中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 孙茂文, 王涛, 孙元, 杨玉莹, 仇华吉. 2021
- Maowen Sun, Tao Wang, Yuan Sun, Yuying Yang, Hua-Ji Qiu. 2021
- 非洲猪瘟病毒的免疫逃逸策略
- Immunoevasion strategies of African swine fever virus
- 微生物学报, 61(2): 249-262
- Acta Microbiologica Sinica, 61(2): 249-262
-
文章历史
- 收稿日期:2020-03-19
- 修回日期:2020-05-21
- 网络出版日期:2020-07-31
2. 中国农业科学院哈尔滨兽医研究所兽医生物技术国家重点实验室, 黑龙江 哈尔滨 150069
2. State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, Heilongjiang Province, China
非洲猪瘟(African swine fever,ASF)是由非洲猪瘟相关病毒科非洲猪瘟病毒属的成员——非洲猪瘟病毒(African swine fever virus,ASFV)引起的一种急性、热性、广泛出血性的高度接触传染性疾病。家猪和野猪均易感,强毒株致死率可达100%,自然弱毒株呈亚临床或慢性感染,并可对部分强毒株攻击提供一定的保护[1]。上世纪20年代初ASF在肯尼亚首次被发现,此后主要在撒哈拉以南的非洲地区流行。上世纪中叶传入欧洲,随后传至南美洲地区。2007年格鲁吉亚暴发ASF并迅速波及俄罗斯、立陶宛等多个欧洲国家。2018年8月初,辽宁某猪场暴发我国首例ASF疫情[2],这也是亚洲的首例报道。截至目前,我国31个省、市、自治区共计暴发176例ASF疫情(http://www.moa.gov.cn/gk/yjgl_1/yqfb/),给我国养猪业造成巨大经济损失。
ASF被世界动物卫生组织列为必须报告的动物疫病,我国将其列为一类动物传染病[3]。ASF被发现至今近一个世纪,无商品化疫苗和有效的治疗性药物,仅部分国家和地区采取严格的生物安全措施根除了此病[4]。ASFV生物学特性复杂,编码部分复制非必需蛋白参与免疫逃逸。由于ASFV大部分基因功能未知,制约了疫苗研发和配套检测技术的发展。本文对ASFV逃逸天然免疫和适应性免疫应答等方面进行了总结,着重讨论了ASFV免疫逃逸基因及其编码蛋白的功能,旨在加深对ASFV免疫逃逸策略的认识,为ASFV的致病机制与疫苗研发提供思路。
1 非洲猪瘟病毒概述ASFV曾先后被归为虹彩病毒科和痘病毒科。根据DNA序列分析、病毒结构与复制方式的差异,国际病毒学分类委员会(International Committee on Taxonomy of Viruses,ICTV)将ASFV列为非洲猪瘟相关病毒科非洲猪瘟病毒属,是目前已知的唯一虫媒DNA病毒[5]。ASFV病毒粒子直径约260–300 nm,是有囊膜的二十面体线性双链DNA(Double-stranded DNA,dsDNA)病毒,属于核质大DNA病毒(Nucleo-cytoplasmic large DNA viruses,NCLDV)超家族[6]。ASFV成熟病毒粒子自内向外分别是基因组、核心壳、内膜、衣壳和囊膜,基因组长170–194 kb[5],其长度差异主要源于多基因家族(Multigene families,MGFs)基因拷贝数的变化。ASFV编码54种结构蛋白和100多种非结构蛋白[3],参与病毒基因组的复制、DNA修复、转录、病毒组装及免疫逃逸等。根据B646L基因核苷酸序列差异已发现ASFV有24种基因型[7]。格鲁吉亚、俄罗斯、中国、东南亚和东欧地区流行的ASFV毒株主要是基因Ⅱ型,其他基因型主要流行于非洲和南美洲等。
家猪、野猪和钝缘蜱是ASFV的天然宿主。疣猪、钝缘蜱等自然宿主感染后无明显临床表现,是本病的传播媒介之一。ASFV通过水平传播,暂未证实可经垂直传播[8]。水平传播主要是通过直接接触和间接接触:(1)直接接触,病猪/带毒猪或康复猪作为病原携带者与易感猪接触,例如舔舐、同槽采食和饮水(最低感染剂量1 TCID50)等[9-10]。(2)间接接触,软蜱、受污染的饲料、2 m之内的气溶胶[11]、猪肉产品、运输车辆或人员流动等。虽然目前未证实ASFV可在硬蜱体内复制或作为传播媒介,但猪误食接触ASFV污染血液的苍蝇也可能会造成感染[12]。鉴于ASFV的上述特性,落实生物安全防控是疫苗应用前的最有效措施。
2 ASFV逃逸天然免疫应答针对入侵的病原微生物,机体可通过一系列精细的免疫应答机制消灭和清除病原体,与此同时,病原微生物也进化出多种免疫逃逸策略。ASFV编码与宿主细胞相互作用的免疫逃逸相关蛋白,通过调控IFN产生、炎症反应、细胞凋亡、自噬及宿主蛋白合成等生物学过程(图 1),干预宿主细胞正常生命周期和细胞因子分泌等,进而抑制宿主的天然免疫应答。
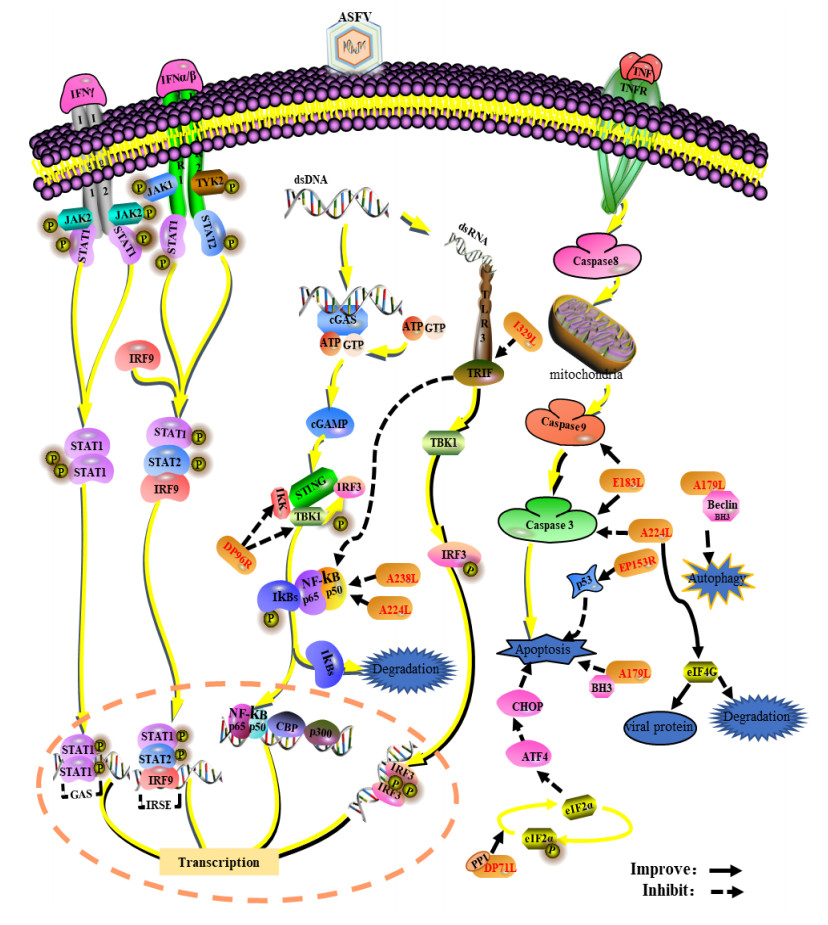
|
| 图 1 ASFV免疫逃逸的机制 Figure 1 Immunoevasion mechanisms of ASFV. ASFV regulates such biological processes as IFN production, inflammatory response, apoptosis, autophagy and host protein synthesis by interacting with host cells, suppress the host innate immune response to ultimately achieve immune escape. |
2.1 ASFV的细胞嗜性
ASFV具有严格的细胞嗜性,主要感染单核-巨噬细胞,也能感染内皮细胞、树突状细胞和外周血单核细胞等[13-18]。CD163是成熟巨噬细胞的表面标志,曾被认为是介导ASFV感染的受体[19]。但ASFV Georgia 2007/1株分别感染CD163–猪和正常猪后,在临床症状、死亡率、病理变化和病毒血症等方面无明显差异,从而排除了CD163是介导ASFV Georgia 2007/1株感染的受体[20]。在ASFV弱毒株(NHV/P68)和强毒株(Armenia/07、E70)感染下,比较细胞系(IPAM-WT、IPAM-CD163、C∆2+、WSL)和猪肺泡巨噬细胞(Porcine alveolar macrophages,PAMs)的敏感性及细胞膜受体,也未发现与ASFV感染相关的特异性受体[21]。完整的ASFV感染周期包括吸附、内化、复制、组装与释放,ASFV无法感染部分细胞系也与早期ASFV翻译和/或DNA复制的抑制有关[22]。因此,结合ASFV的细胞侵入途径和感染周期,认为靶细胞膜受体是实现ASFV感染的关键因子,但并非唯一条件。
2.2 ASFV与干扰素IFN-α/β是天然免疫发挥抗病毒作用的重要细胞因子。ASFV体外感染巨噬细胞时,弱毒株诱导IFN-α/β的能力明显强于强毒株[23-25]。在ASFV体外感染巨噬细胞试验中也发现,除了诱导细胞因子表达差异外,ASFV在巨噬细胞中的复制能力也有差异。ASFV强毒株(BA71、Georgia 2007、OUR T88/1)和弱毒株(OUR T88/3)感染经IFN-α预处理24 h的PAM,IFN-α能抑制弱毒株的复制而对强毒株无影响[26]。目前对ASFV强弱毒株参与IFN调控机制仍不清楚,但Raquel等[27]认为ASFV诱导IFN-β生成的差异与环GMP-AMP合成酶-干扰素基因刺激因子(Cyclic GMP-AMP synthase-stimulator of interferon genes,cGAS-STING)通路有关。
ASFV编码的MGF360和MGF505/530基因抑制干扰素表达,是目前构建基因缺失疫苗的主要靶基因。A276R作为MGF360的成员,能够抑制核转录因子-κB (Nuclear factor kappa B,NF-κB)、干扰素调节因子3(Interferon regulatory factor 3,IRF3)活化,但抑制IFN-β的转录/翻译与NF-κB途径无关[28]。I329L是Toll样受体3 (Toll-like receptors 3,TLR3)的同源蛋白,与接头蛋白TRIF作用抑制NF-κB、IRF3的活化来下调IFN-β表达[28-29]。DP96R通过其C端抑制cGAS-STING-TBK1介导的NF-κB活化,进而抑制Ⅰ型IFN的生成[30]。此外,在ASFV感染早期,与酪氨酸激酶-信号转导子及转录激活子(Janus kinase-signal transducer and activator of transcription,JAK-STAT)通路相关的JAK2、STAT1和CREB表达上调[24]。A528R能通过抑制Poly(I:C)诱导的IRF3和NF-κB活性或JAK-STAT途径抑制IFN-β生成[28]。除Ⅰ型IFN外,NK细胞、巨噬细胞、T淋巴细胞或早期产生的细胞因子(如IL-12、IL-18)诱导淋巴细胞分泌IFN-γ。IFN-γ与相应受体结合,激活相应淋巴细胞进而抑制ASFV在巨噬细胞中的复制和可能间接影响ASFV与宿主间反应,特别是感染早期[31-32]。IFN是一类具有免疫调节和抗病毒作用的细胞因子,抑制Ⅰ型IFN产生及其抗病毒效应是实现ASFV免疫逃逸的重要策略之一。目前ASFV有超过半数的蛋白功能未知,对相关蛋白调控干扰素生成的分子机制探究也是解析ASFV致病机制的有效手段之一。
2.3 ASFV与炎症反应急性ASFV感染时,淋巴细胞耗减、中性粒细胞减少、未成熟的免疫细胞和非典型淋巴细胞积聚,以及TNF-α、IL-6/8/1β/17/23、粒细胞集落刺激因子和C反应蛋白含量增加[33-34]。ASFV体外感染巨噬细胞后与体内试验数据类似,TNF超家族(FASLG、LTA、LTB、TNFSF4/10/13B/18)、TNF-α和IL-1β/17A等促炎性细胞因子上调表达及抗炎性细胞因子IL-10/10RA下调表达[35]。A238L、L83L是目前已知参与炎症反应调控的蛋白。A238L能抑制NF-κB及其介导的钙调磷酸酶活性。A238L通过抑制蛋白激酶C-θ介导p300氨基末端反式激活域活性升高,调控NF-κB、NFATc2和c-Jun的活化,影响下游COX-2和TNF-α的表达[36-38]。它也能抑制p65/RelA乙酰化和p300的反式激活,下调诱导型NO合酶(Inducible nitric oxide synthase,iNOS)表达[39]。L83L是非毒力基因,编码与IL-1β结合的高度保守的早期蛋白[40],但L83L的具体功能及作用机制还不明确。因此,需要深入研究ASFV编码蛋白调控机体炎症反应的分子和机制,这有助于ASFV致病机制的研究。
2.4 ASFV与细胞凋亡细胞凋亡是一种细胞的程序性死亡,也是宿主天然免疫与适应性免疫的重要防御策略。ASFV编码相关蛋白来调控细胞凋亡,如A224L、E183L、DP71L、EP153R和A179L等。A224L是凋亡抑制基因(Inhibitor of apoptosis genes,IAP)家族成员,能结合天冬氨酸特异性的胱氨酸蛋白酶3 (Cysteine aspartic acid specific protease 3,Caspase-3)的蛋白水解片段,抑制Caspase-3的激活及其蛋白酶活性[41]。它也能抑制TNF-α诱导的凋亡和活化NF-кB,诱导IAP、Bcl-2家族发挥抗凋亡作用,但可能被A238L拮抗。EP153R编码具有红细胞吸附特性的C型凝集素样蛋白,能诱导和/或维持病毒CD2v与相应细胞受体作用,也能抑制宿主细胞p53蛋白的反式活性来抑制细胞凋亡[42-43]。A179L是一个高度保守的蛋白,在ASFV感染早、晚期均有表达。作为Bcl-2家族成员,它与单个BH3蛋白亚家族成员(Bid、Bim和Puma亲和力最高,其次为Hrk、Noxa、Bmf,与Bik、Bad的亲和力最低[44])结合抑制下游蛋白Bax、Bak活化或结合Bcl-2等抗凋亡蛋白,从而抑制细胞凋亡。此外,ASFV也能抑制内质网应激(Endoplasmic reticulum stress,ERS)途径诱导的细胞凋亡。CHOP作为促凋亡蛋白,是ERS诱导细胞凋亡的重要分子,而PKP样内质网激酶(PKP-like ER kinase,PERK)-真核起始因子2α (Eukaryotic initiation factor 2α,eIF2α)-ATF4途径又是诱导CHOP表达所必需[45]。该过程eIF2α发生磷酸化,而DP71L则通过募集蛋白磷酸化酶1 (Protein phosphatase 1,PP1)引起eIF2α去磷酸化,抑制ERS诱导的细胞凋亡[46]。
E183L编码结构蛋白p54,参与病毒吸附和转运。它通过自身149–161位氨基酸动力蛋白结合基序激活线粒体凋亡途径[47]。ASFV除了编码与促凋亡相关的蛋白,还诱导巨噬细胞分泌半乳糖凝集素3和TNF-α,造成旁淋巴细胞凋亡[24, 48]。为了确保ASFV复制、增殖及扩散,ASFV编码蛋白参与调控细胞凋亡,这也是实现其免疫逃逸的重要策略。
2.5 ASFV与自噬自噬是细胞凋亡以外的另一种重要的病原清除机制,能直接降解病原并将其产物为宿主细胞自身所用。Beclin-1是重要的自噬调节因子,单纯疱疹病毒1型(Herpes simplex virus 1,HSV-1)的ICP 34.5与Beclin-1作用抑制自噬。ASFV编码的DP71L是ICP 34.5的类似物,却不能抑制自噬,但ASFV编码的A179L能抑制自噬小体的形成[49]。Suresh等[50]解析A179L结合Beclin-1 BH3基序晶体结构发现,A179L通过相同的配体结合槽与Beclin-1和Bcl-2结合,Beclin-1的K115与A179L的D80和E76离子通道作用,这两个氨基酸的突变可能会降低A179L与Beclin-1的结合。定点突变配体结合槽可以抑制Beclin-1与A179L结合,导致A179L抑制自噬小体形成的能力丧失[50]。
研究人员设计了能增加基因覆盖率和减少探针冗余的DNA芯片,ASFV Georgia 2007株体外感染巨噬细胞后发现,ATG2A、ATG9A、ATG101、ATG4B、BNIP3、GADD45A和ATG7等自噬相关基因分别在感染3 h和6 h后下调表达,而核蛋白1 (Nuclear protein 1,NUPR1)上调表达[35]。A179L是目前ASFV中唯一鉴定出的自噬相关蛋白,解析A179L与Beclin-1结合的晶体结构特点,为研究A179L抑制自噬的机理、小分子抑制剂开发及自噬在ASFV感染中的作用提供了依据,但自噬在ASFV致病机制中的作用有待深入研究。
2.6 ASFV调控宿主蛋白合成ASFV编码与RNA转录和修饰相关的蛋白,但仍依赖宿主的蛋白合成系统。ASFV通过募集翻译相关因子、线粒体重分布于“病毒工厂”和降解宿主mRNA等,抑制宿主蛋白合成。eIF4F由eIF4E、eIF4A和eIF4G组成,是诱导宿主mRNA翻译起始的关键因子。BA71V感染Vero细胞14 h后,eIF4A、p53和eIF4E等显著降低[51]。间接免疫荧光试验发现,胞质中的eIF4GI、eIF4E、eIF3b、eIF2α、eEF2、核糖体P蛋白和线粒体被募集至“病毒工厂”附近,且16 h后胞质中无明显荧光分布[52]。虽然Caspase-3水解eIF4G来抑制宿主蛋白合成,但与ASFV抑制宿主蛋白的合成无关[51, 53]。
ASFV mRNA与宿主mRNA结构相似,具有5′帽子和3′poly(A)尾结构,依赖帽子结构介导的翻译起始[54]。D250R是一种早期蛋白,也是ASFV目前已知的唯一有脱帽酶活性的蛋白,能够特异性结合核糖体蛋白L23a[51]。在ASFV感染晚期,D250R主要位于“病毒工厂”,对宿主mRNA有选择性且能抑制病毒/宿主蛋白合成[51]。推测其介导RNA释放,避免dsRNA聚集和病毒蛋白合成过度等激活免疫应答。消耗宿主系统翻译相关物质及改变其亚细胞定位,影响宿主细胞生理功能和促进病毒蛋白合成,为病毒增殖及扩散提供了条件。
3 ASFV逃逸适应性免疫应答 3.1 抗原递呈抗原递呈过程分为内源性加工递呈途径(主要组织相容性复合体I类(Major histocompatibility complex I,MHC-I)途径)、外源性加工递呈途径(MHC-Ⅱ类途径)和交叉抗原递呈途径。调控抗原递呈途径的关键细胞/分子也是ASFV的免疫逃逸策略之一。
ASFV弱毒株(BA71V、NH/P68)体外感染树突状细胞或巨噬细胞时,能下调细胞表面MHC-I的表达,但ASFV强毒株(22653/14)无此效应[55-56]。在ASFV强毒株(L60)感染猪的脾脏中,MHC-I/II的表达水平降低[14]。ASFV Georgia 2007株能下调MHC-II抗原加工的关键因子DMA、DMB及上调其抑制因子DOA、DOB表达,与前期体内试验的数据类似[14, 35]。虽然对ASFV下调MHC表达的分子机制不明,但认为ASFV通过改变TGN46的定位,破坏高尔基体反面网状结构(Trans-Golgi network,TGN)来降低MHC-I向膜表面递呈的能力或者下调蛋白酶体、溶酶体等实现对MHC-I/II抗原递呈能力的调控[35, 57]。EP153R主要影响MHC-I从内质网向细胞膜的分泌,进而抑制MHC-I的表达[58],但对影响MHC-I向细胞膜的具体转运机制不明。随着对ASFV调控MHC表达差异和转运机制的理解,会进一步加深对ASFV致病机制的认识。
3.2 ASFV与体液免疫抗体作为体液免疫的主要效应分子,特别是中和抗体对抑制病毒感染至关重要。p54、p72和p30抗体分别抑制病毒吸附和内化,但它们并不是典型的中和抗体,因为猪体内存在相应的抗体时,也不能提供完全保护[59-61]。利用ASFV Pr4株(104 TCID50)攻击经杆状病毒表达p30、p54、p72、p22的免疫猪,临床症状延迟出现和病毒血症降低,但攻毒后4 d与未免疫组无明显差异,且攻毒后7–10 d死亡[61]。感染ASFV E75株康复猪的血清对在Vero和巨噬细胞培养的强毒株E75、E70、L60、Malawi Lil 20/1和低代次适应毒株E75CV/V3有86%–97%的中和效果,但不能中和L60/V、DR-I/V、DR-II/V和HT/V等高代次适应毒株的感染,推测病毒在细胞传代过程中会丧失某些与康复猪血清反应相关的成分,并证实ASFV外膜的脂质成分,特别是磷脂酰肌醇对ASFV与康复猪血清的反应至关重要[62-63]。此外,ASFV诱导封闭抗体的产生,可能也是体液免疫无法提供完全保护及部分康复猪出现持续带毒的原因[64]。是否存在非中和抗原竞争性抑制中和抗原诱导抗体的产生以及抗原决定簇间相互拮抗还不清楚,但ASFV复杂的结构特点、编码蛋白的多样性以及缺乏典型中和抗体是制约体液免疫研究的原因之一。
ASF发病急、病死率高可能与免疫细胞系统性损伤及免疫抑制有关,但相关机制研究甚少。尽管传统灭活疫苗的抗ASFV效果不佳,但抗体能延迟发病、降低病毒血症和提供部分保护,在ASF疫苗研发过程中仍是作为免疫保护评价的重要指标。
3.3 ASFV与细胞免疫上世纪80年代,发现ASFV诱导细胞毒性T淋巴细胞(Cytotoxic T lymphocytes,CTLs)应答。灭活疫苗不能提供有效的免疫保护,而CD8+ T淋巴细胞参与抗ASFV感染,说明细胞免疫在抗ASFV感染的免疫保护中发挥重要作用[65-68]。p30作为重要的免疫保护性抗原,能诱导明显的抗体和CTLs应答。利用sHA、p54和p30构建DNA疫苗pCMV-sHAPQ免疫猪后,并未对ASFV E75株的攻击(104 HAU50)提供保护[69]。随后设计了泛素化的融合质粒pCMV-UbsHAPQ,采用相同攻毒剂量和方式(肌注),在无抗体的情况下,诱导了强烈的特异性T细胞应答并提供部分保护[69]。同样在未检测到抗体的情况下,免疫DNA文库的猪对ASFV E75株的攻击提供60%的免疫保护[70]。说明诱导CTLs应答和泛素化修饰的DNA疫苗能增强免疫保护,并且细胞免疫与抗ASFV感染具有相关性。
近期研究表明,亚单位疫苗的免疫保护效果与抗原数量、递送/载体系统、免疫剂量等有关,且部分免疫后的猪可能出现了抗体依赖性增强的现象[1, 68, 71]。不同抗原间是否存在协同或干扰也不明确。研究人员利用康复猪血清和免疫血清,结合ELISA和ELISpot筛选出了部分能诱导体液和细胞免疫应答的抗原,但部分暂未进行攻毒试验或免疫保护效果不佳[67, 71-74]。虽然亚单位疫苗可针对性地组合抗原,但盲目的抗原组合是不可取的。需要进一步鉴定新的保护性抗原(特别是交叉保护性抗原),选择科学的组合方式、制定新的免疫策略及递送系统等以期提高免疫保护效果。
4 结语和展望ASF作为一种“百年老病”,我们对它的认知却像是一种新病。ASFV编码54种结构蛋白和100多种非结构蛋白,参与病毒复制、转录、DNA修复和免疫逃逸等[3](图 2)。ASFV通过对相关信号转导通路、靶细胞及其他相关细胞的正常生命周期、生理功能等严格调控,从而逃逸天然免疫系统的识别与清除。此外,对宿主免疫系统、细胞的系统性损伤也导致了宿主难以诱导有效的适应性免疫应答。

|
| 图 2 ASFV基因及其功能参与ASFV结构、DNA复制及修复、RNA转录、免疫逃逸/毒力和功能未知基因 Figure 2 ASFV genes and their functions. Taking part in ASFV viral structure, DNA replication and repair, RNA transcription, immunoevasion/virulence and function unknown genes. |
从宿主方面来看,淋巴细胞耗减、中性粒细胞减少是急性ASF感染的主要特点[33],至今仍不清楚ASFV致病的分子机制。ASF的致死率与猪自身情况及毒株遗传背景等因素有关,从日龄和感染剂量两个方面模拟ASFV感染发现,γσT细胞和IL-10与ASF的致死率呈负相关[75]。ASFV感染后,γσT细胞可能会弥补单核-巨噬细胞出现的抗原递呈障碍,提高抗原递呈能力[31]。最新研究表明,ASFV自然弱毒株(OUR T88/3)和基因缺失弱毒株(Benin△MGF)分别以104 TCID50剂量免疫后130 d,不能对Benin 97/1株(104 TCID50)的攻击提供保护,认为免疫后未产生长久的免疫保护可能与调节性T细胞和IL-10的增加有关[76]。因此,γσT细胞、调节性T细胞和IL-10会影响机体的抗ASFV感染能力,探究相应的分子机制和挖掘更多与ASFV免疫调节相关的分子,有利于提高疫苗的保护效果和增加对ASFV致病机制的认识。此外,ASFV也诱导旁淋巴细胞凋亡,但具体方式和机制并不明确。ASFV编码的A179L蛋白通过相同的配体结合槽抑制细胞凋亡和自噬小体形成[50],ASFV是如何准确调控这两个重要的防御机制也是一个值得思考的问题。
从病毒方面来看,目前对ASFV本身的认知存在巨大的空白,严重制约了疫苗研发和相关机制研究。ASFV编码多种免疫逃逸相关蛋白,如细胞凋亡相关蛋白A224L、A179L、EP153R、DP71L和E183L,自噬相关蛋白A179L,调控蛋白合成的DP71L、A224L、D250R,调控MHC表达的EP153R。阐明ASFV编码蛋白的功能及其免疫逃逸策略,有助于我们理解ASFV的致病机制。敲除毒力和/或免疫逃逸相关基因构建基因缺失疫苗,或者针对性地构建提高体液、细胞免疫应答水平的亚单位疫苗,能最大程度地保证疫苗安全性和免疫保护效果,但基因的选择和抗原组合都是疫苗研发的难点。
在ASF众多的疫苗研发策略中,目前只有减毒活疫苗免疫猪后能够提供完全保护,被认为是短期内最有希望成功的疫苗。越来越多的研究也已表明减毒活疫苗免疫后诱导的特异性抗体及CTLs水平与免疫保护之间有较高的相关性。因此,有效激活机体体液和细胞免疫应答,建立长久的免疫记忆对于未来ASF疫苗研发至关重要。虽然ASFV-G-△I177L与HLJ/-18-7GD候选疫苗株毒力完全致弱[77-78],为ASF防控带来曙光,但安全性仍有待进一步研究。多组学分析、质谱分析和电镜技术等为研究ASFV提供了技术支持。系统全面地分析ASFV毒力/免疫逃逸基因及其功能,为新型疫苗研发及抗ASFV药物设计提供依据和靶点。疫苗是防控ASF亟需的工具,但它只是一种预防手段。消灭传染源,落实生物安全防控措施,做好环境控制、饲料营养和饲养管理,才是防控ASF以及其他传染病的关键。
| [1] | Lokhandwala S, Petrovan V, Popescu L, Sangewar N, Elijah C, Stoian A, Olcha M, Ennen L, Bray J, Bishop RP, Waghela SD, Sheahan M, Rowland RRR, Mwangi W. Adenovirus-vectored African swine fever virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Veterinary Microbiology, 2019, 235: 10-20. DOI:10.1016/j.vetmic.2019.06.006 |
| [2] | Zhou XT, Li N, Luo YZ, Liu Y, Miao FM, Chen T, Zhang SF, Cao PL, Li XD, Tian KG, Qiu HJ, Hu RL. Emergence of African swine fever in China, 2018. Transboundary and Emerging Diseases, 2018, 65(6): 1482-1484. DOI:10.1111/tbed.12989 |
| [3] |
Wang T, Sun Y, Luo YZ, Qiu HJ. Prevention, control and vaccine development of African swine fever:challenges and countermeasures. Chinese Journal of Biotechnology, 2018, 34(12): 1931-1942.
(in Chinese) 王涛, 孙元, 罗玉子, 仇华吉. 非洲猪瘟防控及疫苗研发:挑战与对策. 生物工程学报, 2018, 34(12): 1931-1942. |
| [4] | Sánchez-Vizcaíno JM, Mur L, Martínez-López B. African swine fever:an epidemiological update. Transboundary and Emerging Diseases, 2012, 59: 27-35. DOI:10.1111/j.1865-1682.2011.01293.x |
| [5] | Alonso C, Borca M, Dixon L, Revilla Y, Rodriguez F, Escribano JM. ICTV virus taxonomy profile:Asfarviridae. Journal of General Virology, 2018, 99(5): 613-614. DOI:10.1099/jgv.0.001049 |
| [6] | Wang N, Zhao DM, Wang JL, Zhang YL Wang M, Gao Y, Li F, Wang JF, Bu ZG, Rao ZH, Wang XX. Architecture of African swine fever virus and implications for viral assembly. Science, 2019, 366(6465): 640-644. DOI:10.1126/science.aaz1439 |
| [7] | Quembo CJ, Jori F, Vosloo W, Heath L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transboundary and Emerging Diseases, 2018, 65(2): 420-431. DOI:10.1111/tbed.12700 |
| [8] | Penrith ML, Vosloo W. Review of African swine fever:transmission, spread and control. Journal of the South African Veterinary Association, 2009, 80(2): 58-62. |
| [9] | Eblé PL, Hagenaars TJ, Weesendorp E, Quak S, Moonen-Leusen HW, Loeffen WLA. Transmission of African swine fever virus via carrier (survivor) pigs does occur. Veterinary Microbiology, 2019, 237: 108345. DOI:10.1016/j.vetmic.2019.06.018 |
| [10] | Niederwerder MC, Stoian AMM, Rowland RRR, Dritz SS, Petrovan V, Constance LA, Gebhardt JT, Olcha M, Jones CK, Woodworth JC, Fang Y, Liang J, Hefley TJ. Infectious dose of African swine fever virus when consumed naturally in liquid or feed. Emerging Infectious Diseases, 2019, 25(5): 891-897. DOI:10.3201/eid2505.181495 |
| [11] | Wilkinson PJ, Donaldson AI, Greig A, Bruce W. Transmission studies with African swine fever virus:Infections of pigs by airborne virus. Journal of Comparative Pathology, 1977, 87(3): 487-495. DOI:10.1016/0021-9975(77)90037-8 |
| [12] | Olesen AS, Lohse L, Hansen MF, Boklund A, Halasa T, Belsham GJ, Rasmussen TB, Bøtner A, Bødker R. Infection of pigs with African swine fever virus via ingestion of stable flies (Stomoxys calcitrans). Transboundary and Emerging Diseases, 2018, 65(5): 1152-1157. DOI:10.1111/tbed.12918 |
| [13] | Wardley RC, Wilkinson PJ. The association of African swine fever virus with blood components of infected pigs. Archives of Virology, 1977, 55(4): 327-334. DOI:10.1007/BF01315054 |
| [14] | González-Juarrero M, Lunney JK, Sánchez-Vizcaíno JM, Mebus C. Modulation of splenic macrophages, and swine leukocyte antigen (SLA) and viral antigen expression following African swine fever virus (ASFV) inoculation. Archives of Virology, 1992, 123(1/2): 145-156. |
| [15] | Gómez-Villamandos JC, Bautista MJ, Sánchez-Cordón PJ, Carrasco L. Pathology of African swine fever:the role of monocyte-macrophage. Virus Research, 2013, 173(1): 140-149. DOI:10.1016/j.virusres.2013.01.017 |
| [16] | Gregg DA, Schlafer DH, Mebus CA. African swine fever virus infection of skin-derived dendritic cells in vitro causes interference with subsequent foot-and-mouth disease virus infection. Journal of Veterinary Diagnostic Investigation, 1995, 7(1): 44-51. DOI:10.1177/104063879500700106 |
| [17] | Sánchez-Cordón PJ, Romero-Trevejo JL, Pedrera M, Sánchez-Vizcaíno JM, Bautista MJ, Gómez-Villamandos JC. Role of hepatic macrophages during the viral haemorrhagic fever induced by African swine fever virus. Histology and Histopathology, 2008, 23(6): 683-691. |
| [18] | Carrillo C, Borca MV, Afonso CL, Onisk DV, Rock DL. Long-term persistent infection of swine monocytes/macrophages with African swine fever virus. Journal of Virology, 1994, 68(1): 580-583. DOI:10.1128/JVI.68.1.580-583.1994 |
| [19] | Sánchez-Torres C, Gómez-Puertas P, Gómez-del-Moral M, Alonso F, Escribano JM, Ezquerra A, Domínguez J. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Archives of Virology, 2003, 148(12): 2307-2323. DOI:10.1007/s00705-003-0188-4 |
| [20] | Popescu L, Gaudreault NN, Whitworth KM, Murgia MV, Nietfeld JC, Mileham A, Samuel M, Wells KD, Prather RS, Rowland RRR. Genetically edited pigs lacking CD163 show no resistance following infection with the African swine fever virus isolate, Georgia 2007/1. Virology, 2017, 501: 102-106. DOI:10.1016/j.virol.2016.11.012 |
| [21] | Sánchez EG, Riera E, Nogal M, Gallardo C, Fernández P, Bello-Morales R, López-Guerrero JA, Chitko-McKown CG, Richt JA, Revilla Y. Phenotyping and susceptibility of established porcine cells lines to African swine fever virus infection and viral production. Scientific Reports, 2017, 7(1): 10369. DOI:10.1038/s41598-017-09948-x |
| [22] | Carrascosa AL, Bustos MJ, Galindo I, Viñuela E. Virus-specific cell receptors are necessary, but not sufficient, to confer cell susceptibility to African swine fever virus. Archives of Virology, 1999, 144(7): 1309-1321. DOI:10.1007/s007050050589 |
| [23] | Gil S, Sepúlveda N, Albina E, Leitão A, Martins C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (INFα, TNFα and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Archives of Virology, 2008, 153(10): 1845-1854. DOI:10.1007/s00705-008-0196-5 |
| [24] | Zhang FQ, Hopwood P, Abrams CC, Downing A, Murray F, Talbot R, Archibald A, Lowden S, Dixon LK. Macrophage transcriptional responses following in vitro infection with a highly virulent African swine fever virus isolate. Journal of Virology, 2006, 80(21): 10514-10521. DOI:10.1128/JVI.00485-06 |
| [25] | Afonso CL, Piccone ME, Zaffuto KM, Neilan J, Kutish GF, Lu Z, Balinsky CA, Gibb TR, Bean TJ, Zsak L, Rock DL. African swine fever virus multigene family 360 and 530 genes affect host interferon response. Journal of Virology, 2004, 78(4): 1858-1864. DOI:10.1128/JVI.78.4.1858-1864.2004 |
| [26] | Golding JP, Goatley L, Goodbourn S, Dixon LK, Taylor G, Netherton CL. Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology, 2016, 493: 154-161. DOI:10.1016/j.virol.2016.03.019 |
| [27] | García-Belmonte R, Pérez-Núñez D, Pittau M, Richt JA, Revilla Y. African swine fever virus Armenia/07 virulent strain controls interferon beta production through the cGAS-STING pathway. Journal of Virology, 2019, 93(12): e02298-18. |
| [28] | Correia S, Ventura S, Parkhouse RM. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Research, 2013, 173(1): 87-100. DOI:10.1016/j.virusres.2012.10.013 |
| [29] | de Oliveira VL, Almeida SCP, Soares HR, Crespo A, Marshall-Clarke S, Parkhouse RME. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Archives of Virology, 2011, 156(4): 597-609. DOI:10.1007/s00705-010-0894-7 |
| [30] | Wang XX, Wu J, Wu YT, Chen HJ, Zhang SF, Li JX, Xin T, Jia H, Hou SH, Jiang YT, Zhu HF, Guo XY. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochemical and Biophysical Research Communications, 2018, 506(3): 437-443. DOI:10.1016/j.bbrc.2018.10.103 |
| [31] | Takamatsu HH, Denyer MS, Lacasta A, Stirling CMA, Argilaguet JM, Netherton CL, Oura CAL, Martins C, Rodríguez F. Cellular immunity in ASFV responses. Virus Research, 2013, 173(1): 110-121. DOI:10.1016/j.virusres.2012.11.009 |
| [32] | Esparza I, González JC, Viñuela E. Effect of interferon-α, interferon-γ and tumour necrosis factor on African swine fever virus replication in porcine monocytes and macrophages. Journal of General Virology, 1988, 69(12): 2973-2980. DOI:10.1099/0022-1317-69-12-2973 |
| [33] | Zakaryan H, Cholakyans V, Simonyan L, Misakyan A, Karalova E, Chavushyan A, Karalyan Z. A study of lymphoid organs and serum proinflammatory cytokines in pigs infected with African swine fever virus genotype II. Archives of Virology, 2015, 160(6): 1407-1414. DOI:10.1007/s00705-015-2401-7 |
| [34] | Karalyan Z, Voskanyan H, Ter-Pogossyan Z, Saroyan D, Karalova E. IL-23/IL-17/G-CSF pathway is associated with granulocyte recruitment to the lung during African swine fever. Veterinary Immunology and Immunopathology, 2016, 179: 58-62. DOI:10.1016/j.vetimm.2016.08.005 |
| [35] | Zhu JJ, Ramanathan P, Bishop EA, O'Donnell V, Gladue DP, Borca MV. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS One, 2019, 14(11): e0223955. DOI:10.1371/journal.pone.0223955 |
| [36] | Granja AG, Nogal ML, Hurtado C, Vila V, Carrascosa AL, Salas ML, Fresno M, Revilla Y. The viral protein A238L inhibits cyclooxygenase-2 expression through a nuclear factor of activated T cell-dependent transactivation pathway. Journal of Biological Chemistry, 2004, 279(51): 53736-53746. DOI:10.1074/jbc.M406620200 |
| [37] | Granja AG, Nogal ML, Hurtado C, del Aguila C, Carrascosa AL, Salas ML, Fresno M, Revilla Y. The viral protein A238L inhibits TNF-α expression through a CBP/p300 transcriptional coactivators pathway. Journal of Immunology, 2006, 176(1): 451-462. DOI:10.4049/jimmunol.176.1.451 |
| [38] | Granja AG, Perkins ND, Revilla Y. A238L inhibits NF-ATc2, NF-κB, and c-Jun activation through a novel mechanism involving protein kinase C-θ-mediated up-regulation of the amino-terminal transactivation domain of p300. Journal of Immunology, 2008, 180(4): 2429-2442. DOI:10.4049/jimmunol.180.4.2429 |
| [39] | Granja AG, Sabina P, Salas ML, Fresno M, Revilla Y. Regulation of inducible nitric oxide synthase expression by viral A238L-mediated inhibition of p65/RelA acetylation and p300 transactivation. Journal of Virology, 2006, 80(21): 10487-10496. DOI:10.1128/JVI.00862-06 |
| [40] | Borca MV, O'Donnell V, Holinka LG, Ramírez-Medina E, Clark BA, Vuono EA, Berggren K, Alfano M, Carey LB, Richt JA, Risatti GR, Gladue DP. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Research, 2018, 249: 116-123. DOI:10.1016/j.virusres.2018.03.017 |
| [41] | Nogal ML, González de Buitrago G, Rodríguez C, Cubelos B, Carrascosa AL, Salas ML, Revilla Y. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. Journal of Virology, 2001, 75(6): 2535-2543. DOI:10.1128/JVI.75.6.2535-2543.2001 |
| [42] | Galindo I, Almazán F, Bustos MJ, Viñuela E, Carrascosa AL. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology, 2000, 266(2): 340-351. DOI:10.1006/viro.1999.0080 |
| [43] | Hurtado C, Granja AG, Bustos MJ, Nogal ML, González de Buitrago G, de Yébenes VG, Salas ML, Revilla Y, Carrascosa AL. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology, 2004, 326(1): 160-170. DOI:10.1016/j.virol.2004.05.019 |
| [44] | Banjara S, Caria S, Dixon LK, Hinds MG, Kvansakul M. Structural insight into African swine fever virus A179L-mediated inhibition of apoptosis. Journal of Virology, 2017, 91(6): e02228-16. |
| [45] | Fels DR, Koumenis C. The PERK/eIF2α/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biology & Therapy, 2006, 5(7): 723-728. |
| [46] | Zhang FQ, Moon A, Childs K, Goodbourn S, Dixon LK. The African swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2α and inhibits CHOP induction but is dispensable for these activities during virus infection. Journal of Virology, 2010, 84(20): 10681-10689. DOI:10.1128/JVI.01027-10 |
| [47] | Hernáez B, Díaz-Gil G, García-Gallo M, Quetglas JI, Rodríguez-Crespo I, Dixon L, Escribano JM, Alonso C. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Letters, 2004, 569(1/3): 224-228. |
| [48] | Gómez del Moral M, Ortuño E, Fernández-Zapatero P, Alonso F, Alonso C, Ezquerra A, Domínguez J. African swine fever virus infection induces tumor necrosis factor alpha production:implications in pathogenesis. Journal of Virology, 1999, 73(3): 2173-2180. DOI:10.1128/JVI.73.3.2173-2180.1999 |
| [49] | Hernaez B, Cabezas M, Muñoz-Moreno R, Galindo I, Cuesta-Geijo MA, Alonso C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Current Molecular Medicine, 2013, 13(2): 305-316. DOI:10.2174/156652413804810736 |
| [50] | Banjara S, Shimmon GL, Dixon LK, Netherton CL, Hinds MG, Kvansakul M. Crystal structure of African swine fever virus A179L with the autophagy regulator Beclin. Viruses, 2019, 11(9): 789. DOI:10.3390/v11090789 |
| [51] | Quintas A, Pérez-Núñez D, Sánchez EG, Nogal ML, Hentze MW, Castelló A, Revilla Y. Characterization of the African swine fever virus decapping enzyme during infection. Journal of Virology, 2017, 91(24): e00990-17. |
| [52] | Castelló A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y. Regulation of host translational machinery by African swine fever virus. PLoS Pathogens, 2009, 5(8): e1000562. DOI:10.1371/journal.ppat.1000562 |
| [53] | Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Molecular and Cellular Biology, 1998, 18(12): 7565-7574. DOI:10.1128/MCB.18.12.7565 |
| [54] | Salas ML, Kuznar J, Viñuela E. Polyadenylation, methylation, and capping of the RNA synthesized in vitro by African swine fever virus. Virology, 1981, 113(2): 484-491. DOI:10.1016/0042-6822(81)90176-8 |
| [55] | Franzoni G, Graham SP, Giudici SD, Bonelli P, Pilo G, Anfossi AG, Pittau M, Nicolussi PS, Laddomada A, Oggiano A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Veterinary Microbiology, 2017, 198: 88-98. DOI:10.1016/j.vetmic.2016.12.010 |
| [56] | Franzoni G, Graham SP, Sanna G, Angioi P, Fiori MS, Anfossi A, Amadori M, Dei Giudici S, Oggiano A. Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Veterinary Microbiology, 2018, 216: 190-197. DOI:10.1016/j.vetmic.2018.02.021 |
| [57] | Netherton CL, McCrossan MC, Denyer M, Ponnambalam S, Armstrong J, Takamatsu HH, Wileman TE. African swine fever virus causes microtubule-dependent dispersal of the trans-Golgi network and slows delivery of membrane protein to the plasma membrane. Journal of Virology, 2006, 80(22): 11385-11392. DOI:10.1128/JVI.00439-06 |
| [58] | Hurtado C, Bustos MJ, Granja AG, de León P, Sabina P, López-Viñas E, Gómez-Puertas P, Revilla Y, Carrascosa AL. The African swine fever virus lectin EP153R modulates the surface membrane expression of MHC class I antigens. Archives of Virology, 2011, 156(2): 219-234. DOI:10.1007/s00705-010-0846-2 |
| [59] | Gómez-Puertas P, Rodríguez F, Oviedo JM, Brun A, Alonso C, Escribano JM. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology, 1998, 243(2): 461-471. DOI:10.1006/viro.1998.9068 |
| [60] | Barderas MG, Rodríguez F, Gómez-Puertas P, Avilés M, Beitia F, Alonso C, Escribano JM. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Archives of Virology, 2001, 146(9): 1681-1691. DOI:10.1007/s007050170056 |
| [61] | Neilan JG, Zsak L, Lu Z, Burrage TG, Kutish GF, Rock DL. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology, 2004, 319(2): 337-342. DOI:10.1016/j.virol.2003.11.011 |
| [62] | Zsak L, Onisk DV, Afonso CL, Rock DL. Virulent African swine fever virus isolates are neutralized by swine immune serum and by monoclonal antibodies recognizing a 72-kDa viral protein. Virology, 1993, 196(2): 596-602. DOI:10.1006/viro.1993.1515 |
| [63] | Gómez-Puertas P, Oviedo JM, Rodríguez F, Coll J, Escribano JM. Neutralization susceptibility of African swine fever virus is dependent on the phospholipid composition of viral particles. Virology, 1997, 228(2): 180-189. DOI:10.1006/viro.1996.8391 |
| [64] | Escribano JM, Galindo I, Alonso C. Antibody-mediated neutralization of African swine fever virus:myths and facts. Virus Research, 2013, 173(1): 101-109. DOI:10.1016/j.virusres.2012.10.012 |
| [65] | Jenson JS, Childerstone A, Takamatsu HH, Dixon LK, Parkhouse RME. The cellular immune recognition of proteins expressed by an African swine fever virus random genomic library. Journal of Immunological Methods, 2000, 242(1/2): 33-42. |
| [66] | Oura CAL, Denyer MS, Takamatsu H, Parkhouse RME. In vivo depletion of CD8+ T lymphocytes abrogates protective immunity to African swine fever virus. Journal of General Virology, 2005, 86(9): 2445-2450. DOI:10.1099/vir.0.81038-0 |
| [67] | Stone SS, Hess WR. Antibody response to inactivated preparations of African swine fever virus in pigs. American Journal of Veterinary Research, 1967, 28(123): 475-481. |
| [68] | Blome S, Gabriel C, Beer M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine, 2014, 32(31): 3879-3882. DOI:10.1016/j.vaccine.2014.05.051 |
| [69] | Argilaguet JM, Pérez-Martín E, Nofrarías M, Gallardo C, Accensi F, Lacasta A, Mora M, Ballester M, Galindo-Cardiel I, López-Soria S, Escribano JM, Reche PA, Rodríguez F. DNA vaccination partially protects against African swine fever virus lethal challenge in the absence of antibodies. PLoS One, 2012, 7(9): e40942. DOI:10.1371/journal.pone.0040942 |
| [70] | Lacasta A, Ballester M, Monteagudo PL, Rodríguez JM, Salas ML, Accensi F, Pina-Pedrero S, Bensaid A, Argilaguet J, López-Soria S, Hutet E, Le Potier MF, Rodríguez F. Expression library immunization can confer protection against lethal challenge with African swine fever virus. Journal of Virology, 2014, 88(22): 13322-13332. DOI:10.1128/JVI.01893-14 |
| [71] | Cadenas-Fernández E, Sánchez-Vizcaíno JM, Kosowska A, Rivera B, Mayoral-Alegre F, Rodríguez-Bertos A, Yao JX, Bray J, Lokhandwala S, Mwangi W, Barasona JA. Adenovirus-vectored African swine fever virus antigens cocktail is not protective against virulent Arm07 isolate in Eurasian wild boar. Pathogens, 2020, 9(3): 171. DOI:10.3390/pathogens9030171 |
| [72] | Kollnberger SD, Gutierrez-Castañeda B, Foster-Cuevas M, Corteyn A, Parkhouse RME. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. Journal of General Virology, 2002, 83(6): 1331-1342. DOI:10.1099/0022-1317-83-6-1331 |
| [73] | Netherton CL, Goatley LC, Reis AL, Portugal R, Nash RH, Morgan SB, Gault L, Nieto R, Norlin V, Gallardo C, Ho CS, Sánchez-Cordón PJ, Taylor G, Dixon LK. Identification and immunogenicity of African swine fever virus antigens. Frontiers in Immunology, 2019, 10: 1318. DOI:10.3389/fimmu.2019.01318 |
| [74] | Lokhandwala S, Waghela SD, Bray J, Martin CL, Sangewar N, Charendoff C, Shetti R, Ashley C, Chen CH, Berghman LR, Mwangi D, Dominowski PJ, Foss DL, Rai S, Vora S, Gabbert L, Burrage TG, Brake D, Neilan J, Mwangi W. Induction of robust immune responses in swine by using a cocktail of adenovirus-vectored African swine fever virus antigens. Clinical and Vaccine Immunology, 2016, 23(11): 888-900. DOI:10.1128/CVI.00395-16 |
| [75] | Post J, Weesendorp E, Montoya M, Loeffen WL. Influence of age and dose of African swine fever virus infections on clinical outcome and blood parameters in pigs. Viral Immunology, 2017, 30(1): 58-69. DOI:10.1089/vim.2016.0121 |
| [76] | Sánchez-Cordón PJ, Jabbar T, Chapman D, Dixon LK, Montoya M. Absence of long-term protection in domestic pigs immunized with attenuated African swine fever virus isolate OURT88/3 or BeninΔMFG correlates with increased levels of regulatory T cells and IL-10. Journal of Virology, 2020, 94(14): e00350-20. |
| [77] | Chen WY, Zhao DM, He XJ, Liu RQ, Wang ZL, Zhang XF, Li F, Shan D, Chen HF, Zhang JW, Wang LL, Wen ZY, Wang XJ, Guan YT, Liu JX, Bu ZG. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Science China Life Sciences, 2020, 63(5): 623-634. DOI:10.1007/s11427-020-1657-9 |
| [78] | Borca MV, Ramirez-Medina E, Silva E, Vuono E, Rai A, Pruitt S, Holinka LG, Velazquez-Salinas L, Zhu J, Gladue DP. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. Journal of Virology, 2020, 94(7): e02017-19. |
 2021, Vol. 61
2021, Vol. 61