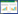中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张道锋, 史贤明. 2021
- Daofeng Zhang, Xianming Shi. 2021
- 银白色葡萄球菌的发现、分布与致病性
- Emergence, distribution and pathogenicity of Staphylococcus argenteus
- 微生物学报, 61(2): 263-278
- Acta Microbiologica Sinica, 61(2): 263-278
-
文章历史
- 收稿日期:2020-03-21
- 修回日期:2020-05-27
- 网络出版日期:2020-06-17
2. 上海交通大学农业与生物学院, 中美食品安全联合研究中心, 微生物代谢国家重点实验室, 上海 200240
2. MOST-USDA Joint Research Center for Food Safety, School of Agriculture and Biology & State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200240, China
葡萄球菌属(Staphylococcus)是人类和其他温血动物体表、皮脂腺体和黏膜等部位的主要菌群之一,在自然环境中分布广泛[1]。很多凝固酶阳性葡萄球菌(coagulase-positive Staphylococci,CoPS)可以引起人体感染,其中,以金黄色葡萄球菌(S. aureus)最为知名。金黄色葡萄球菌是人类体表、前鼻腔的正常菌群,也是条件致病菌,部分菌株具有很强的致病性和传染性,还可引发社区感染[2]。同时,由于分布广泛、与动植物关系密切,金黄色葡萄球菌也是引发食物中毒、威胁食品安全的重要菌群之一[3]。因为在临床感染和食品安全领域的重要性,金黄色葡萄球菌得到了广泛而深入的研究。目前(2020年2月),金黄色葡萄球菌的多位点序列分型(multilocus sequence typing,MLST)数据库已收录5800多个ST (sequence type)型(https://pubmlst.org/saureus/);已发布的基因组数据超过11000份(https://www.ncbi.nlm.nih.gov/genome/)。2015年,Tong等将金黄色葡萄球菌种内的2个远源支系独立出来,建立为2个新的物种,分别命名为银白色葡萄球菌(S. argenteus)和施韦策葡萄球菌(S. schweitzeri)[4],这3个物种构成了新的金黄色葡萄球菌复合群(S. aureus complex,SAC),共用同一个MLST数据库。这一分类学提议随后被著名病原微生物学期刊Journal of Clinical Microbiology认可并向病原微生物学界推广[5]。2019年,欧洲临床微生物与传染病学会(European Society of Clinical Microbiology and Infectious Diseases,ESCMID)下属的葡萄球菌和葡萄球菌病研究组(Study Group for Staphylococci and Staphylococcal Diseases,ESGS)针对实践中银白色葡萄球菌的诊断、监测、治疗和预防等问题提出了建议[6]。
银白色葡萄球菌的绝大多数菌株分离自人体,可以引起与金黄色葡萄球菌类似的感染症状,比如皮肤与软组织感染、菌血症和败血症等,甚至导致死亡[6-8]。同时,银白色葡萄球菌也含有葡萄球菌肠毒素(staphylococcal enterotoxin,SE)基因,可以导致食物中毒[9-11]。目前,该物种已在20多个国家被分离鉴定,呈现出全球分布局势[6, 9]。比较基因组学分析发现[12-13],银白色葡萄球菌含有金黄色葡萄球菌 > 75%的毒力基因,且绝大多数氨基酸序列一致性 > 85%;ST2250谱系已经发生国际间传播。与银白色葡萄球菌类似,施韦策葡萄球菌也含有金黄色葡萄球菌 > 75%的毒力基因,甚至有些特征与金黄色葡萄球菌更加相似[4, 12]。然而,目前该物种的分离株只在非洲被分离获得,主要源自灵长类[14-15]和蝙蝠(Rousettus aegyptiacus和Eidolon helvum)[16-18]。虽然体外实验表明施韦策葡萄球菌对人体细胞具有毒性[19-20],也发现了人源分离株,但尚未证实可以感染人类[21-22]。
从已有的文献资料来看,这3个物种16S rRNA基因序列几乎一致(相似性 > 99.7%),表型特征非常相似,基因组水平的差异也比一般的种间差异要小(图 1)。最明显的2个差异是[4, 23]:1)银白色葡萄球菌因为缺少操纵子crtOPQMN (可合成staphyloxanthin等黄色色素)均形成白色菌落,而金黄色葡萄球菌和施韦策葡萄球菌均含有crtOPQMN;2)银白色葡萄球菌和施韦策葡萄球菌的肽聚糖类为A3α A11.8型(L-Lys–L-Ala–Gly4–5),而金黄色葡萄球菌为A3α A11.2型(L-Lys–Gly4–5)。这2个新种的出现,加深了人们对金黄色葡萄球菌相关类群多样性的认识,为研究金黄色葡萄球菌的进化和致病性机制提供了新的参比对象。因为与人类健康相关,且国内关注较少,本文从发现过程、分布、致病性和鉴定方法等方面,重点对银白色葡萄球菌的国内外研究现状进行综述。
|
| 图 1 银白色葡萄球菌在属内的分类地位 Figure 1 Phylogeny of S. argenteus among the genus Staphylococcus. A: phylogenetic tree based on 16S rRNA gene sequences from the type strains of the representative staphylococci. All the sequences were downloaded from the EzBioCloud server (https://www.ezbiocloud.net/), and the tree was inferred using the Neighbor-Joining method and the Kimura 2-parameter method in MEGA X[24]. B: maximum-likelihood phylogenetic tree based on concatenated deduced amino acid sequences of 1375 single copy core genes among S. aureus complex (SAC) and S. simiae. The tree was reconstructed based on our previous study[12]. The branch lengths are indicated on the branches. |
1 银白色葡萄球菌的发现过程
在银白色葡萄球菌被建立为独立的物种之前,相关菌株通常被作为金黄色葡萄球菌对待。这主要是因为银白色葡萄球菌与金黄色葡萄球菌的表型特征极其相似,常规的生理生化方法难以将二者区分[6, 23]。但是通过对核心基因组上的基因序列进行分析,可以区分这2个物种(图 1-B)。MLST是进行金黄色葡萄球菌分子分型等多样性调查时常用的指标之一,金黄色葡萄球菌的MLST数据库也被相关研究者广泛采用。因此,本文主要通过MLST数据追溯早期与银白色葡萄球菌相关的文献资料。
金黄色葡萄球菌的MLST方法于2000年正式被建立,使用的7个管家基因分别是arcC、aroE、gmk、glpF、pta、tpi和yqiL[25]。2002年,Okuma等[26]在分析35株耐甲氧西林金黄色葡萄球菌(methicillin-resistant S. aureus,MRSA)时报道了一株与常见金黄色葡萄球菌谱系具有明显差异的菌株(属于ST75)。该菌株分离自澳大利亚Darwin地区,在当时并没有引起特别的注意。随后,与ST75密切相关的菌株相继在澳大利亚、英国、柬埔寨、斐济、法国、法属圭亚那、新西兰、泰国、特立尼达&多巴哥共和国、比利时、新加坡、马来西亚、加蓬、中国、以色列、丹麦、缅甸、老挝、日本、瑞士、阿治曼、瑞典等20多个国家和地区被报道(表 1,表 2)。绝大多数菌株分离自东南亚和澳大利亚北部[9]。
| Country | Source | Lineage | SCCmec | pvl | References |
| Asia | |||||
| Ajman | Human nares | CC2250, CC2198, CC2596 | – | +/– | [27] |
| Cambodia | Human nares | ST1223 | n.d. | n.d. | [28] |
| India | Hospital | ST2731a | + | [29] | |
| Israel | Hospital | ST2250 | n.d. | n.d. | [13] |
| Japan | Hospital, food | ST2250, ST2198, ST1223, ST3951 | –b | –b | [10-11, 30-32] |
| Laos | Hospital | ST1223, ST2250 | +/– | – | [33] |
| Malaysia | Hospital | ST2250 | +b | –b | [13] |
| Myanmar | Hospital, healthy human | ST2198, ST2250, ST2854, ST4625 | –b | +b | [34-35] |
| Singapore | Hospital | ST2250 | n.d. | n.d. | [13] |
| Thailand | Hospital, rabbit, raw milk |
ST1223, ST2198, ST2250, ST2854, ST2793, ST4210a, ST4211a, ST4630a, ST4631a, ST4632a, ST4638a |
+/–b | +/– | [7-8, 13, 36-38] |
| Africa | |||||
| Gabon | Gorilla, hospital | ST2198, ST2617a | –b | – | [39-40] |
| Nigeria | Bat | ST3952, ST3960, ST3961, ST3963, ST3980, ST4326 | n.d. | – | [18] |
| Tunisia | Sheep | ST2056a | n.d. | – | [41] |
| America | |||||
| Dominican Republic | Hospital | ST1793a | n.d. | n.d. | [42] |
| French Guiana | Human nares | ST1223 | n.d. | – | [43] |
| Trinidad & Tobago | Hospital | CC1223, CC2250 | – | – | [44] |
| U.S.A | Hospital | ST2250 | – | – | [45] |
| Europe | |||||
| Belgium | Healthy human | ST2250, ST3240a | +/– | – | [46] |
| Denmark | Hospital | ST1223, ST2250, ST2793, ST2854 | +/– b | –b | [47] |
| France | Hospital | ST2250 | –b | – | [48] |
| Germany | Hospital | ST1267a | [49] | ||
| Norway | Hospital | ST2793, ST1223 | + | n.d. | [50] |
| Sweden | Hospital | ST2250, ST1223, ST2793 | +b | +/– b | [51-53] |
| U.K. | Hospital | ST2793 | + | – | [4] |
| Oceanica | |||||
| Australia | Hospital | ST75a, ST258a, ST883a, ST1223, ST1303a, ST1304a, ST1823, ST1824, ST1848, ST1849, ST1850, ST2198, ST2793 |
+/– | – | [9, 54] |
| Fiji | Hospital | CC75 | –b | n.d. | [55-56] |
| New Zealand | Hospital | CC75 | n.d. | n.d. | [55] |
| CC: clonal complex; n.d.: no data available; pvl: genes encoding Panton-Valentine leucocidin (PVL); ST: sequence type. +: positive; –: negative. a: STs with 1–3 loci closely related to S. aureus (identity > 97%); b: summarized on the results of partial isolates because the other had no data available. | |||||
| Location | Source | Lineage | SCCmec | pvl | References |
| Chengdu | Fish product, RTE food | ST1610a, ST3484a | –b | – | [60-61] |
| Chongqing | Hospital, chicken | ST2250 | +/– | –b | [62-63] |
| Guangzhou | RTE food | ST3504a, ST2196a, ST3482*, ST2483a | n.d. | – | [60] |
| Haikou | Hospital | ST2196a, ST4435a | +/– | n.d. | [64] |
| Hongkong | Fish product | ST1685a | – | n.d. | [61] |
| Ningbo | n.d. | ST2250, ST3261 | – | – | [23] |
| Sanya | RTE food | ST3485a | n.d. | – | [60] |
| Shanghai | Hospital, healthy human, pork | ST2250, ST4297a | – | – | [23, 65] |
| Shijiazhuang | Fish product | ST2196a | – | n.d. | [61] |
| Taiwan | Hospital | ST2250, ST2793, ST1223, ST2198 | – | n.d. | [66-67] |
| Xiamen | Slaughter house, rte food | ST2483a, ST3387a, ST3388a | +b | – | [60, 68] |
| Zhanjiang | RTE food | ST2483a | n.d. | – | [60] |
| CC: clonal complex; n.d.: no data available; pvl: genes encoding Panton-Valentine leucocidin (PVL); RTE: ready-to-eat; ST: sequence type. +: positive; –: negative. a: STs with 1–3 loci closely related to S. aureus (identity > 97%); b: summarized on the results of partial isolates because the other had no data available. | |||||
在相关的菌株被发现之初,一些学者就注意到ST75相关的类群CC75 (clonal complex 75)与典型的金黄色葡萄球菌在基因序列上存在显著的差异。McDonald等[57]于2006年建立了一种基于MLST基因位点单核苷酸多态性(single- nucleotide polymorphism,SNP)的实时定量PCR (real-time PCR)方法来鉴定ST75相关的菌株。银白色葡萄球菌可以使用金黄色葡萄球菌的MLST方法进行分型,但是由于基因序列差异较大,使用标准的金黄色葡萄球菌引物时,经常会出现一些问题,尤其是aroE和glpF。2009年,Ruimy等[28]和Ng等[58]分别针对这2个基因位点重新设计引物来完成人体来源CC75相关菌株的MLST。他们也指出:aroE和glpF位点不易扩增可能会导致相关的菌株被误认为是无害的葡萄球菌而未被报道,而之前报道的ST75、ST805、ST883和ST1304在这2个位点上可能是不正确的(表 1,图 2)。另外,Ng等[58]也发现,CC75相关的菌株在基于gap、rpoB、sodA、tuf和hsp60等管家基因构建的系统发育树上与典型的金黄色葡萄球菌存在显著差异。目前来看,Ng等的猜测很可能是对的,因为2009年之后(尤其是银白色葡萄球菌被建立为独立的物种之后),几乎再没有属于这4个ST分离株的报道;银白色葡萄球菌已测序基因组(> 150个)的菌株均不属于这4个ST,也未发现在aroE和glpF位点上与金黄色葡萄球菌一致性 > 97%的基因组。但是,除了aroE和glpF,在其他位点也存在类似问题的ST仍有不少被报道(图 2)。
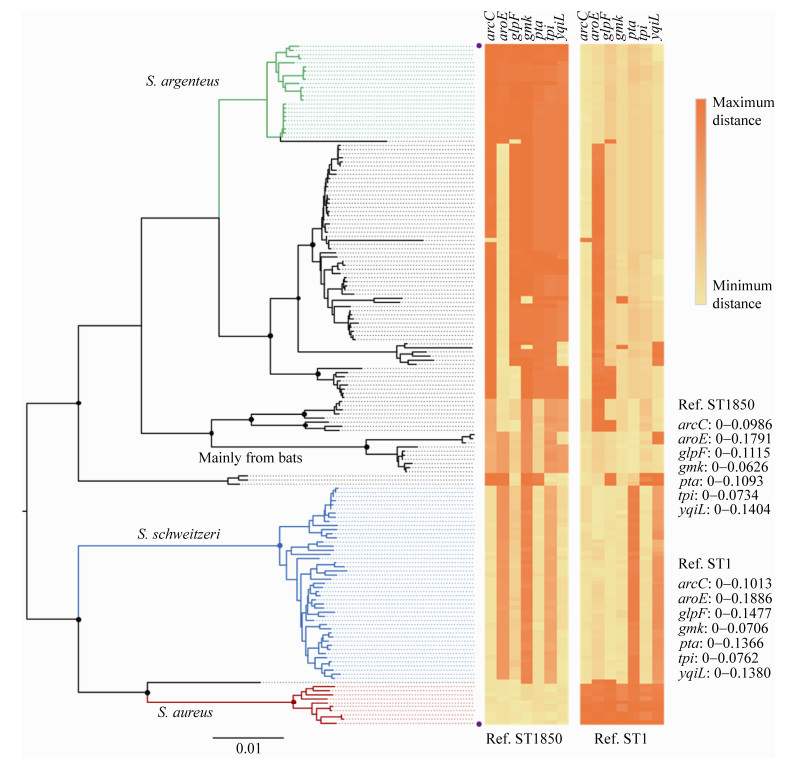
|
| 图 2 银白色葡萄球菌相关ST的多样性及其序列分化 Figure 2 Diversity and divergence of sequence types (ST) associated with S. argenteus. The Neighbor-Joining tree was constructed based on 165 STs downloaded from the S. aureus MLST database (https://pubmlst.org/saureus/), including 10 well-known S. aureus STs (the red clade), 23 confirmed S. argenteus STs (green), 47 confirmed S. schweitzeri STs (blue), 7 STs mainly from bats, and 78 contentious STs containing 1–3 loci closely related to S. aureus (identity > 97%). The STs contain 1–3 loci closely related to S. argenteus and/or S. schweitzeri were not included in the analysis. The Kimura 2-parameter method and bootstrap test (1000 replicates) were employed to infer the evolutionary relationships in MEGA X[24], and the percentage (> 70%) of replicate trees in which the associated taxa clustered together are indicated with black dots on the nodes. The distances of each loci from each ST compared with ST1850 (upper) and ST1 (nether), which are marked with purple dots on the right of the tree tips, are shown on the right. The distances were calculated based on nucleotide sequences using Kimura 2-parameter method in MEGA X[24]. A high-resolution version of this figure with tip labels (STs) is available on request from the author DF Zhang (zdf@hhu.edu.cn). |
2011年,Holt等[59]报道了CC75相关菌株MSHR1132T (=DSM 28299T =CGMCC 1.13854T)的全基因组,基因组分析结果表明:菌株MSHR1132T与典型金黄色葡萄球菌的平均核苷酸分歧值为10%,而金黄色葡萄球菌种内通常 < 2%;附属基因组上的一些遗传元件也出现在MSHR1132T上,如vSaα,vSaβ,SCCmec等;MSHR1132T缺少可以合成黄色色素的操纵子crtOPQMN,含有CRISPR-Cas (clustered regularly interspaced short palindromic repeat-associated system)系统。根据这些结果,Holt等认为MSHR1132T和CC75相关的菌株应该代表一个独立的物种,并建议命名为S. argenteus。2015年,Tong等[4]开展了系统而全面的分类鉴定工作,将CC75相关的类群建立为一个新的物种,正式命名为S. argenteus (argenteus意为silvery)。Tong等[4]选用了6株银白色葡萄球菌、6株施韦策葡萄球菌和18株金黄色葡萄球菌开展生理生化鉴定工作,结果表明这3个物种在底物利用和产水解酶类等方面不存在类群水平的差异,但是种间的ANI (average nucleotide identity)和DDH (DNA-DNA hybridization)值< 95%和 < 70%。显然,银白色葡萄球菌和施韦策葡萄球菌在基因组水平确实应该代表独立的物种。由于除了crtOPQMN和细胞壁肽聚糖类型(见上文)之外未发现其他明显的差异,这3个物种也被统称为S. aureus complex (SAC)[12, 17-19]。Becker等[6]也建议在单独描述银白色葡萄球菌和施韦策葡萄球菌时应该注明,它们是金黄色葡萄球菌复合群成员(member of S. aureus complex)。
银白色葡萄球菌在我国南方地区也有广泛分布,但是除了台湾地区之外,其他城市分离到的菌株均较少(表 2)。Zhang等[23]使用普通PCR方法对一个非核糖体多肽合成酶(nonribosomal peptide synthetase,NRPS)基因上的特异片段进行扩增,通过比较PCR产物片段的大小,成功地从宁波和上海分离的839株“金黄色葡萄球菌”中筛选出6株(0.7%)银白色葡萄球菌。这6株菌是2005–2014年间从食品、人体体表或临床样本中分离获得,属于ST3261 (1株)和ST2250 (5株)。Cao等[69]在研究CRISPR-Cas系统时,报道了一株分离自上海的临床菌株SH3及其基因组序列,随后Zhang等[12]的基因组分析结果表明SH3属于银白色葡萄球菌ST2250。Jiang等[62]报道了2014年重庆地区一名64岁女性髋关节被银白色葡萄球菌反复感染的病例,而导致感染的菌株XNO106 (ST2250)可以像一些金黄色葡萄球菌一样形成小菌落变种(small colony variants,SCV)以提高自身的耐药性。Chen等[66]针对台湾医学中心2010–2012年间的394株菌血症“金黄色葡萄球菌”分离株的回顾性研究表明,47株(11.93%)属于银白色葡萄球菌,均对甲氧西林敏感。与甲氧西林敏感金黄色葡萄球菌(methicillin-susceptible S. aureus,MSSA)相比,银白色葡萄球菌表现出更高比例的继发性多菌感染、血小板减少、下呼吸道感染和短期死亡。Chu等[67]从进行血液透析的病人血液中分离获得的73株菌中,10株属于银白色葡萄球菌。Li等[63]从重庆采集的鸡肉样品中分离得到6株银白色葡萄球菌ST2250菌株,均含有Ⅲ型SCCmec元件。另外,我国其他地区报道的10多个疑似银白色葡萄球菌ST中,均含有1–3个与金黄色葡萄球菌高度相似的等位基因(表 2,图 2)。
2 银白色葡萄球菌的鉴定方法银白色葡萄球菌在表型上明显区别于金黄色葡萄球菌和施韦策葡萄球菌的特点是:目前发现的所有银白色葡萄球菌在常见的培养基上均形成白色菌落。这是因为这些菌株缺少可以合成黄色色素的操纵子crtOPQMN[4, 23, 59]。然而,金黄色葡萄球菌大约也有10%的分离株会形成白色菌落,这通常是因为基因突变或者代谢通路被抑制[23]。由于缺少显著的表型差异,使得银白色葡萄球菌与金黄色葡萄球菌的区分、鉴定无法依靠传统的生理生化方法或相关的自动化鉴定系统来完成。目前,有效的区分、鉴定方法主要是基于基因序列差异的分子生物学手段或者MALDI-TOF MS (matrix-assisted laser desorption/ ionization time-of flight mass spectrometry)。MALDI-TOF MS的方法能否区分银白色葡萄球菌和金黄色葡萄球菌(以及施韦策葡萄球菌),取决于所使用的比对数据库的版本,或者需要检测人员具有足够的经验来识别结果中的特异信号[6, 39, 70]。
虽然,已知的银白色葡萄球菌和部分金黄色葡萄球菌均可以形成白色菌落,但是在筛选银白色葡萄球菌时通过白色菌落这一特点可以排除绝大多数金黄色葡萄球菌和施韦策葡萄球菌。Zhang等[23]根据菌落颜色和一个NRPS基因的特异位点设计了一种“两步法”来区分、鉴定这3个物种。金黄色葡萄球菌在这个NRPS基因的特异区段,相比另外两个物种出现了一段约180 bp的缺失。因此,针对这个区段进行PCR扩增并进行琼脂糖凝胶电泳即可得到以下结果:金黄色葡萄球菌产生一个较短的PCR产物(约160 bp);银白色葡萄球菌和施韦策葡萄球菌产生一个较长的产物(约340 bp)。将菌落颜色和NRPS的PCR产物长短结合起来,即可区分这3个物种:形成黄色菌落、产生较短PCR产物的是金黄色葡萄球菌;形成黄色菌落、产生较长PCR产物的是施韦策葡萄球菌;形成白色菌落、产生较短PCR产物的是金黄色葡萄球菌;形成白色菌落、产生较长PCR产物的是银白色葡萄球菌。需要注意的是,该方法是针对初步鉴定为“金黄色葡萄球菌”的分离株设计,比如经过Baird-Parker琼脂平板筛选出来的菌株[23]。
鉴定银白色葡萄球菌最可靠的方法是进行保守基因的序列测定,比如gap、rpoB、sodA、tuf和hsp60[4, 58, 63],并且通常只需要使用一个基因进行比较分析即可。作为银白色葡萄球菌鉴定的“黄金标准”,MLST不仅可以反映种内多样性,也提供了菌株之间进化关系的基本框架。由于核心基因组上的显著差异,使用标准的金黄色葡萄球菌MLST方法时,银白色葡萄球菌在aroE和glpf (有时也包括yqiL)位点会出现PCR扩增困难的情况,这是因为这些基因的种间变异程度较大(图 2)。Ruimy等[28]、Ng等[58]和Thaipadungpanit等[8]分别针对这2个基因位点单独设计引物来完成CC75相关菌株的MLST。但是在不确定分离株属于哪个物种时,不同物种使用不同的引物就会显得繁琐、低效。Zhang等[23]设计了对SAC 3个物种普遍适用的aroE和glpF引物,简化了涉及多物种时的MLST方法。使用标准引物和PCR反应条件检测nuc1时,部分银白色葡萄球菌和所有的施韦策葡萄球菌会出现无法扩增的情况,但是如果设计出通用引物,nuc1基因测序的方法在理论上也可以区分SAC 3个物种[4, 71]。spa分型也被认为是一种有望区分SAC 3个物种的方法[6],但是尚缺少标准的判断依据,且部分银白色葡萄球菌会出现无法扩增相应片段的情况[43, 68]。另外,基于RT-qPCR的方法也需要更多菌株以进一步验证其可靠性[57, 72]。一些金黄色葡萄球菌检验的国家标准,如GB 4789.10-2016、GB/T 7918.5-1987和GB/T 14926.5-2001,只涉及生理生化特征,理论上也可以将银白色葡萄球菌作为疑似的金黄色葡萄球菌检出,并监测其安全隐患。涉及核酸特征的一些地方标准和行业标准,是否能检出银白色葡萄球菌,还有待进一步验证。
3 银白色葡萄球菌的致病性和毒力基因由于相关的报道较少,银白色葡萄球菌在临床感染、食品安全和畜牧养殖领域的重要性尚不清楚。在早期,研究者认为银白色葡萄球菌的毒力比金黄色葡萄球菌低,临床感染后患者的表现也不同[7, 59, 73]。而最近的一些研究发现,在医院感染、发病率和致死率方面二者并没有明显区别[8, 66]。目前,已发现银白色葡萄球菌可以导致的感染有皮肤和软组织感染(包括化脓、坏死性筋膜炎)[57-59]、骨骼及关节感染[8, 48, 62, 74]、血液感染[7-8, 48, 59]和毒素型食物中毒[10-11]等,可以污染的食品有猪肉、鸡肉、鱼肉和即食食品(表 2),也可以污染食品加工与生产相关的环境[35, 68]或被健康人群携带[28, 43]。从这些方面来看,银白色葡萄球菌和金黄色葡萄球菌并没有明显的区别。
葡萄球菌黄素(staphyloxanthin)是金黄色葡萄球菌表现出黄色表型的主要化合物,由出现在同一个操纵子crtOPQMN上的5个基因和aldH基因参与合成[75]。因为具有抗氧化和抗嗜中性粒细胞作用,抑制葡萄球菌黄素合成通路会导致典型金黄色葡萄球菌毒力的下降[76]。然而将crtOPQMN导入菌株MSHR1132T后,该菌株除了抗氧化能力提高之外,对小鼠的皮肤感染能力并没有显著变化,对兔子防御肽耐受能力和导致心脏内膜炎能力反而减弱[73, 77]。因此,银白色葡萄球菌较弱的致病性并不仅仅是因为缺少葡萄球菌黄素,很可能还涉及其他方面的机制。
Zhang等[12]通过比较基因组学分析对银白色葡萄球菌的毒力基因进行预测,结果发现在111个金黄色葡萄球菌毒力基因中,85个(76.6%)出现在银白色葡萄球菌中,56个(50.5%)基因的种间核苷酸一致性表现出显著差异(P < 0.01)。对金黄色葡萄球菌非常重要的一些毒力基因(簇)也出现在银白色葡萄球菌的基因组上,比如:对菌膜形成非常重要的icaA-D;编码Ⅶ型分泌系统的esaA-C、essA-C和esxAB;吸附血红素相关基因isdA-G和srtB;基因岛νSaα和νSaβ;通常携带毒力基因和耐药基因的原噬菌体;以及溶血素、荚膜多糖和葡萄球菌肠毒素(staphylococcal enterotoxins,SE)等相关基因[6, 9, 12]。已知的27种SE (也被称为葡萄球菌超抗原)基因已有12种出现在银白色葡萄球菌中[9, 19]。早期的分离株均不携带杀白细胞毒素(pantone-valentine leucocidin,PVL)基因,而后来发现的部分菌株却含有相关基因[7, 27, 36-37, 48] (表 1)。其他大部分尚未在银白色葡萄球菌中发现的毒力基因,通常是出现在金黄色葡萄球菌的可移动元件上,且很容易获得或丢失(主要是SE和其他外毒素基因)。而基因组分析也表明了一些可移动元件在SAC物种间转移的可能性,如CRISPR-Cas系统、SCCmec元件、原噬菌体等[12, 47, 68]。
从已有的数据来看,银白色葡萄球菌的耐药性整体比金黄色葡萄球菌弱,除了blaZ介导的盘尼西林耐药性之外,其他耐药性比率较低,如四环素、氨基糖苷类、克林霉素和红霉素等[7, 36, 48, 57, 66]。在甲氧西林耐药性方面,除了从MRSA中筛选得到的菌株外,耐甲氧西林银白色葡萄球菌的比率并不高(表 1)。已发现银白色葡萄球菌含有的SCCmec元件主要类型为Ⅳ型[46-47, 51, 57-78],少量菌株含有Ⅱ型[52]、Ⅲ型[63]或Ⅴ/Ⅶ型[51, 68]。
4 银白色葡萄球菌的原始生境病原菌溯源对了解其致病性形成、传播过程以及防控措施的制定至关重要。银白色葡萄球菌的原始生境是后续相关研究急需要解答的重要问题之一。除了东南亚和澳大利亚,其他地区分离得到的银白色葡萄球菌比率均较低,绝大多数报道只涉及个位数的菌株[9]。即使2015年之后,一些研究者已经注意到这种病原体,这种情况依然没有改变。值得注意的是,目前已报道的分离株主要来自临床样品或人体携带情况调查(表 1)。因此,很可能银白色葡萄球菌的原始生境并不是人体相关的环境[9],它也不是一个“古老”的人类病原菌。一些学者认为,银白色葡萄球菌很可能是来源于家畜相关的环境,其主要的谱系ST1223和ST2250是经过宿主适应性进化才具备感染人类的能力[9, 12-13]。支持性的数据主要来自对ST2250的研究。首先,sel26和sel27 (编码SEl26和SEl27)出现在所有的ST2250菌株基因组上,但是在金黄色葡萄球菌中出现频率很低(3/248);Zhang等筛选到的3株菌均分离自生牛乳,而不同SAC物种来源的SEl26和SEl27对不同宿主的活性也存在差异[19]。其次,普遍存在于ST2250基因组上的CRISPR-Cas系统、四环素耐药基因tet (L)和重金属抗性基因,也主要存在于家畜相关的金黄色葡萄球菌中[13]。再次,主要分离自绵羊和山羊的金黄色葡萄球菌厌氧亚种(S. aureus subsp. anaerobius)也缺少crtOPQMN[9]。另外,Moradigaravand等[13]使用分子钟推算结果表明,银白色葡萄球菌ST2250大约是在15年前传入泰国。
另外,一些蝙蝠来源的菌株被鉴定为银白色葡萄球菌,这些ST非常稀有,与其他银白色葡萄球菌的ST型相关,但是存在较大差异[18] (图 2)。银白色葡萄球菌的传播和起源是否与蝙蝠有关还需要进一步研究。但是,这从另外一个方面表明,野生动物相关的环境可能存在着更加多样的SAC类群。
5 总结和展望银白色葡萄球菌是2015年正式被建立的病原新物种,与金黄色葡萄球菌共用同一个MLST数据库,属于金黄色葡萄球菌复合群。该物种呈现出全球分布局势,已在我国10多个城市被分离培养。因为被正式命名的时间较晚,并且与金黄色葡萄球菌缺少足够的差异,银白色葡萄球菌尚未被广泛认识。分子水平的差异可能导致大量相关菌株被鉴定为金黄色葡萄球菌或其他细菌,而被错误地报道或未被报道。这些问题导致现有银白色葡萄球菌的文献资料较少,人们对这个类群的认识也非常有限。
银白色葡萄球菌可以导致多种人体感染,可以污染食品,也可以被健康人群携带,这些表现均与金黄色葡萄球菌类似,但是其耐药性偏弱。因此,ESCNID认为[6],在医疗卫生行业的实践中没有必要将二者进行区分。但是在基础研究领域,比如分子流行病学调查,有必要将它们明确区分,以尽快摸清其分布与传播规律。涉及分子水平的一些实验方法,也需要尽快升级换代,以满足SAC不同物种的诊断和研究需求。我国对银白色葡萄球菌的研究已经获得了一些结果,但是非常有限,尤其是临床感染方面的研究较少。泰国和我国台湾地区已经发现了一定规模的银白色葡萄球菌感染,我国大陆地区(尤其是南方)可能也存在着一定程度的流行,因此有必要对银白色葡萄球菌开展进一步的调查研究。而银白色葡萄球菌的起源和原始生境问题,则需要不同国家地区、不同领域的微生物学者共同努力才能解答。
| [1] | Schleifer KH, Bell JA, Staphylococcus Rosenbach 1884, 18AL (Nom. Cons. Opin. 17 Jud. Comm. 1958, 153.)//Vos PD, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editor. Bergey's Manual of Systematic Bacteriology. second edition (Volume Three). New York: Springer, 2009. |
| [2] | Wertheim HF, Melles DC, Vos MC, Van Leeuwen W, Van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. The Lancet Infectious Diseases, 2005, 5(12): 751-762. DOI:10.1016/S1473-3099(05)70295-4 |
| [3] | Hennekinne JA, De Buyser ML, Dragacci S. Staphylococcus aureus and its food poisoning toxins:characterization and outbreak investigation. FEMS Microbiology Reviews, 2012, 36(4): 815-836. DOI:10.1111/j.1574-6976.2011.00311.x |
| [4] | Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(1): 15-22. |
| [5] | Munson E, Carroll KC. What's in a name? New bacterial species and changes to taxonomic status from 2012 through 2015. Journal of Clinical Microbiology, 2017, 55(1): 24-42. DOI:10.1128/JCM.01379-16 |
| [6] | Becker K, Schaumburg F, Kearns A, Larsen AR, Lindsay JA, Skov RL, Westh H. Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri:a position paper of members of the ESCMID Study Group for Staphylococci and Staphylococcal Diseases (ESGS). Clinical Microbiology and Infection, 2019, 25(9): 1064-1070. DOI:10.1016/j.cmi.2019.02.028 |
| [7] | Chantratita N, Wikraiphat C, Tandhavanant S, Wongsuvan G, Ariyaprasert P, Suntornsut P, Thaipadungpanit J, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Anukunananchai J, Phiphitaporn S, Srisamang P, Chetchotisakd P, West TE, Peacock SJ. Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand:a prospective multicentre observational study. Clinical Microbiology and Infection, 2016, 22(5): 458.e11-458.e19. DOI:10.1016/j.cmi.2016.01.008 |
| [8] | Thaipadungpanit J, Amornchai P, Nickerson EK, Wongsuvan G, Wuthiekanun V, Limmathurotsakul D, Peacock SJ. Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. Journal of Clinical Microbiology, 2015, 53(3): 1005-1008. DOI:10.1128/JCM.03049-14 |
| [9] | Shi XM, Zhang DF. Staphylococcus argenteus:an emerging foodborne pathogen?. Current Opinion in Food Science, 2018, 20: 76-81. DOI:10.1016/j.cofs.2018.03.015 |
| [10] | Suzuki Y, Kubota H, Ono HK, Kobayashi M, Murauchi K, Kato R, Hirai A, Sadamasu K. Food poisoning outbreak in Tokyo, Japan caused by Staphylococcus argenteus. International Journal of Food Microbiology, 2017, 262: 31-37. DOI:10.1016/j.ijfoodmicro.2017.09.005 |
| [11] | Wakabayashi Y, Umeda K, Yonogi S, Nakamura H, Yamamoto K, Kumeda Y, Kawatsu K. Staphylococcal food poisoning caused by Staphylococcus argenteus harboring staphylococcal enterotoxin genes. International Journal of Food Microbiology, 2018, 265: 23-29. DOI:10.1016/j.ijfoodmicro.2017.10.022 |
| [12] | Zhang DF, Zhi XY, Zhang J, Paoli GC, Cui Y, Shi CL, Shi XM. Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus. BMC Genomics, 2017, 18(1): 808. DOI:10.1186/s12864-017-4149-9 |
| [13] | Moradigaravand D, Jamrozy D, Mostowy R, Anderson A, Nickerson EK, Thaipadungpanit J, Wuthiekanun V, Limmathurotsakul D, Tandhavanant S, Wikraiphat C, Wongsuvan G, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Eoin West T, Blane B, Parkhill J, Chantratita N, Peacock SJ. Evolution of the Staphylococcus argenteus ST2250 clone in Northeastern Thailand is linked with the acquisition of livestock-associated staphylococcal genes. mBio, 2017, 8(4): e00802-17. |
| [14] | Schaumburg F, Alabi AS, Köck R, Mellmann A, Kremsner PG, Boesch C, Becker K, Leendertz FH, Peters G. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environmental Microbiology Report, 2012, 4(1): 141-146. DOI:10.1111/j.1758-2229.2011.00316.x |
| [15] | Schaumburg F, Pauly M, Anoh E, Mossoun A, Wiersma L, Schubert G, Flammen A, Alabi AS, Muyembe-Tamfum JJ, Grobusch MP, Karhemere S, Akoua-Koffi C, Couacy-Hymann E, Kremsner PG, Mellmann A, Becker K, Leendertz FH, Peters G. Staphylococcus aureus complex from animals and humans in three remote African regions. Clinical Microbiology and Infection, 2015, 21(4): 345.e1-345.e8. DOI:10.1016/j.cmi.2014.12.001 |
| [16] | Akobi B, Aboderin O, Sasaki T, Shittu A. Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiology, 2012, 12: 279. DOI:10.1186/1471-2180-12-279 |
| [17] | Held J, Gmeiner M, Mordmuller B, Matsiégui PB, Schaer J, Eckerle I, Weber N, Matuschewski K, Bletz S, Schaumburg F. Bats are rare reservoirs of Staphylococcus aureus complex in Gabon. Infection, Genetics and Evolution, 2017, 47: 118-120. DOI:10.1016/j.meegid.2016.11.022 |
| [18] | Olatimehin A, Shittu AO, Onwugamba FC, Mellmann A, Becker K, Schaumburg F. Staphylococcus aureus complex in the straw-colored fruit bat (Eidolon helvum) in Nigeria. Frontiers in Microbiology, 2018, 9: 162. DOI:10.3389/fmicb.2018.00162 |
| [19] | Zhang DF, Yang XY, Zhang J, Qin XJ, Huang XZ, Cui Y, Zhou M, Shi CL, French NP, Shi XM. Identification and characterization of two novel superantigens among Staphylococcus aureus complex. International Journal of Medical Microbiology, 2018, 308(4): 438-446. DOI:10.1016/j.ijmm.2018.03.002 |
| [20] | Johansson C, Rautelin H, Kaden R. Staphylococcus argenteus and Staphylococcus schweitzeri are cytotoxic to human cells in vitro due to high expression of alpha-hemolysin Hla. Virulence, 2019, 10(1): 502-510. DOI:10.1080/21505594.2019.1620062 |
| [21] | Ngoa UA, Schaumburg F, Adegnika AA, Kösters K, Möller T, Fernandes JF, Alabi A, Issifou S, Becker K, Grobusch MP, Kremsner PG, Lell B. Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Tropica, 2012, 124(1): 42-47. DOI:10.1016/j.actatropica.2012.06.005 |
| [22] | Okuda KV, Toepfner N, Alabi AS, Arnold B, Bélard S, Falke U, Menschner L, Monecke S, Ruppelt-Lorz A, Berner R. Molecular epidemiology of Staphylococcus aureus from Lambaréné, Gabon. European Journal of Clinical Microbiology & Infectious Diseases, 2016, 35(12): 1963-1973. |
| [23] | Zhang DF, Xu XB, Song QF, Bai YL, Zhang Y, Song MH, Shi CL, Shi XM. Identification of Staphylococcus argenteus in eastern China based on a nonribosomal peptide synthetase (NRPS) gene. Future Microbiology, 2016, 11(9): 1113-1121. DOI:10.2217/fmb-2016-0017 |
| [24] | Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096 |
| [25] | Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology, 2000, 38(3): 1008-1015. DOI:10.1128/JCM.38.3.1008-1015.2000 |
| [26] | Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitorn C, Ito T, Hiramatsu K. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. Journal of Clinical Microbiology, 2002, 40(11): 4289-4294. DOI:10.1128/JCM.40.11.4289-4294.2002 |
| [27] | Senok A, Nassar R, Kaklamanos EG, Belhoul K, Abu Fanas S, Nassar M, Azar AJ, Müller E, Reissig A, Gawlik D, Monecke S, Ehricht R. Molecular characterization of Staphylococcus aureus isolates associated with nasal colonization and environmental contamination in academic dental clinics. Microbial Drug Resistance, 2020, 26(6): 661-669. DOI:10.1089/mdr.2019.0318 |
| [28] | Ruimy R, Armand-Lefevre L, Barbier F, Ruppé E, Cocojaru R, Mesli Y, Maiga A, Benkalfat M, Benchouk S, Hassaine H, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Feil EJ. Comparisons between geographically diverse samples of carried Staphylococcus aureus. Journal of Bacteriology, 2009, 191(18): 5577-5583. DOI:10.1128/JB.00493-09 |
| [29] | Rajan V, Schoenfelder SMK, Ziebuhr W, Gopal S. Genotyping of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) in a tertiary care centre in Mysore, South India:ST2371-SCCmec Ⅳ emerges as the major clone. Infection, Genetics and Evolution, 2015, 34: 230-235. DOI:10.1016/j.meegid.2015.05.032 |
| [30] | Kitagawa H, Ohge H, Hisatsune J, Masuda K, Aziz F, Hara T, Kuroo Y, Sugai M. Low incidence of Staphylococcus argenteus bacteremia in Hiroshima, Japan. Journal of Infection and Chemotherapy, 2020, 26(1): 140-143. DOI:10.1016/j.jiac.2019.07.011 |
| [31] | Ohnishi T, Shinjoh M, Ohara H, Kawai T, Kamimaki I, Mizushima R, Kamada K, Itakura Y, Iguchi S, Uzawa Y, Yoshida A, Kikuchi K. Purulent lymphadenitis caused by Staphylococcus argenteus, representing the first Japanese case of Staphylococcus argenteus (multilocus sequence type 2250) infection in a 12-year-old boy. Journal of Infection and Chemotherapy, 2018, 24(11): 925-927. DOI:10.1016/j.jiac.2018.03.018 |
| [32] | Aung MS, Urushibara N, Kawaguchiya M, Sumi A, Takahashi S, Ike M, Ito M, Habadera S, Kobayashi N. Molecular epidemiological characterization of Staphylococcus argenteus clinical isolates in Japan:identification of three clones (ST1223, ST2198, and ST2550) and a novel staphylocoagulase genotype XV. Microorganisms, 2019, 7(10): 389. DOI:10.3390/microorganisms7100389 |
| [33] | Yeap AD, Woods K, Dance DAB, Pichon B, Rattanavong S, Davong V, Phetsouvanh R, Newton PN, Shetty N, Kearns AM. Molecular epidemiology of Staphylococcus aureus skin and soft tissue infections in the Lao People's Democratic Republic. The American Journal of Tropical Medicine and Hygiene, 2017, 97(2): 423-428. DOI:10.4269/ajtmh.16-0746 |
| [34] | Aung MS, San T, San N, Oo WM, Ko PM, Thet KT, Urushibara N, Kawaguchiya M, Sumi A, Kobayashi N. Molecular characterization of Staphylococcus argenteus in Myanmar:identification of novel genotypes/clusters in staphylocoagulase, protein A, alpha-haemolysin and other virulence factors. Journal of Medical Microbiology, 2019, 68(1): 95-104. DOI:10.1099/jmm.0.000869 |
| [35] | Aung MS, San T, Aye MM, Mya S, Maw WW, Zan KN, Htut WHW, Kawaguchiya M, Urushibara N, Kobayashi N. Prevalence and genetic characteristics of Staphylococcus aureus and Staphylococcus argenteus isolates harboring Panton-Valentine Leukocidin, enterotoxins, and TSST-1 genes from food handlers in Myanmar. Toxins, 2017, 9(8): 241. DOI:10.3390/toxins9080241 |
| [36] | Indrawattana N, Pumipuntu N, Suriyakhun N, Jangsangthong A, Kulpeanprasit S, Chantratita N, Sookrung N, Chaicumpa W, Buranasinsup S. Staphylococcus argenteus from rabbits in Thailand. Microbiologyopen, 2019, 8(4): e00665. DOI:10.1002/mbo3.665 |
| [37] | Pumipuntu N, Tunyong W, Chantratita N, Diraphat P, Pumirat P, Sookrung N, Chaicumpa W, Indrawattana N. Staphylococcus spp. associated with subclinical bovine mastitis in central and northeast provinces of Thailand. PeerJ, 2019, 7(1): e6587. |
| [38] | Piewngam P, Zheng Y, Nguyen TH, Dickey SW, Joo HS, Villaruz AE, Glose KA, Fisher EL, Hunt RL, Li B, Chiou J, Pharkjaksu S, Khongthong S, Cheung GYC, Kiratisin P, Otto M. Pathogen elimination by probiotic Bacillus via signalling interference. Nature, 2018, 562(7728): 532-537. DOI:10.1038/s41586-018-0616-y |
| [39] | Schuster D, Rickmeyer J, Gajdiss M, Thye T, Lorenzen S, Reif M, Josten M, Szekat C, Melo LDR, Schmithausen RM, Liégeois F, Sahl HG, Gonzalez JPJ, Nagel M, Bierbaum G. Differentiation of Staphylococcus argenteus (formerly:Staphylococcus aureus clonal complex 75) by mass spectrometry from S. aureus using the first strain isolated from a wild African great ape. International Journal of Medical Microbiology, 2017, 307(1): 57-63. DOI:10.1016/j.ijmm.2016.11.003 |
| [40] | Schaumburg F, Alabi AS, Mombo-Ngoma G, Kaba H, Zoleko RM, Diop DA, Mackanga JR, Basra A, Gonzalez R, Menendez C, Grobusch MP, Kremsner PG, Köck R, Peters G, Ramharter M, Becker K. Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clinical Microbiology and Infection, 2014, 20(6): O390-O396. DOI:10.1111/1469-0691.12417 |
| [41] | Gharsa H, Ben Slama K, Lozano C, Gómez-Sanz E, Klibi N, Ben Sallem R, Gómez P, Zarazaga M, Boudabous A, Torres C. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Veterinary Microbiology, 2012, 156(3/4): 367-373. |
| [42] | Uhlemann AC, Dumortier C, Hafer C, Taylor BS, Sánchez J, Rodriguez-Taveras C, Leon P, Rojas R, Olive C, Lowy FD. Molecular characterization of Staphylococcus aureus from outpatients in the Caribbean reveals the presence of pandemic clones. European Journal of Clinical Microbiology & Infectious Diseases, 2012, 31(4): 505-511. |
| [43] | Ruimy R, Angebault C, Djossou F, Dupont C, Epelboin L, Jarraud S, Armand Lefevre L, Bes M, Lixandru BE, Bertine M, El Miniai A, Renard M, Bettinger RM, Lescat M, Clermont O, Peroz G, Lina G, Tavakol M, Vandenesch F, Van Belkum A, Rousset F, Andremont A. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans?. The Journal of Infectious Diseases, 2010, 202(6): 924-934. DOI:10.1086/655901 |
| [44] | Monecke S, Stieber B, Roberts R, Akpaka PE, Slickers P, Ehricht R. Population structure of Staphylococcus aureus from Trinidad & Tobago. PLoS One, 2014, 9(2): e89120. DOI:10.1371/journal.pone.0089120 |
| [45] | Neyaz L, Karki AB, Fakhr MK. The whole-genome sequence of plasmid-bearing Staphylococcus argenteus strain B3-25B from retail beef liver encodes the type Ⅶ secretion system and several virulence factors. Microbiology Resource Announcements, 2019, 8(45): e00962-19. |
| [46] | Argudín MA, Dodémont M, Vandendriessche S, Rottiers S, Tribes C, Roisin S, De Mendonça R, Nonhoff C, Deplano A, Denis O. Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. European Journal of Clinical Microbiology & Infectious Diseases, 2016, 35(6): 1017-1022. |
| [47] | Hansen TA, Bartels MD, Høgh SV, Dons LE, Pedersen M, Jensen TG, Kemp M, Skov MN, Gumpert H, Worning P, Westh H. Whole genome sequencing of Danish Staphylococcus argenteus reveals a genetically diverse collection with clear separation from Staphylococcus aureus. Frontiers in Microbiology, 2017, 8: 1512. DOI:10.3389/fmicb.2017.01512 |
| [48] | Dupieux C, Blondé R, Bouchiat C, Meugnier H, Bes M, Laurent S, Vandenesch F, Laurent F, Tristan A. Community-acquired infections due to Staphylococcus argenteus lineage isolates harbouring the Panton-Valentine leucocidin, France, 2014. Eurosurveillance, 2015, 20(23): 21154. |
| [49] | Monecke S, Müller E, Buechler J, Rejman J, Stieber B, Akpaka PE, Bandt D, Burris R, Coombs G, Hidalgo-Arroyo GA, Hughes P, Kearns A, Abós SM, Pichon B, Skakni L, Söderquist B, Ehricht R. Rapid detection of Panton-Valentine leukocidin in Staphylococcus aureus cultures by use of a lateral flow assay based on monoclonal antibodies. Journal of Clinical Microbiology, 2013, 51(2): 487-495. DOI:10.1128/JCM.02285-12 |
| [50] | Tunsjo HS, Kalyanasundaram S, Charnock C, Leegaard TM, Moen AEF. Challenges in the identification of methicillin-resistant Staphylococcus argenteus by routine diagnostics. APMIS, 2018, 126(6): 533-537. DOI:10.1111/apm.12843 |
| [51] | Giske CG, Dyrkell F, Arnellos D, Vestberg N, Hermansson Panna S, Fröding I, Ullberg M, Fang H. Transmission events and antimicrobial susceptibilities of methicillin-resistant Staphylococcus argenteus in Stockholm. Clinical Microbiology and Infection, 2019, 25(10): 1289.e5-1289.e8. DOI:10.1016/j.cmi.2019.06.003 |
| [52] | Kaden R, Engstrand L, Rautelin H, Johansson C. Which methods are appropriate for the detection of Staphylococcus argenteus and is it worthwhile to distinguish S. argenteus from S. aureus?. Infection and Drug Resistance, 2018, 11: 2335-2344. DOI:10.2147/IDR.S179390 |
| [53] | Tång Hallbäck E, Karami N, Adlerberth I, Cardew S, Ohlén M, Engström Jakobsson H, Svensson Stadler L. Methicillin-resistant Staphylococcus argenteus misidentified as methicillin-resistant Staphylococcus aureus emerging in western Sweden. Journal of Medical Microbiology, 2018, 67(7): 968-971. DOI:10.1099/jmm.0.000760 |
| [54] | Coombs GW, Monecke S, Pearson JC, Tan HL, Chew YK, Wilson L, Ehricht R, O'Brien FG, Christiansen KJ. Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiology, 2011, 11: 215. DOI:10.1186/1471-2180-11-215 |
| [55] | Ritchie SR, Thomas MG, Rainey PB. The genetic structure of Staphylococcus aureus populations from the Southwest Pacific. PLoS One, 2014, 9(7): e100300. DOI:10.1371/journal.pone.0100300 |
| [56] | Jenney A, Holt D, Ritika R, Southwell P, Pravin S, Buadromo E, Carapetis J, Tong S, Steer A. The clinical and molecular epidemiology of Staphylococcus aureus infections in Fiji. BMC Infectious Diseases, 2014, 14: 160. DOI:10.1186/1471-2334-14-160 |
| [57] | McDonald M, Dougall A, Holt D, Huygens F, Oppedisano F, Giffard PM, Inman-Bamber J, Stephens AJ, Towers R, Carapetis JR, Currie BJ. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote Aboriginal communities. Journal of Clinical Microbiology, 2006, 44(10): 3720-3727. DOI:10.1128/JCM.00836-06 |
| [58] | Ng JWS, Holt DC, Lilliebridge RA, Stephens AJ, Huygens F, Tong SYC, Currie BJ, Giffard PM. Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. Journal of Clinical Microbiology, 2009, 47(7): 2295-2300. DOI:10.1128/JCM.00122-09 |
| [59] | Holt DC, Holden MTG, Tong SYC, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biology and Evolution, 2011, 3: 881-895. DOI:10.1093/gbe/evr078 |
| [60] | Yang XJ, Yu SB, Wu QP, Zhang JM, Wu S, Rong DL. Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Frontiers in Microbiology, 2018, 9: 197. DOI:10.3389/fmicb.2018.00197 |
| [61] | Rong DL, Wu QP, Xu MF, Zhang JM, Yu SB. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Staphylococcus aureus from retail aquatic products in China. Frontiers in Microbiology, 2017, 8: 714. DOI:10.3389/fmicb.2017.00714 |
| [62] | Jiang B, You B, Tan L, Yu SP, Li H, Bai GQ, Li S, Rao XC, Xie Z, Shi XM, Peng YZ, Hu XM. Clinical Staphylococcus argenteus develops to small colony variants to promote persistent infection. Frontiers in Microbiology, 2018, 9: 1347. DOI:10.3389/fmicb.2018.01347 |
| [63] | Li QC, Li Y, Tang YY, Meng C, Ingmer H, Jiao XN. Prevalence and characterization of Staphylococcus aureus and Staphylococcus argenteus in chicken from retail markets in China. Food Control, 2019, 96: 158-164. DOI:10.1016/j.foodcont.2018.08.030 |
| [64] | Li XH, Huang T, Xu K, Li CL, Li YR. Molecular characteristics and virulence gene profiles of Staphylococcus aureus isolates in Hainan, China. BMC Infectious Diseases, 2019, 19(1): 873. DOI:10.1186/s12879-019-4547-5 |
| [65] | He L, Zheng HX, Wang YN, Le KY, Liu Q, Shang J, Dai YX, Meng HW, Wang X, Li TM, Gao QQ, Qin JX, Lu HY, Otto M, Li M. Detection and analysis of methicillin-resistant human-adapted sequence type 398 allows insight into community-associated methicillin-resistant Staphylococcus aureus evolution. Genome Medicine, 2018, 10(1): 5. DOI:10.1186/s13073-018-0514-9 |
| [66] | Chen SY, Lee H, Wang XM, Lee TF, Liao CH, Teng LJ, Hsueh PR. High mortality impact of Staphylococcus argenteus on patients with community-onset staphylococcal bacteraemia. International Journal of Antimicrobial Agents, 2018, 52(6): 747-753. DOI:10.1016/j.ijantimicag.2018.08.017 |
| [67] | Chu C, Wong MY, Tseng YH, Lin CL, Tung CW, Kao CC, Huang YK. Vascular access infection by Staphylococcus aureus from removed dialysis accesses. MicrobiologyOpen, 2019, 8(8): e00800. |
| [68] | Zhou WY, Li XH, Shi L, Wang HH, Yan H. Novel SCCmec type XⅡ methicillin-resistant Staphylococcus aureus isolates identified from a swine production and processing chain. Veterinary Microbiology, 2018, 225: 105-113. DOI:10.1016/j.vetmic.2018.09.007 |
| [69] | Cao LY, Gao CH, Zhu JD, Zhao LP, Wu QF, Li M, Sun BL. Identification and functional study of type Ⅲ-A CRISPR-Cas systems in clinical isolates of Staphylococcus aureus. International Journal of Medical Microbiology, 2016, 306(8): 686-696. DOI:10.1016/j.ijmm.2016.08.005 |
| [70] | Chen SY, Lee H, Teng SH, Wang XM, Lee TF, Huang YC, Liao CH, Teng LJ, Hsueh PR. Accurate differentiation of novel Staphylococcus argenteus from Staphylococcus aureus using MALDI-TOF MS. Future Microbiology, 2018, 13(9): 997-1006. DOI:10.2217/fmb-2018-0015 |
| [71] | Schaumburg F, Pauly M, Schubert G, Shittu A, Tong S, Leendertz F, Peters G, Becker K. Characterization of a novel thermostable nuclease homolog (NucM) in a highly divergent Staphylococcus aureus clade. Journal of Clinical Microbiology, 2014, 52(11): 4036-4038. DOI:10.1128/JCM.02327-14 |
| [72] | Bogestam K, Vondracek M, Karlsson M, Fang H, Giske CG. Introduction of a hydrolysis probe PCR assay for high-throughput screening of methicillin-resistant Staphylococcus aureus with the ability to include or exclude detection of Staphylococcus argenteus. PLoS One, 2018, 13(2): e0192782. DOI:10.1371/journal.pone.0192782 |
| [73] | Tong SY, Sharma-Kuinkel BK, Thaden JT, Whitney AR, Yang SJ, Mishra NN, Rude T, Lilliebridge RA, Selim MA, Ahn SH, Holt DC, Giffard PM, Bayer AS, Deleo FR, Fowler Jr VG. Virulence of endemic nonpigmented northern Australian Staphylococcus aureus clone (clonal complex 75, S. argenteus) is not augmented by staphyloxanthin. The Journal of Infectious Diseases, 2013, 208(3): 520-527. DOI:10.1093/infdis/jit173 |
| [74] | Rigaill J, Grattard F, Grange S, Forest F, Haddad E, Carricajo A, Tristan A, Laurent F, Botelho-Nevers E, Verhoeven PO. Community-acquired Staphylococcus argenteus sequence type 2250 bone and joint infection, France, 2017. Emerging Infectious Diseases, 2018, 24(10): 1958-1961. DOI:10.3201/eid2410.180727 |
| [75] | Kim SH, Lee PC. Functional expression and extension of staphylococcal staphyloxanthin biosynthetic pathway in Escherichia coli. Journal of Biological Chemistry, 2012, 287(26): 21575-21583. DOI:10.1074/jbc.M112.343020 |
| [76] |
Chen FF, Di HX, Lan LF. Small molecules targeting Staphylococcus aureus virulence. Chinese Science Bulletin, 2013, 58(36): 3743-3752.
(in Chinese) 陈菲菲, 狄红霞, 蓝乐夫. 金黄色葡萄球菌重要毒力因子的功能及其抑制剂研究进展. 科学通报, 2013, 58(36): 3743-3752. |
| [77] | Xiong YQ, Yang SJ, Tong SYC, Alvarez DN, Mishra NN. The role of Staphylococcal carotenogenesis in resistance to host defense peptides and in vivo virulence in experimental endocarditis model. Pathogens and Disease, 2015, 73(3): ftv056. |
| [78] | Monecke S, Kanig H, Rudolph W, Müller E, Coombs G, Hotzel H, Slickers P, Ehricht R. Characterisation of Australian MRSA strains ST75- and ST883-MRSA-Ⅳ and analysis of their accessory gene regulator locus. PLoS One, 2010, 5(11): e14025. DOI:10.1371/journal.pone.0014025 |
 2021, Vol. 61
2021, Vol. 61