标识别和降解。首先,Cascade识别相匹配靶标DNA,促进crRNA和靶DNA结合。随后,招募具有剪切功能的蛋白(如Cas3)在特定靶位点剪切DNA[19]。但Cascades复合物效应蛋白组成复杂,限制了1类CRISPR/Cas系统在基因编辑中的应用。根据Makarova对CRISPR/Cas的分类,结核杆菌的CRISPR/Cas系统属于1类Ⅲ-A型[20]。
2类CRISPR/Cas系统包括Ⅱ、Ⅴ、Ⅵ和9个亚型,这一系统是理想的基因组编辑工具[12]。Cas9蛋白和Cas12a (cpf1)蛋白分别属于Ⅱ型[21]及Ⅴ型系统[22]。Cas13蛋白属于Ⅵ型[23]。
2012年,Jennifer等发现脓链球菌(Streptococcus pyogenes)的Cas9蛋白(SpCas9)在crRNA与tracrRNA形成的复合物引导下靶向切割DNA[11]。2015年发现的新CRISPR核酸内切酶Cas12a由单链crRNA引导在特定位点靶向切割DNA,产生的平末端造成的基因编辑更有优势。与Cas9相比,Cas12a识别富含胸腺嘧啶(T) (CTN或TTN)的PAM,Cas12a分子量更小,需要的crRNA序列更短,在某些细菌中进行编辑的细胞毒性更低[22],比如谷氨酸棒状杆菌[24]、耻垢分枝杆菌、结核杆菌[25]。与Cas9相比,Cas12a对人细胞U2OS进行定点突变的脱靶率更低,特异性更强[26]。
2 结核杆菌内源CRISPR/Cas系统与免疫防疫结核杆菌也存在CRISPR/Cas免疫系统[27]。CRISPR/Cas系统介导的抗噬菌体或质粒的免疫可分为3个阶段(图 1),下面就结核杆菌中存在的Ⅲ-A型CRISPR/Cas系统来介绍这种免疫机制。第一步是适应期,噬菌体或质粒原间隔区(Proto-Spacer)插入到第一个重复序列之后,Cas1和Cas2在此过程中起关键作用[28]。第二步是表达阶段,重复间隔序列被转录成长的前体pre-crRNA,被Cas6等Cas蛋白加工成短的成熟的crRNAs。Cas6作为核酸内切酶,其295和297位的甘氨酸构成有助于与RNA的结合,pre-crRNA最终形成单个间隔序列[28]。第三步是干扰阶段,成熟的crRNA和Cas蛋白(Cas6、Csm1、Csm2、Csm3、Csm4、Csm5)组装形成crRNAP (crRNA-CRISPR/ Cas protein),识别并切割入侵的噬菌体或质粒[28-29]。2020年Wei等发现结核杆菌中敲除Cas基因簇后,质粒的转化率增加,Ⅲ-A型CRISPR/Cas系统的存在可以增强结核杆菌的生存能力[27]。
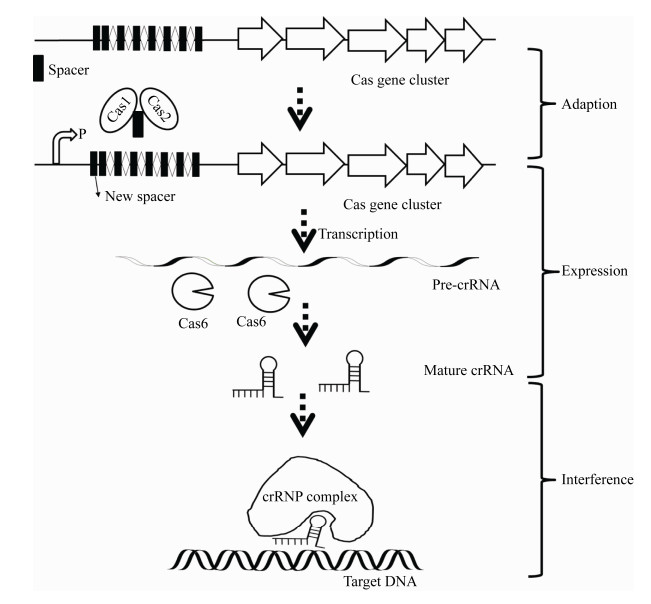
|
| 图 1 结核分枝杆菌Ⅲ-A型CRISPR/Cas系统的作用机制 Figure 1 Overview of the M. tuberculosis Ⅲ-A CRISPR-Cas system action. |
3 分枝杆菌CRISPR和Cas比较基因组学研究
结核杆菌H37Rv全基因组有4411529个碱基,约4000个可读框,但是大部分可读框的功能未知[30]。结核杆菌基因组中含有多个相同的36 bp的重复序列[31]。这类串联的重复序列只存在于临床分离结核杆菌的特定区域[32]。2012年我们课题组通过比较基因组分析,发现Mycobacterium tuberculosis H37Rv、M. tuberculosis H37Ra、M. tuberculosis F11、M. tuberculosis CDC1551、M. tuberculosis KZN 1435、M. bovis AF2122/97、M. bovis BCG str. Pasteur 1173P2、M. bovis BCG str. Tokyo 172、Mycobacterium sp. MCS、Mycobacterium sp. KMS、Mycobacterium sp. JLS、M. avium 104、M. gilvum PYR-GCK、M. marinum M等14种分枝杆菌基因组中都存在CRISPR结构,牛分枝杆菌和结核杆菌中发现有完整的CRISPR/Cas系统(图 2)。结核杆菌的2个CRISPR的重复序列的数目分别是24和18,牛分枝杆菌中2个CRISPR的重复序列的数目分别是25和17,两者都包含非重复的间隔序列、分散的短重复序列、前导序列和Cas蛋白,其中间隔区序列50%相似,暗示结核杆菌和M. bovis之间有共同的噬菌体[33]。结核杆菌和牛分枝杆菌的重复序列与Cas6结合序列一样,但不能形成稳定的二级结构。麻风分枝杆菌是例外,其重复序列能形成经典的茎环结构[33]。

|
| 图 2 结核分枝杆菌、牛分枝杆菌、鸟分枝杆菌中的CRISPR/Cas组成 Figure 2 CRISPR loci and Cas genes in three mycobacterial genomes. In M. tuberculosis, two tandem CRISPRs flanked by Cas gene cluster whose names were Cas2, Cas1, Csm6, Csm5, Csm4, Csm3, Csm2, Csm1, and Cas6. Cas1 and Cas2 are presumably crucial for spacer acquisition[34]. Cas6 is a metal ion-independent nucleases (Ca2+ and Mn2+)[35]. |
结核杆菌CRISPR阵列包含多个36 bp串联直接重复序列(Direct repeats,DR),DR具有保守序列(TGAGGTGCGGCGTGAGCGCGGGT),被94个35–41 bp的间隔子(Spacer)隔开。间隔子序列和排列次序在结核杆菌复合群中相对保守[31]。DR长度和间隔子随结核杆菌菌株而异。这可能是插入序列IS6110转座到DR区,通过邻近或者远端DR之间同源重组,或者复制滑动(Replication slippage)导致间隔子序列缺失所致[34, 36]。
重复序列如CRISPR位点也用于致病菌流行病学调查中的基因分型、系统发育和群体遗传学[35, 37]。根据CRISPR位点间隔区寡核苷酸分型确定主要流行株及其传播途径[38-39]。2019年Liu等对7株临床耐药结核杆菌进行了全基因组测序,对基因变异位点进行了比较基因组分析,发现其具有不同间隔的CRISPR位点,提示它们可能在结核杆菌基因组改变中起重要作用[40]。基于膜反向杂交的间隔区核苷酸分型也用于对临床分离结核杆菌进行调查[41-42]。
结核杆菌3个与致病性有关的基因岛中,基因岛MPI-1编码CRISPR/Cas有关的基因,提示CRISPR/Cas可能与结核杆菌的致病性有关[43]。我们总结了结核杆菌CRISPR/Cas相关基因功能(表 1)。
| Protein | Gene | Function | References |
| Cas2 | Rv2816c | Associated with cell wall synthesis in MTB | [44] |
| Cas1 | Rv2817c | Cas1 presumably crucial for spacer acquisition | [16] |
| Csm4 | Rv2820c | Involved in virulence of MTB, and inhibited host defense function |
[45] |
| Csm5 | Rv2819c | Involved in the virulence of MTB | [46] |
| Csm2 | Rv2822c | Stabilized the active structure of the Csm complex to facilitate the reaction |
[47] |
| Cas6 | Rv2824c | Cleaved the repeat RNA is ion dependent | [33-48] |
| Csm6 | Rv2818c | Ancillary ribonuclease, which responsible for cleavage of RNA to enhance immunity |
[49] |
3.1 Cas2
Cas2是在间隔区获得中起重要作用的保守的蛋白,为CRISPR系统功能正常发挥提供保障。结核杆菌不同菌株中Cas2基因核苷酸的相似性为100%。嗜肺军团菌(Legionella pneumophila) Cas2蛋白在侵染宿主哈氏变形虫(Hartmannella)和棘阿米巴原虫(Acanthamoeba)以及逃避巨噬细胞免疫过程中发挥重要作用[50]。结核杆菌Cas2 (Rv2816c)在致病分枝杆菌中非常保守。我们课题组在耻垢分枝杆菌中异源表达Cas2 (Rv2816c),构建Rv2816c重组耻垢分枝杆菌。该重组菌对多种胁迫敏感,在巨噬细胞中的存活能力降低[44],但具体机制尚有待研究。
3.2 Cas1CRISPR/Cas系统中,来自不同细菌的Cas1的生化特性不同。来自大肠杆菌的Cas1与DNA修复有关[51]。结核杆菌Cas1 (Rv2817c)蛋白是一种保守的金属依赖性核酸内切酶,Cas1蛋白通过和其他Cas蛋白形成复合物从而获取外源基因的前间隔区序列,抵御外源DNA入侵[16]。57.14%的临床菌株缺失Cas1基因,提示Cas1可能与结核杆菌的毒力有关。耻垢分枝杆菌中过表达Rv2817c后DNA损伤修复功能受损,对抗结核药物更敏感,胁迫应答能力降低[52]。比较3株CRISPR区间隔区和重复区完全缺失的菌株发现,缺失Cas1和DR的结核杆菌临床菌株对DNA损伤更加敏感,染色体不能正常分离[53]。
3.3 Csm4在耻垢分枝杆菌中异源表达结核杆菌CRISPR相关蛋白Csm4 (Rv2820c),得到的Rv2820c重组耻垢分枝杆菌感染巨噬细胞,其胞内存活力降低,iNOS表达增加,产生更多NO[45]。比较从结核性脑膜炎患者分离的3株生长快速的结核杆菌的基因组发现Csm4发生移码突变,导致氨基酸改变,编码产物缩短为原长度的40%。Csm4截短菌株的毒力和胞内存活力增强[54]。结核杆菌北京株中截短的Csm4在耻垢分枝杆菌中异源表达,得到的Csm4重组耻垢分枝杆菌毒力增强,侵染巨噬细胞后产生更多NO,对宿主免疫攻击的抵抗力增加[55]。
3.4 Csm5Csm5在H37Ra中的表达水平比H37Rv中低,提示Csm5可能与结核杆菌的毒力有关[56]。Csm5是Cas蛋白复合物的重要组成部分,Csm5靠近crRNA 3′端,暗示Csm5可能有助于crRNA成熟和结合靶基因。作为RNA结合蛋白,Csm5晶体结构提示Csm5亚基的铁氧还蛋白样(Ferredoxin-like)折叠的结构多样性强,是Ⅲ型CRISPR/Cas系统crRNA与靶RNA结合的结构基础[46]。
3.5 Csm2目前已经解析了表皮葡萄球菌Ⅲ-A CRISPR/Cas的Csm2的晶体结构。晶体结构中的保守赖氨酸残基对结合靶基因非常关键。Csm2稳定Csm复合物的活性结构来促进反应[47]。结核杆菌Csm2的晶体结构尚无报道。
3.6 Cas6结核杆菌的1类Ⅲ-A CRISPR/Cas中,成熟的crRNA-CRISPR/Cas蛋白复合物识别并结合特定DNA序列,Cas6 (Rv2824c)蛋白切割外源DNA,抵御外源干扰入侵。H99A和G295A/G297A突变证实了Cas6蛋白的活性位点,对于剪切靶点非常重要[27]。
3.7 Csm6Ⅲ-A CRISPR/Cas系统中CRISPR- RNA效应复合物可以检测入侵的RNA,触发一系列防御反应。环寡腺苷酸(Cyclic oligoadenylate,cOA)尤其是环状-6腺苷酸(Cyclic hexa-adenylate,cA6),可以激活Csx1/Csm6家族效应蛋白,后者非特异性降解入侵RNA,增强细菌免疫力[49]。
综上,Ⅲ-A型CRISPR/Cas系统可帮助结核杆菌抵御外源入侵遗传物质,也可以调节结核杆菌基因表达。有的临床分离株CRISPR/Cas系统缺失或不完整,但毒性和在巨噬细胞中的存活率却增加,这也提示CRISPR/Cas系统与菌株进化有关[53]。5株表面光滑的结核杆菌(Smooth tuberculosis,STB)的全基因组序列中,CRISPR/Cas系统不完整。这些菌株在感染后的小鼠中的持留能力和毒力较弱,提示CRISPR/Cas系统与毒力有关[57]。7株临床结核杆菌分离株的CRISPR/Cas系统的间隔区存在差异,但药物敏感性没有差异。这些也提示CRISPR/Cas的多样性和功能之间关系很复杂[40]。
4 CRISPR与分枝杆菌基因编辑 4.1 基于dCas9的基因编辑结核杆菌生长缓慢,遗传操作受限,高效的结核杆菌遗传操作系统是深入认识结核杆菌生物学,尤其是持留和耐药分子机理的重要基础。
CRISPRi技术广泛应用于真核及原核生物的基因编辑[58],包括结核杆菌。基于dCas9的分枝杆菌基因编辑技术主要是优化来源于化脓链球菌的Cas9密码子,突变Cas9蛋白的D10A/H840A位点,获得了dCas9蛋白。通过在分枝杆菌体内表达dCas9和sgRNA,构建了一套能简单高效地抑制分枝杆菌基因表达的CRISPRi系统,原理如图 3-A。该系统能有效抑制靶基因转录,验证特定基因是否为结核杆菌的必需基因并进行功能分析[59]。目前,这个方法测试了结核杆菌几个必需基因如sigH (Rv3323)、pknB (Rv0014)、inhA (Rv1484)、dfrA (Rv2763c)、wag31 (Rv2145c)和ftsZ (Rv2150c),发现CRISPRi能明显抑制这些基因的表达,导致生长抑制或改变结核杆菌对小分子抑制剂的敏感性和细胞形态。该系统的优势是不改变细菌染色体结构[60]。dCas9偶联荧光素酶基因的系统可快速检测临床标本中的结核杆菌,缩短诊断时间[61]。筛选Cas9的11个同源基因发现,用来源于嗜热链球菌的Cas9进行CRISPRi可降低Cas9的细胞毒性,提高CRISPRi对分枝杆菌表达调控的效率。高效率基因敲除可用于鉴定结核杆菌必需基因、发现新药物靶标[62]。

|
| 图 3 Ⅱ型CRISPR/ Cas系统在分枝杆菌中的应用 Figure 3 The application of Ⅱ CRISPR/Cas in mycobacteria. A: CRISPR/dCas9 system is most deeply studied CRISPR systems, used for gene transcription inhibition; B: CRISPR/Cas12a system used for gene editing in M. tuberculosis. HA1: homology arms 1; HA2: homology arms 2; M1: marker 1. |
4.2 基于Cas12a的基因编辑
CRISPR/Cas12a在耻垢分枝杆菌中的基因编辑,主要通过同源重组修复,有效进行基因编辑如图 3-B[25]。基于Cas12a/guide RNA (gRNA)的平台,可以快速、准确鉴定分枝杆菌种类。该方法通过设计靶向rpoB的物种特异性的gRNA探针,可以区分结核杆菌和其他非结核分枝杆菌如M. abscessus、M. intracellulare、M. avium、M. kansasii、M. gordonae和M. fortuitum。该方法准确区分了73个临床样本中的72个[63]。在大肠杆菌中引入结核杆菌的NHEJ系统后能修复DNA双链断裂,但效率很低[64]。CRISPR/Cas12a与NHEJ系统偶联在耻垢分枝杆菌的基因编辑效率高达70%[65]。通过改进NHEJ系统后的与CRISPR/Cas12a偶联,可有效地在结核分枝杆菌中产生大规模的基因突变[15]。
5 CRISPR系统与结核病防控随着CRISPR/Cas编辑技术的发展,在结核杆菌中也有应用,有助于深入分析其基因功能。比如CRISPRi文库可以在结核杆菌中进行全基因组筛选,鉴定必需基因和新药物靶点[66]。用CRISPRi验证了MmpL3作为结核杆菌药物靶点的可行性[67]。
5.1 外源CRISPR/Cas系统CRISPR/Cas可以高效靶向抗生素耐药基因,crRNAs可以有效地引导Cas9靶向大肠杆菌染色体上所有拷贝,导致DNA断裂,细胞不能通过同源重组修复,从而导致细胞死亡,通过II-A类CRISPR/Cas系统编辑噬菌体基因组,侵染宿主后可进行基因组编辑(图 4-A)。若crRNAs导致目标定位效率低,通过表达Mu噬菌体Gam蛋白可进一步阻断同源重组,确保细胞死亡[64]。利用CRISPR/Cas9系统靶向超广谱耐β-内酰胺类抗生素的大肠杆菌的耐药基因,恢复其对抗生素的敏感性,具体机制如图 4-A[68]。利用CRISPR/Cas9系统清除大肠杆菌的耐药NDM-1质粒,使产NDM-1耐药模式菌恢复对亚胺培南和其他β-内酰胺类抗生素的敏感性,具体机制如图 4-B[69]。基于CRISPR系统的方法可以解决现有抗生素的缺陷,即不具有广谱性,特异性杀死有害细菌,这样可以减少选择压力[70]。这些方法也可能在结核杆菌中应用。

|
| 图 4 基于CRISPR/Cas系统的噬菌体基因组编辑、裂解耐药基因和耐药质粒 Figure 4 Killing of microbial by a phagemid delivered CRISPR system. RNA-guided nucleases targeting specific DNA sequences (genome 4-A or plasmid 4-B) are delivered efficiently to microbial population using bacteriophage. |
目前已经开发了基于CRISPR/Cas12a的结核杆菌快速检测方法,对179例患者样品来源的结核杆菌进行测试的敏感性为79%。这极大缩短临床结核病诊断周期[71]。基于CRISPRi的结核杆菌文库能覆盖结核杆菌的基因组,该文库由具有不同的sgRNAs组成,有助于发现新的药物靶点,对于抗结核药物的开发具有重要意义[72]。
5.2 内源CRISPR/Cas系统通过开发内源性的CRISPR/Cas系统来抵抗微生物,即通过激活微生物中自身的CRISPR/Cas系统,基于序列信息定量去除个别细菌,为治疗耐药菌感染提供了新方向。通过E. coli K-12内源性的I-E型CRISPR/Cas系统可以抑制相关基因的表达[73]。运用大肠杆菌内源性I-E型CRISPR/Cas系统可以导致细菌程序性死亡[74]。利用Lactobacillus crispatus内源性I-E型CRISPR/Cas系统进行基因编辑,靶向3个位点,导致99%以上的细胞死亡[75]。结核杆菌内源CRISPR/Cas系统开发也可能用于基因抑制,使耐药结核杆菌恢复敏感性。敲除基因组中Cas6,插入启动子激活Cascade,转录成熟的crRNA与Cascade结合在特定位点与DNA结合,阻断RNA聚合酶转录,从而抑制结核杆菌中基因表达,具体原理如图 5,结核杆菌缺失Rv2837c (cnpB)后内源性CRISPR/Cas系统活性增加。Rv2837c (CnpB)蛋白通过寡核糖核酸酶(Orn)控制结核分枝杆菌CRISPR/Cas系统的表达,为控制内源性CRISPR/Cas系统的转录提供了新的方向[76]。

|
| 图 5 结核杆菌内源CRISPR/Cas系统可用于基因抑制 Figure 5 Repurposing the Type Ⅲ-A CRISPR-Cas system in tuberculosis for programmable gene repression. |
5.3 CRISPR/Cas系统改造噬菌体
CRISPR/Cas系统还可作为噬菌体基因组编辑工具,改造后的噬菌体可以杀死耐药细菌。噬菌体可以作为CRISPR/Cas载体,使CRISPR/Cas系统在致病菌中表达,靶向剪切抗性质粒或基因组,恢复抗生素敏感性,阻断耐药基因的水平转移(图 4)。利用非致病的分枝杆菌递送裂解性分枝杆菌噬菌体TM4可以杀死巨噬细胞内的结核杆菌和鸟分枝杆菌等致病菌[77]。这一递送噬菌体的体系可以显著降低小鼠脾脏细胞内的细菌数量。利用温和噬菌体“一步法”包装CRISPR/Cas9系统可以高效靶向清除细菌耐药质粒,减少细菌耐药质粒在环境中的释放和积累,阻断耐药质粒在细菌间的接合转移,减少耐药细菌传播[69]。
但噬菌体也演化出多种策略逃避宿主细菌的CRISPR/Cas系统的作用[78-79]。比如anti-CRISPR蛋白(AcrVIAs蛋白)帮助噬菌体成功逃逸CRISPR-Cas13a系统,这为疾病诊断和治疗提供了一个开关,基因编缉完成后,可以消除Cas蛋白剪切活性[80]。同时临床几种噬菌体联用,或噬菌体与抗生素联用,可以避免耐药细菌的出现,目前有用非敏感型抗生素与噬菌体联用在临床成功治疗超级耐药菌引发的尿路感染[81-82]。
分枝杆菌噬菌体治疗耐药结核杆菌、检测是否存在结核杆菌或者耐药结核杆菌的工作也有报道。比如6种特定噬菌体对结核杆菌细胞壁的脂阿拉伯甘露聚糖(Lipoarabinomannan)有特异性血清学反应,可开发作为临床诊断结核病的工具[83]。分枝杆菌噬菌体可有效治疗豚鼠的结核杆菌感染,但疗效不如异烟肼[84]。基于噬菌体的治疗虽然取得一定的成果,但离临床应用还有很长距离,值得研究。
6 总结和展望随着CRISPR/Cas技术的发展和结核杆菌CRISPR/Cas系统的发现和功能研究,为结核杆菌基础生物学性质研究提供了新工具。CRISPR/Cas编辑系统在结核杆菌中应用,因具有高度的靶标基因序列特异性,对于揭示结核杆菌的关键致病基因,发现新型抗结核药物靶标和开发疫苗方面具有重要意义。CRISPR/Cas系统与分枝杆菌噬菌体联合,用于结核病防控也是未来值得进一步研究的方向。
| [1] | The Lancet. End the tuberculosis emergency:a promise is not enough. The Lancet, 2019, 394(10208): 1482. |
| [2] | Wright A, Zignol M, Van Deun A, Falzon D, Gerdes SR, Feldman K, Hoffner S, Drobniewski F, Barrera L, Van Soolingen D, Boulabhal F, Paramasivan CN, Kam KM, Mitarai S, Nunn P, Raviglione M. Epidemiology of antituberculosis drug resistance 2002-07:an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. The Lancet, 2009, 373(9678): 1861-1873. DOI:10.1016/S0140-6736(09)60331-7 |
| [3] | Sharma K, Verma R, Advani J, Chatterjee O, Solanki HS, Sharma A, Varma S, Modi M, Ray P, Mukherjee KK, Sharma M, Dhillion MS, Suar M, Chatterjee A, Pandey A, Prasad TSK, Gowda H. Whole genome sequencing of Mycobacterium tuberculosis Isolates from extrapulmonary sites. OMICS:A Journal of Integrative Biology, 2017, 21(7): 413-425. DOI:10.1089/omi.2017.0070 |
| [4] | Makarova KS, Koonin EV. Annotation and classification of CRISPR-Cas systems//Lundgren M, Charpentier E, Fineran PC. CRISPR. New York: Humana Press, 2015: 47-75. |
| [5] | Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-cas systems:all the way there and back. Genome Biology and Evolution, 2017, 9(10): 2812-2825. DOI:10.1093/gbe/evx192 |
| [6] | Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007, 315(5819): 1709-1712. DOI:10.1126/science.1138140 |
| [7] | Bikard D, Barrangou R. Using CRISPR-Cas systems as antimicrobials. Current Opinion in Microbiology, 2017, 37: 155-160. DOI:10.1016/j.mib.2017.08.005 |
| [8] | Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature Biotechnology, 2014, 32(11): 1141-1145. DOI:10.1038/nbt.3011 |
| [9] | Bondy-Denomy J, Davidson AR. To acquire or resist:the complex biological effects of CRISPR-Cas systems. Trends in Microbiology, 2014, 22(4): 218-225. DOI:10.1016/j.tim.2014.01.007 |
| [10] | Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology, 1987, 169(12): 5429-5433. DOI:10.1128/JB.169.12.5429-5433.1987 |
| [11] | Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology, 2002, 43(6): 1565-1575. DOI:10.1046/j.1365-2958.2002.02839.x |
| [12] | Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-823. DOI:10.1126/science.1231143 |
| [13] | Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Research, 2012, 22(7): 1327-1333. DOI:10.1101/gr.138792.112 |
| [14] | Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 2013, 8(11): 2281-2308. DOI:10.1038/nprot.2013.143 |
| [15] | Yan MY, Li SS, Ding XY, Guo XP, Jin Q, Sun YC. A CRISPR-Assisted nonhomologous end-joining strategy for efficient genome editing in Mycobacterium tuberculosis. mBio, 2020, 11(1): e02364-19. |
| [16] | Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nature Reviews Microbiology, 2015, 13(11): 722-736. DOI:10.1038/nrmicro3569 |
| [17] | Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV. Evolutionary classification of CRISPR-Cas systems:a burst of class 2 and derived variants. Nature Reviews Microbiology, 2020, 18(2): 67-83. DOI:10.1038/s41579-019-0299-x |
| [18] | Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Current Opinion in Microbiology, 2017, 37: 67-78. DOI:10.1016/j.mib.2017.05.008 |
| [19] | Xiao YB, Luo M, Dolan AE, Liao MF, Ke AL. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science, 2018, 361(6397): eaat0839. DOI:10.1126/science.aat0839 |
| [20] | Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology, 2011, 9(6): 467-477. DOI:10.1038/nrmicro2577 |
| [21] | Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 2013, 152(5): 1173-1183. DOI:10.1016/j.cell.2013.02.022 |
| [22] | Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 2015, 163(3): 759-771. DOI:10.1016/j.cell.2015.09.038 |
| [23] | Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science, 2016, 353(6299): aaf5573. DOI:10.1126/science.aaf5573 |
| [24] | Liu W, Tang DD, Wang HJ, Lian JZ, Huang L, Xu ZN. Combined genome editing and transcriptional repression for metabolic pathway engineering in Corynebacterium glutamicum using a catalytically active Cas12a. Applied Microbiology and Biotechnology, 2019, 103(21/22): 8911-8922. DOI:10.1007/s00253-019-10118-4 |
| [25] | Yan MY, Yan HQ, Ren GX, Zhao JP, Guo XP, Sun YC. CRISPR-Cas12a-Assisted recombineering in bacteria. Applied and Environmental Microbiology, 2017, 83(17): e00947-17. |
| [26] | Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nature Biotechnology, 2016, 34(8): 869-874. DOI:10.1038/nbt.3620 |
| [27] | Wei WJ, Zhang S, Fleming J, Chen Y, Li ZH, Fan SS, Liu Y, Wang W, Wang T, Liu Y, Ren BG, Wang M, Jiao JJ, Chen YY, Zhou Y, Zhou YF, Gu SJ, Zhang XL, Wan L, Chen T, Zhou L, Chen Y, Zhang XE, Li CY, Zhang HT, Bi LJ. Mycobacterium tuberculosis type Ⅲ-A CRISPR/Cas system crRNA and its maturation have atypical features. FASEB Journal, 2019, 33(1): 1496-1509. DOI:10.1096/fj.201800557RR |
| [28] | Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic characterization of antiplasmid immunity through a type Ⅲ-A CRISPR-Cas system. Journal of Bacteriology, 2014, 196(2): 310-317. DOI:10.1128/JB.01130-13 |
| [29] | Choudhary E, Lunge A, Agarwal N. Strategies of genome editing in mycobacteria:achievements and challenges. Tuberculosis, 2016, 98: 132-138. DOI:10.1016/j.tube.2016.03.005 |
| [30] | Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry Ⅲ CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 1998, 393(6685): 537-544. DOI:10.1038/31159 |
| [31] | Groenen PMA, Bunschoten AE, van Soolingen D, van Embden JDA. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Molecular Microbiology, 1993, 10(5): 1057-1065. DOI:10.1111/j.1365-2958.1993.tb00976.x |
| [32] | Brudey K, Gutierrez MC, Vincent V, Parsons LM, Salfinger M, Rastogi N, Sola C. Mycobacterium africanum genotyping using novel spacer oligonucleotides in the direct repeat locus. Journal of Clinical Microbiology, 2004, 42(11): 5053-5057. DOI:10.1128/JCM.42.11.5053-5057.2004 |
| [33] | He LM, Fan XY, Xie JP. Comparative genomic structures of Mycobacterium CRISPR-Cas. Journal of Cellular Biochemistry, 2012, 113(7): 2464-2473. DOI:10.1002/jcb.24121 |
| [34] | Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria:DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One, 2009, 4(11): e7815. DOI:10.1371/journal.pone.0007815 |
| [35] | Mokrousov I, Rastogi N. Spacer-based macroarrays for CRISPR genotyping. Methods in Molecular Biology, 2015, 1311: 111-131. |
| [36] | Botelho A, Canto A, Leão C, Cunha MV. Clustered regularly interspaced short palindromic repeats (CRISPRs) analysis of members of the Mycobacterium tuberculosis complex. Methods in Molecular Biology, 2015, 1247: 373-389. |
| [37] | Mokrousov I, Vyazovaya A, Narvskaya O. Mycobacterium tuberculosis latin american-mediterranean family and its sublineages in the light of robust evolutionary markers. Journal of Bacteriology, 2014, 196(10): 1833-1841. DOI:10.1128/JB.01485-13 |
| [38] | Borile C, Labarre M, Franz S, Sola C, Refrégier G. Using affinity propagation for identifying subspecies among clonal organisms: lessons from M. tuberculosis. BMC Bioinformatics, 2011, 12: 224. |
| [39] | Grissa I, Bouchon P, Pourcel C, Vergnaud G. On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie, 2008, 90(4): 660-668. DOI:10.1016/j.biochi.2007.07.014 |
| [40] | Liu F, Hu YF, Wang Q, Li HM, Gao GF, Liu CH, Zhu BL. Comparative genomic analysis of Mycobacterium tuberculosis clinical isolates. BMC Genomics, 2014, 15: 469. DOI:10.1186/1471-2164-15-469 |
| [41] | Abadia E, Zhang J, Ritacco V, Kremer K, Ruimy R, Rigouts L, Gomes HM, Elias AR, Fauville-Dufaux M, Stoffels K, Rasolofo-Razanamparany V, De Viedma DG, Herranz M, Al-Hajoj S, Rastogi N, Garzelli C, Tortoli E, Suffys PN, Van Soolingen D, Refregier G, Sola C. The use of microbead-based spoligotyping for Mycobacterium tuberculosis complex to evaluate the quality of the conventional method:providing guidelines for Quality Assurance when working on membranes. BMC Infectious Diseases, 2011, 11: 110. DOI:10.1186/1471-2334-11-110 |
| [42] | Shariat N, Dudley EG. CRISPRs:molecular signatures used for pathogen subtyping. Applied and Environmental Microbiology, 2014, 80(2): 430-439. DOI:10.1128/AEM.02790-13 |
| [43] | Xie J, Zhou FF, Xu GG, Mai GQ, Hu J, Wang GQ, Li F. Genome-wide screening of pathogenicity islands in Mycobacterium tuberculosis based on the genomic barcode visualization. Molecular Biology Reports, 2014, 41(9): 5883-5889. DOI:10.1007/s11033-014-3463-4 |
| [44] | Huang QQ, Luo HP, Liu MQ, Zeng J, Abdalla AE, Duan XK, Li QM, Xie JP. The effect of Mycobacterium tuberculosis CRISPR-associated Cas2(Rv2816c) on stress response genes expression, morphology and macrophage survival of Mycobacterium smegmatis. Infection, Genetics and Evolution, 2016, 40: 295-301. DOI:10.1016/j.meegid.2015.10.019 |
| [45] |
Zhai XQ, Bao L, Luo T, Peng X, Sun CF, Yang GP. Effect of the expression of iNOS Induced by Mycobacterium tuberculosis CRISPR-associated Csm4(Rv2820c) on intracellular viability of Mycobacterium smegmatis. Journal of Sichuan University (Medical Science Edition), 2018, 49(3): 319-324.
(in Chinese) 翟小倩, 鲍朗, 罗涛, 彭璇, 孙长峰, 杨国平. 结核杆菌CRISPR-associated Csm4(Rv2820c)诱导iNOS表达对耻垢杆菌胞内存活的影响. 四川大学学报(医学版), 2018, 49(3): 319-324. |
| [46] | An Y, Park KH, Lee M, Kim TJ, Woo EJ. Crystal structure of the Csm5 subunit of the type Ⅲ-A CRISPR-Cas system. Biochemical and Biophysical Research Communications, 2020, 523(1): 112-116. DOI:10.1016/j.bbrc.2019.12.046 |
| [47] | Takeshita D, Sato M, Inanaga H, Numata T. Crystal structures of Csm2 and Csm3 in the Type Ⅲ-A CRISPR-Cas effector complex. Journal of Molecular Biology, 2019, 431(4): 748-763. DOI:10.1016/j.jmb.2019.01.009 |
| [48] | Mokrousov I, Limeschenko E, Vyazovaya A, Narvskaya O. Corynebacterium diphtheriae spoligotyping based on combined use of two CRISPR loci. Biotechnology Journal, 2007, 2(7): 901-906. DOI:10.1002/biot.200700035 |
| [49] | Grüschow S, Athukoralage JS, Graham S, Hoogeboom T, White MF. Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Research, 2019, 47(17): 9259-9270. DOI:10.1093/nar/gkz676 |
| [50] | Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio, 2013, 4(2): e00074-13. |
| [51] | Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV, Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Molecular Microbiology, 2011, 79(2): 484-502. DOI:10.1111/j.1365-2958.2010.07465.x |
| [52] | Wei JW, Lu N, Li ZY, Wu XY, Jiang T, Xu L, Yang C, Guo S. The Mycobacterium tuberculosis CRISPR-Associated Cas1 Involves persistence and tolerance to anti-tubercular Drugs. BioMed Research International, 2019, 2019: 7861695. |
| [53] | Freidlin PJ, Nissan I, Luria A, Goldblatt D, Schaffer L, Kaidar-Shwartz H, Chemtob D, Dveyrin Z, Head SR, Rorman E. Structure and variation of CRISPR and CRISPR-flanking regions in deleted-direct repeat region Mycobacterium tuberculosis complex strains. BMC Genomics, 2017, 18(1): 168. DOI:10.1186/s12864-017-3560-6 |
| [54] | Lam JT, Yuen KY, Ho PL, Weng XH, Zhang WH, Chen S, Yam WC. Truncated Rv2820c enhances mycobacterial virulence ex vivo and in vivo. Microbial Pathogenesis, 2011, 50(6): 331-335. DOI:10.1016/j.micpath.2011.02.008 |
| [55] | Zhai XQ, Luo T, Peng X, Ma PJ, Wang CH, Zhang CX, Suo J, Bao L. The truncated Rv2820c of Mycobacterium tuberculosis Beijing family augments intracellular survival of M. smegmatis by altering cytokine profile and inhibiting NO generation. Infection, Genetics and Evolution, 2018, 59: 75-83. DOI:10.1016/j.meegid.2018.01.027 |
| [56] | Rindi L, Lari N, Garzelli C. Genes of Mycobacterium tuberculosis H37Rv downregulated in the attenuated strain H37Ra are restricted to M. tuberculosis complex species. The New Microbiologica, 2001, 24(3): 289-294. |
| [57] | Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie AS, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez MC, Leclerc C, Bentley SD, Stinear TP, Brisse S, Medigue C, Parkhill J, Cruveiller S, Brosch R. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nature Genetics, 2013, 45(2): 172-179. DOI:10.1038/ng.2517 |
| [58] | Ouellette SP. Feasibility of a conditional knockout system for Chlamydia based on CRISPR interference. Frontiers in Cellular and Infection Microbiology, 2018, 8: 59. DOI:10.3389/fcimb.2018.00059 |
| [59] | Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nature Communications, 2015, 6: 6267. DOI:10.1038/ncomms7267 |
| [60] | Singh AK, Carette X, Potluri LP, Sharp JD, Xu RF, Prisic S, Husson RN. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Research, 2016, 44(18): e143. DOI:10.1093/nar/gkw625 |
| [61] | Zhang YH, Qian L, Wei WJ, Wang Y, Wang BN, Lin PP, Liu WC, Xu LZ, Li X, Liu DM, Cheng SD, Li JF, Ye YX, Li H, Zhang XH, Dong YM, Zhao XJ, Liu CH, Zhang HM, Ouyang Q, Lou CB. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synthetic Biology, 2017, 6(2): 211-216. DOI:10.1021/acssynbio.6b00215 |
| [62] | Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, Fortune SM. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nature Microbiology, 2017, 2: 16274. DOI:10.1038/nmicrobiol.2016.274 |
| [63] | Xiao GH, He X, Zhang S, Liu YY, Liang ZH, Liu HM, Zhang JJ, Ou M, Cai SH, Lai WJ, Zhang TY, Ren LL, Zhang GL. Cas12a/Guide RNA-Based platform for rapid and accurate identification of major Mycobacterium species. Journal of Clinical Microbiology, 2020, 58(2): e01368-19. |
| [64] | Cui L, Bikard D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Research, 2016, 44(9): 4243-4251. DOI:10.1093/nar/gkw223 |
| [65] | Sun BB, Yang JJ, Yang S, Ye RD, Chen DJ, Jiang Y. A CRISPR-Cpf1-Assisted Non-Homologous end joining genome editing system of Mycobacterium smegmatis. Biotechnology Journal, 2018, 13(9): 1700588. DOI:10.1002/biot.201700588 |
| [66] | Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. CRISPR/Cas9-The ultimate weapon to battle infectious diseases?. Cellular Microbiology, 2017, 19(2): e12693. DOI:10.1111/cmi.12693 |
| [67] | McNeil MB, Cook GM. Utilization of CRISPR interference to validate MmpL3 as a drug target in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy, 2019, 63(8): e00629-19. |
| [68] | Kim JS, Cho DH, Park M, Chung WJ, Shin D, Ko KS, Kweon DH. CRISPR/Cas9-Mediated Re-Sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum β-Lactamases. Journal of Microbiology and Biotechnology, 2016, 26(2): 394-401. DOI:10.4014/jmb.1508.08080 |
| [69] | Liu HB, Zhu BH, Liang BB, Xu XB, Qiu SF, Jia LL, Li P, Yang L, Li YR, Xiang Y, Xie J, Wang LG, Yang CJ, Sun YS, Song HB. A novel mcr-1 variant carried by an IncI2-Type plasmid identified from a multidrug resistant enterotoxigenic Escherichia coli. Frontiers in Microbiology, 2018, 9: 815. DOI:10.3389/fmicb.2018.00815 |
| [70] | Greene AC. CRISPR-Based antibacterials:transforming bacterial defense into offense:(Trends in Biotechnology 36, 127-130, 2018). Trends in Biotechnology, 2018, 36(12): 1299. DOI:10.1016/j.tibtech.2018.01.009 |
| [71] | Ai JW, Zh ou, X, Xu T, Yang ML, Chen YY, He GQ, Pan N, Cai YW, Li YJ, Wang XR, Su H, Wang T, Zeng WQ, Zhang WH. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerging Microbes & Infections, 2019, 8(1): 1361-1369. |
| [72] | Rock J. Tuberculosis drug discovery in the CRISPR era. PLoS Pathogens, 2019, 15(9): e1007975. DOI:10.1371/journal.ppat.1007975 |
| [73] | Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Research, 2015, 43(1): 674-681. DOI:10.1093/nar/gku971 |
| [74] | Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio, 2014, 5(1): e00928-13. |
| [75] | Hidalgo-Cantabrana C, Goh YJ, Pan M, Sanozky-Dawes R, Barrangou R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(32): 15774-15783. DOI:10.1073/pnas.1905421116 |
| [76] | Zhang Y, Yang J, Bai GC. Regulation of the CRISPR-Associated genes by Rv2837c (CnpB) via an orn-like activity in tuberculosis complex mycobacteria. Journal of Bacteriology, 2018, 200(8): e00743-17. |
| [77] | Broxmeyer L, Sosnowska D, Miltner E, Chacón O, Wagner D, McGarvey J, Barletta RG, Bermudez LE. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium:a model for phage therapy of intracellular bacterial pathogens. The Journal of Infectious Diseases, 2002, 186(8): 1155-1160. DOI:10.1086/343812 |
| [78] | Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science, 2010, 327(5962): 167-170. DOI:10.1126/science.1179555 |
| [79] | van Houte S, Buckling A, Westra ER. Evolutionary ecology of prokaryotic immune mechanisms. Microbiology and Molecular Biology Reviews, 2016, 80(3): 745-763. DOI:10.1128/MMBR.00011-16 |
| [80] | Lin P, Qin SG, Pu QQ, Wang ZH, Wu Q, Gao P, Schettler J, Guo K, Li RP, Li GP, Huang CH, Wei YQ, Gao GF, Jiang JX, Wu M. CRISPR-Cas13 inhibitors block RNA editing in bacteria and mammalian cells. Molecular Cell, 2020, 78(5): 850-861. DOI:10.1016/j.molcel.2020.03.033 |
| [81] | Azam AH, Tanji Y. Bacteriophage-host arm race:an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Applied Microbiology and Biotechnology, 2019, 103(5): 2121-2131. DOI:10.1007/s00253-019-09629-x |
| [82] | Bao J, Wu NN, Zeng YG, Chen LG, Li LL, Yang L, Zhang YY, Guo MQ, Li LS, Li J, Tan DM, Cheng MJ, Gu JM, Qin JH, Liu JZ, Li SR, Pan GQ, Jin X, Yao BX, Guo XK, Zhu TY, Le S. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerging Microbes & Infections, 2020, 9(1): 771-774. |
| [83] | Bua A, Rosu V, Molicotti P, Das Gupta SK, Ahmed N, Zanetti S, Sechi LA. Phages specific for mycobacterial lipoarabinomannan help serodiagnosis of tuberculosis. The New Microbiologica, 2009, 32(3): 293-296. |
| [84] | Sula L, Sulová J, Stolcpartová M. Therapy of experimental tuberculosis in guinea pigs with mycobacterial phages DS-6A, GR-21 T, My-327. Czechoslovak Medicine, 1981, 4(4): 209-214. |
 2021, Vol. 61
2021, Vol. 61