中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 汪雪梅, 翁倩卉, 赵芹芹, 白林泉. 2021
- Xuemei Wang, Qianhui Weng, Qinqin Zhao, Linquan Bai. 2021
- 阿卡波糖生物合成调控蛋白的钓取与功能解析
- Mining and function studies of regulators for acarbose biosynthesis in Actinoplanes sp. SE50/110
- 微生物学报, 61(11): 3667-3685
- Acta Microbiologica Sinica, 61(11): 3667-3685
-
文章历史
- 收稿日期:2021-02-10
- 修回日期:2021-03-03
- 网络出版日期:2021-09-08
目前,糖尿病患者的数量在全球范围内持续增长,其中以2型糖尿病为主,约占患者总数的90%以上[1−2]。临床研究表明,口服降糖药、合理控制饮食以及适当的运动可有效改善餐后血糖浓度,从而能够减缓部分心脑血管病等慢性并发症的发生[3]。目前用于治疗糖尿病的降糖药主要有胰岛素增敏剂、促胰岛素分泌剂、α-葡萄糖苷酶抑制剂等[4]。由于阿卡波糖不仅可以降低血糖水平,还具有预防心血管疾病等功效,自上市以来便成为治疗2型糖尿病的理想药物[5]。在2017年版的《中国2型糖尿病防治指南》中,阿卡波糖被列入治疗2型糖尿病的一线用药。
20世纪70年代,在游动放线菌SE50 (Actinoplanes sp.)的发酵产物中首次分离得到阿卡波糖[6]。其化学结构由氨基环醇、4-氨基-4, 6-脱氧葡萄糖和麦芽糖三部分组成(图 1)。负责阿卡波糖生物合成的相关基因在游动放线菌SE50的染色体上成簇分布,组成acb基因簇[7]。该基因簇(图 5-A)由22个基因构成,长约30 kb,按照基因功能的不同可将已知基因分为2个大类:acbAB和acbVUSRPIJKMLNOC为阿卡波糖生物合成基因;acbWXY和acbFGH为转运蛋白基因[8]。在acb基因簇中没有任何与调控相关的基因,推测存在簇外调控因子对acb基因簇的转录起到调控作用,这为研究其生物合成基因的调控机制带来了挑战。
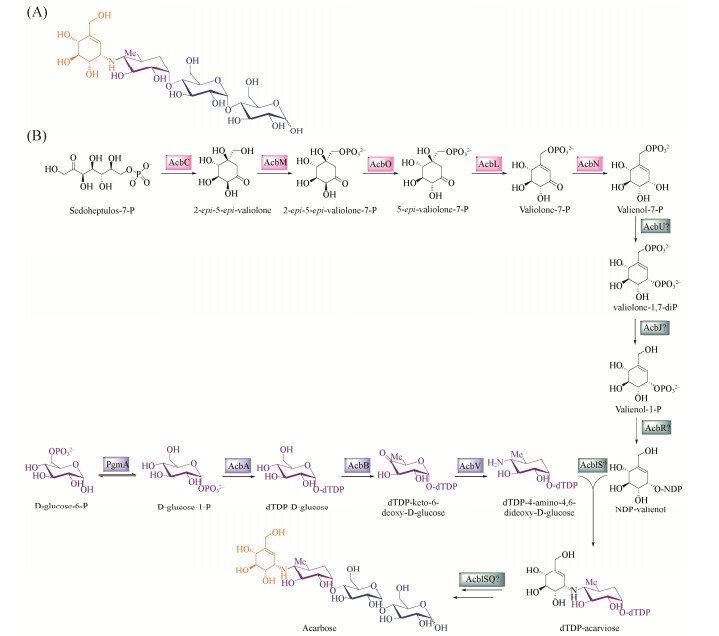
|
| 图 1 阿卡波糖化学结构式及生物合成途径 Figure 1 The structure of acarbose and its biosynthetic pathway. A: The structure of acarbose; B: The biosynthetic pathway of acarbose. |

|
| 图 2 调控基因强化表达对阿卡波糖产量的影响 Figure 2 Effects of the overexpression of regulatory genes on acarbose yield. A: recombinant plasmid pLQ1450 for the overexpression of ACPL_1889; B: verification of WXM-01 by PCR amplification of the kasOp*-ACPL_1889 fragment; C: yield analysis of mutants with gene overexpression. *: significant difference at P < 0.05; **: significant difference at P < 0.01; ***: significant difference at P < 0.001. Error bars, mean±SD (n=3 biological replicates). |

|
| 图 3 调控基因缺失对阿卡波糖产量的影响 Figure 3 Effects of the deletions of regulatory genes on acarbose yield. A: schematic deletion of ACPL_1889; B: verification of WXM-12 by PCR amplification; C: yield analysis of mutants with gene deletion. *: significant difference at P < 0.05; **: significant difference at P < 0.01; ***: significant difference at P < 0.001. Error bars, mean±SD (n=3 biological replicates). |
|
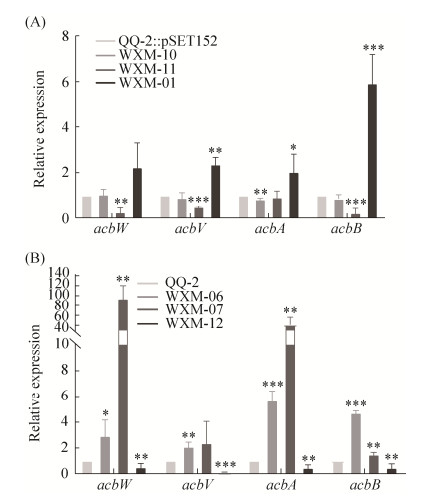
| 图 4 调控基因的强化表达或缺失对阿卡波糖生物合成基因转录水平的影响 Figure 4 Effects of the overexpression or deletions of regulatory genes on the transcription of acarbose biosynthetic genes. A: effects of the overexpression of regulatory genes on the transcription of acarbose biosynthetic genes acbW, acbV, acbA and acbB; B: effects of the deletions of regulatory genes on the transcription of acarbose biosynthetic genes acbW, acbV, acbA and acbB. *: significant difference at P < 0.05; **: significant difference at P < 0.01; ***: significant difference at P < 0.001; Error bars, mean±SD (n=3 biological replicates). |
|
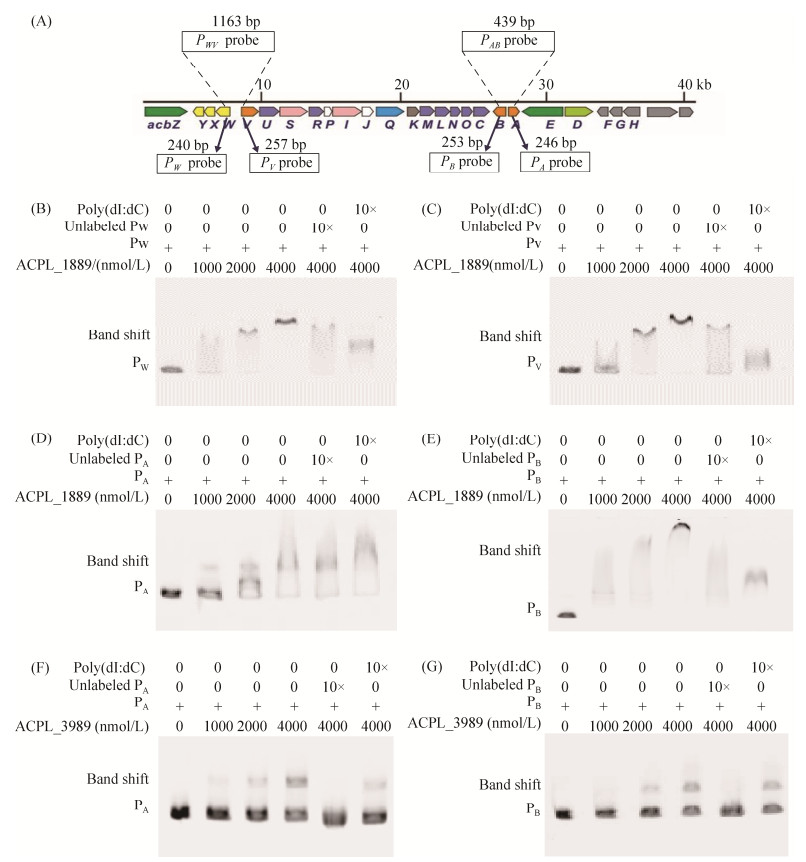
| 图 5 调控蛋白与启动子探针的相互作用 Figure 5 Acarbose biosynthetic gene cluster and interactions between proteins and promoter probes. A: acarbose biosynthetic gene cluster. The PWV and PAB probes for DNA affinity chromatography are indicated. The probes PW, PV, PA, and PB for EMSA are also indicated. B: the interaction between ACPL_1889 and PW. C: the interaction between ACPL_1889 and PV. D: the interaction between ACPL_1889 and PA. E: the interaction between ACPL_1889 and PB. F: the interaction between ACPL_3989 and PA. G: the interaction between ACPL_3989 and PB. Lane 1−4: increasing amounts of proteins (0, 1000, 2000, 4000 nmol/L) were used with 100 ng biotin-labeled probes; lane 5: 1 μg of unlabeled probe was added as specific competition control; lane 6: 1 μg of poly(dI: dC) was added as nonspecific competition control. |
调控因子可以通过直接或间接识别并结合特定的DNA序列,从而调控基因的转录效率[9]。有些调控因子是全局性或多效性调控因子,调控范围比较广泛,如天蓝色链霉菌Streptomyces coelicolor中的Crp、WblA、GlnR,均能参与多条代谢途径的调控[10−14]。途径特异性调控因子通常在基因簇内,能特异性地调控簇内结构基因的转录,如PapR6能特异性地调控普那霉素生物合成,在SE50/110中麦芽糖酶基因amlE受到其上游转录因子AmlR的调控,在Streptomyces sp. FR-008中,fscR1、fscR2、fscR3和fscR4四个调控基因对多烯抗生素杀念菌素的生物合成具有重要调控作用[15−17]。由于放线菌的转录调控系统非常复杂,对于调控因子的寻找及其功能解析比较困难。通过启动子区域DNA亲和层析技术钓取与DNA片段特异结合的蛋白,继而验证其功能,是解析放线菌生物合成调控机制的一种新策略[18]。在恰塔努加链霉菌Streptomyces chattanoogensis L10中,利用wblAch启动子DNA亲和层析技术成功钓取到正调控蛋白AdpAch[19]。因此,本研究以acb基因簇的2个双向启动子PWV、PAB为探针,利用DNA亲和层析技术钓取到与该启动子区域结合的调控蛋白,并通过体内基因强化表达与敲除确定了它们与阿卡波糖产生、阿卡波糖生物合成基因转录水平的相关性,并实现了高产改造,为深入解析阿卡波糖生物合成的转录调控机制及其高产奠定了基础。
1 材料和方法 1.1 材料 1.1.1 菌株、质粒、引物:本研究中所用菌株、质粒和引物见表 1。
| Strains, plasmids, primers | Related features or sequences | Sources or references |
| Actinoplanes spp. | ||
| SE50 | Acarbose producer, wild-type strain | ATCC31042 |
| SE50/110 | Acarbose producer, high-yield strain | ATCC31044 |
| QQ-2 | Actinoplanes sp. SE50/110∆treY | [20] |
| WXM-01 | QQ-2:: ACPL_1889 | This work |
| WXM-02 | QQ-2:: ACPL_4236 | This work |
| WXM-03 | QQ-2:: ACPL_7303 | This work |
| WXM-04 | QQ-2:: ACPL_6479 | This work |
| WXM-05 | QQ-2:: ACPL_8104 | This work |
| WXM-06 | QQ-2∆ACPL_5445 | This work |
| WXM-07 | QQ-2∆ACPL_3989 | This work |
| WXM-08 | QQ-2:: ACPL_7617 | This work |
| WXM-09 | QQ-2:: ACPL_8270 | This work |
| WXM-10 | QQ-2:: ACPL_5445 | This work |
| WXM-11 | QQ-2:: ACPL_3989 | This work |
| WXM-12 | QQ-2∆ACPL_1889 | This work |
| WXM-13 | QQ-2∆ACPL_4236 | This work |
| WXM-14 | QQ-2∆ACPL_7303 | This work |
| WXM-15 | QQ-2∆ACPL_6479 | This work |
| WXM-16 | QQ-2∆ACPL_7617 | This work |
| WXM-17 | QQ-2:: ACPL_1889/ACPL_4236/ACPL_7303/ACPL_6479/ACPL_8104 | This work |
| WXM-18 | WXM-17∆ACPL_5445∆ACPL_3989 | This work |
| Escherichia coli | ||
| DH10B | Cloning host | GIBCO-BRL |
| ET12567(pUZ8002) | recE, dam−, dcm−, hsdS, Cmr, Strr, Tetr, Kmr, for conjugation | [21] |
| BL21(DE3) | Protein expression host | Sangon biotech |
| Plasmids | ||
| pLQ752 | pJTU1278-derived vector with codA gene and the tsr gene replaced by aac(3)IV |
[20] |
| pDR4-K* | aac(3)IV, xylE-neo, kasOp* | [22] |
| pSET152 | oriTRK2, aac(3)IV, 31int, attP | [23] |
| pET30a | oriBR322, Kmr, lacI, His-tag and S-tag coding genes | Novagen |
| pLQ1450 | pSET152-based plasmid for ACPL_1889 overexpression | This work |
| pLQ1451 | pSET152-based plasmid for ACPL_4236 overexpression | This work |
| pLQ1452 | pSET152-based plasmid for ACPL_7303 overexpression | This work |
| pLQ1453 | pSET152-based plasmid for ACPL_6479 overexpression | This work |
| pLQ1454 | pSET152-based plasmid for ACPL_8104 overexpression | This work |
| pLQ1455 | pLQ752-based plasmid for ACPL_5445 inactivation | This work |
| pLQ1456 | pLQ752-based plasmid for ACPL_3989 inactivation | This work |
| pLQ1457 | pSET152-based plasmid for ACPL_7617 overexpression | This work |
| pLQ1458 | pSET152-based plasmid for ACPL_8270 overexpression | This work |
| pLQ1459 | pSET152-based plasmid for ACPL_5445 overexpression | This work |
| pLQ1460 | pSET152-based plasmid for ACPL_3989 overexpression | This work |
| pLQ1461 | pLQ752-based plasmid for ACPL_1889 inactivation | This work |
| pLQ1462 | pLQ752-based plasmid for ACPL_4236 inactivation | This work |
| pLQ1463 | pLQ752-based plasmid for ACPL_7303 inactivation | This work |
| pLQ1464 | pLQ752-based plasmid for ACPL_6479 inactivation | This work |
| pLQ1465 | pLQ752-based plasmid for ACPL_8270 inactivation | This work |
| pLQ1466 | pLQ752-based plasmid for ACPL_8104 inactivation | This work |
| pLQ1467 | pLQ752-based plasmid for ACPL_7617 inactivation | This work |
| pLQ1468 | pET30a-based plasmid for ACPL_1889 expression | This work |
| pLQ1469 | pET30a-based plasmid for ACPL_4236 expression | This work |
| pLQ1470 | pET30a-based plasmid for ACPL_7303 expression | This work |
| pLQ1471 | pET30a-based plasmid for ACPL_6479 expression | This work |
| pLQ1472 | pET30a-based plasmid for ACPL_8104 expression | This work |
| pLQ1473 | pET30a-based plasmid for ACPL_5445 expression | This work |
| pLQ1474 | pET30a-based plasmid for ACPL_3989 expression | This work |
| pLQ1475 | pET30a-based plasmid for ACPL_7617 expression | This work |
| pLQ1476 | pET30a-based plasmid for ACPL_8270 expression | This work |
| pLQ1477 | pSET152-based plasmid for the overexpression of ACPL_1889, ACPL_4236, ACPL_7303, ACPL_6479 and ACPL_8104 | This work |
| Primers | Sequences (5′→3′) | |
| PWV-F | CTCGAAGCTGATGTCGTCGAC | |
| PWV-R | GAAGTCGAGGTATTCCACACC | |
| PAB-F | GAGATTTCTGAGATTGCCTC | |
| PAB-R | CGAGCCGTCACCCAGCAAC | |
| ACPL_1889GB-F | TTGAAGAGGTGACGTCATGGCAGCCACTGGCACAGCT | |
| ACPL_1889GB-R (BamH I) | CTGGATCCTCACGCCTTCTTGCG | |
| ACPL_4236GB-F | TTGAAGAGGTGACGTCATGGGAAGGCCGCGAGCGTT | |
| ACPL_4236GB-R (Not I) | CTGCGGCCGCTCACCGACAACGCTGATCTTGGC | |
| ACPL_7303GB-F | TTGAAGAGGTGACGTCATGAAGCTGGTGACCGCGGT | |
| ACPL_7303GB-R (BamH I) | CTGGATCCTCAGAGGGCGTCGAGGC | |
| ACPL_1889GB-F | TTGAAGAGGTGACGTCATGGCAGCCACTGGCACAGCT | |
| ACPL_1889GB-R (BamH I) | CTGGATCCTCACGCCTTCTTGCG | |
| ACPL_5445GB-F | GTTGAAGAGGTGACGTCGTGCGCGTGAAGTTGATGGC | |
| ACPL_5445GB-R (BamH I) | GTGGATCCTCAGTACTTGCGGGTCGGAA | |
| ACPL_3989GB-F | GTTGAAGAGGTGACGTCGTGTCCATTGACCTGATCGC | |
| ACPL_3989GB-R (BamH I) | GTGGATCCCTAAAGCAGATTGAGGGCCC | |
| ACPL_7617GB-F | TTGAAGAGGTGACGTCGTGTACGTCTCGGCTCGCAC | |
| ACPL_7617GB-R (BamH I) | GTGGATCCCTAGTTCTTCTTTATTGAGG | |
| ACPL_8270GB-F | TTGAAGAGGTGACGTCATGACCGCTGAGCAGCTCATCTCGT | |
| ACPL_8270GB-R (BamH I) | GCGGATCCCTACCGGCCCCGGACCTCGTCGAT | |
| ACPL_8104GB-F | TTGAAGAGGTGACGTCATGGATGAGGTACTGGCGCG | |
| ACPL_8104GB-R (BamH I) | CTGGATCCTCAGACCCGGGCGCGCCGGGCGAGAC | |
| kasOp*-F (Xba I) | GCTCTAGATGTTCACATTCGAACGGTCTC | |
| kasOp*-R | GACGTCACCTCTTCAACTCAG | |
| ACPL_1889-LF (Xba I) | GCTCTAGAAAGGGCATCCTGCTGCTCGA | |
| ACPL_1889-LR (EcoR Ⅰ) | CGGAATTCGAATCCTCACGTCGCAGACA | |
| ACPL_1889-RF (EcoR Ⅰ) | CGGAATTCAAGAAGGCGTGACGGCGGACA | |
| ACPL_1889-RR (Hind Ⅲ) | CCAAGCTTCGTCGAGGTTGTACGACTCG | |
| ACPL_1889-YZ-F | TCTTGGAGAGCCTTCCGGGT | |
| ACPL_1889-YZ-R | TGGAAGCCGGCTATCCGATC | |
| ACPL_4236-LF (Xba I) | GCTCTAGACTGCGATGAACGATGTCGCG | |
| ACPL_4236-LR (EcoR Ⅰ) | CTGGATCCTCATGCCGTCACCCGCCCGGCCTCG | |
| ACPL_4236-RF (EcoR Ⅰ) | GAGAATTCATGCCGGTCTATGACAAACCGATG | |
| ACPL_4236-RR (Hind Ⅲ) | GCAAGCTTTCACCCGGCGGCGGCGGTCA | |
| ACPL_4236-YZ-F | GAGAATTCATGAAAATCTTGGTCACCGG | |
| ACPL_4236-YZ-R | GCAAGCTTTCAGGTCCACCAGGAACGGT | |
| ACPL_7303-LF (Xba I) | GAGAATTCGTGAGCAGGCAGGCCGACCT | |
| ACPL_7303-LR (EcoR Ⅰ) | CGGAATTCGCTTCATGCTCAGTCCTTCC | |
| ACPL_7303-RF (EcoR Ⅰ) | CGGAATTCACGCCCTCTGATCCCATGAC | |
| ACPL_7303-RR (Hind Ⅲ) | CCAAGCTTGATGTCGGGCAGCAGCAAGT | |
| ACPL_7303-YZ-F | ACCGTGTTCTCCCTGGTCGT | |
| ACPL_7303-YZ-R | CAGGTCGAGGTCGCTGTA | |
| ACPL_6479-LF (Xba I) | GCTCTAGACTGTCCAGGTCGTACGAGAC | |
| ACPL_6479-LR (EcoR Ⅰ) | CGGAATTCGTACTGTGCTGTGCCTTCCC | |
| ACPL_6479-RF (EcoR Ⅰ) | CGGAATTCGTTCTTCGCGAGCTGACGCGCT | |
| ACPL_6479-RR (Hind Ⅲ) | CCAAGCTTCCAACGCCGAGTACGAGATG | |
| ACPL_6479-YZ-F | CACGGGACGGGTCGTTAAAG | |
| ACPL_6479-YZ-R | CTACGAGCTGCTCTTCGAGC | |
| ACPL_7617-LF (Xba I) | GCTCTAGAGGAGGATCTCGGTCATGAGG | |
| ACPL_7617-LR (EcoR Ⅰ) | GTGAATTCCTTGACCAGACGTGGGTGC | |
| ACPL_7617-RF (EcoR Ⅰ) | GTGAATTCGAACTCTCCACCACCTCAA | |
| ACPL_7617-RR (Hind Ⅲ) | GCAAGCTTCTCGTCCATCAGCAGCACGTC | |
| ACPL_7617-YZ-F | GAGCCCTGGAACATCAGGTG | |
| ACPL_7617-YZ-R | CGATGTAGGTGGCGTCGATC | |
| ACPL_5445-LF (Xba I) | GCTCTAGAGTTGGACCACCACGTTGGAC | |
| ACPL_5445-LR (EcoR Ⅰ) | GTGAATTCCTGGATCGACTTGGTGTTCG | |
| ACPL_5445-RF (EcoR Ⅰ) | GTGAATTCGTCAAGGACAAGCAGGACGTC | |
| ACPL_5445-RR (Hind Ⅲ) | GCAAGCTTGTGTTGTCCAGCTCCTCGAC | |
| ACPL_5445-YZ-F | CTCTCTTGACGCGCACGATG | |
| ACPL_5445-YZ-R | CTGGTACACGGTGCTGATCC | |
| ACPL_3989-LF (Xba I) | GCACTAGTGGAACTGCAGGAGTCCTTCC | |
| ACPL_3989-LR (EcoR Ⅰ) | GTGAATTCCGTACGACCAGAAGGTGGTC | |
| ACPL_3989-RF (EcoR Ⅰ) | GTGAATTCCCTCGATGTACGTCTCGGTG | |
| ACPL_3989-RR (Hind Ⅲ) | GCAAGCTTGCTCGATGAACAGGTTGGC | |
| ACPL_3989-YZ-F | CGCATGTTCTAGCCGTGAAG | |
| ACPL_3989-YZ-R | GCTGATCGAGACGGACTACC | |
| ACPL_1889-EX-F (EcoR Ⅰ) | CGGAATTCATGGCAGCCACTGGCACAGCT | |
| ACPL_1889-EX-R (Hind Ⅲ) | CCAAGCTTTCACGCCTTCTTGCG | |
| ACPL_3989-EX-F (EcoR Ⅰ) | CTGAATTCGTGTCCATTGACCTGATCGCGGATTCACC | |
| ACPL_3989-EX-R (Hind Ⅲ) | GCAAGCTTCTAAAGCAGATTGAGGGCCCGC | |
| ACPL_4236-EX-F (EcoR Ⅰ) | CGGAATTCATGGGAAGGCCGCGAGCGTT | |
| ACPL_4236-EX-R (Hind Ⅲ) | CCAAGCTTTCACCGACAACGCTGATCTTGGC | |
| ACPL_5445-EX-F (EcoR Ⅰ) | CTGAATTCGTGCGCGTGAAGTTGATGGC | |
| ACPL_5445-EX-R (Hind Ⅲ) | GCAAGCTTTCAGTACTTGCGGGTCGGAAGCG | |
| ACPL_6479-EX-F (EcoR Ⅰ) | CTGAATTCATGCCGTCTGAGTACGCGAAGTCAC | |
| ACPL_6479-EX-R (Hind Ⅲ) | GCAAGCTTTCAGCTCGCGAAGAACGCCCG | |
| ACPL_7303-EX-F (EcoR Ⅰ) | CGGAATTCATGAAGCTGGTGACCGCGGT | |
| ACPL_7303-EX-R (Hind Ⅲ) | GCAAGCTTTCAGAGGGCGTCGAGGC | |
| ACPL_8104-EX-F (EcoR Ⅰ) | CGGAATTCATGGATGAGGTACTGGCGCG | |
| ACPL_8104-EX-R (Hind Ⅲ) | CCAAGCTTTCAGACCCGGGCGCGCCGGGCGAGAC | |
| ACPL_8270-EX-F (EcoR Ⅰ) | CTGAATTCATGACCGCTGAGCAGCTCATCTCGT | |
| ACPL_8270-EX-R (Hind Ⅲ) | GCAAGCTTCTACCGGCCCCGGACCTCGTCGAT | |
| ACPL_7617-EX-F (EcoR Ⅰ) | CTGAATTCGTGTACGTCTCGGCTCGCACGGACTA | |
| ACPL_7617-EX-R (Hind Ⅲ) | GCAAGCTTCTAGTTCTTCTTTATTGAGGTGGTGGAG | |
| hrdB-F | GAGGTCGAGCTCTCCAAGG | |
| hrdB-R | CGATCGAGACGACCAGTCG | |
| acbA-q-F | GTGCTGTCCATCGAGGAGAAAC | |
| acbA-q-R | GTTGACCTCGGTGATCTCCAG | |
| acbB-q-F | CCTCGTTTCAGCTTCGTTCG | |
| acbB-q-R | CGTCCAGTAGCACCTGAGTG | |
| acbW-q-F | CAGTCCTTCGGTGATGATGC | |
| acbW-q-R | GACATCACCAGCATCTGCG | |
| acbV-q-F | GACCACCTCACCACGCTC | |
| acbV-q-R | CACGTCCCAGTGCACCAG | |
| Pw-FAM-F | ACGATCATGCCGGTCGTGC | |
| Pw-R | CGCGACATTGTGGAAACTCGG | |
| Pv-F | CTGAGTGTGCGGCTCGCTG | |
| Pv-FAM-R | CGCCGGTGTCGAAACGAAC | |
| Pa-F | CCGCCGGTGACCAAGATTTT | |
| Pa-FAM-R | TCACCGGTCGAAGCCGTGAG | |
| Pb-FAM-F | GTTACAAAATGGGACCCG | |
| Pb-R | CCAGCAATATTCCGCGCAC | |
1.1.2 主要试剂和仪器:
实验所用限制性内切酶、反转录酶、EMSA试剂盒等购自Thermo Scientific公司;胶回收试剂盒购自Omega公司;各种抗生素购自生工生物工程(上海)股份有限公司;蛋白胨、酵母提取物等培养基相关试剂购自上海国药集团化学试剂有限公司;本研究所用引物和DNA测序由生工生物工程(上海)股份有限公司完成。本研究所用仪器主要包括安捷伦公司的液相色谱、伯乐生命医学产品有限公司的PCR仪、宁波新芝生物科技有限公司的超声破碎仪、德国耶拿分析股份公司的荧光定量PCR仪、GE公司的多功能激光扫描成像仪。
1.1.3 培养基和培养方法:LB培养基(g/L)蛋白胨10,氯化钠10,酵母提取物5。STY菌株活化培养基(g/L):蔗糖30,蛋白胨5,酵母提取物5,酪蛋白水解物1,磷酸氢二钾1,氯化钾0.5,硫酸亚铁0.05,琼脂20。SM培养基(g/L):葡萄糖15,麦芽糖10,麦芽提取物10,甘油10,蛋白胨5,酵母提取物5,磷酸氢二钾1,酪蛋白水解物1。SAM培养基(g/L):黄豆饼粉20,甘露醇20,琼脂20,pH 7.2。种子培养基(g/L):黄豆饼粉40,麦芽糖15,葡萄糖10,甘油10,可溶性淀粉10,碳酸钙2.5,pH 7.2。发酵培养基(g/L):麦芽糖50,葡萄糖30,谷氨酸3,磷酸氢二钾1,氯化铁0.5,黄豆饼粉10,碳酸钙2.5,pH 7.2。游动放线菌具体培养方法均参照文献[20]。
1.2 钓取调控蛋白 1.2.1 游动放线菌总蛋白提取:收集50mL发酵后的游动放线菌菌丝体,6500 r/min离心10 min后以10 mL buffer A (300 mmol/L NaCl,25 mmol/L Tris-HCl,pH 8.0)重悬,清洗2次,再以50 mL binding buffer (10%甘油,500 mmol/L NaCl,20 mmol/L Tris-HCl,1 mmol/L EDTA,pH 8.0)重悬后,使用超声破碎仪进行破碎,超声一个工作周期总时长5 min,超声3 s,暂停5 s,功率为40%,共重复3个工作周期。超声破碎结束后在4 ℃、12000 r/min离心25 min,取上清,重复离心1次,使用3 kDa超滤管超滤,得到浓缩的游动放线菌总蛋白。
1.2.2 DNA亲和层析:通过PCR扩增得到生物素标记的DNA探针,将100 μL亲和层析所用填料(GE公司的Streptavidin-agarose)离心弃去上清,用100 μL binding buffer重悬后与10 μg左右DNA探针混合均匀,离心弃去上清,加入1 mL binding buffer重悬后离心除去未与填料结合的DNA。将结合有探针的填料与150 mg左右的总蛋白均匀混合,同时加入250 μg左右鱼精DNA以减少非特异性蛋白的结合。将整个体系放置于4 ℃冰浴环境,旋转混匀1 h。离心,弃去上清,将沉淀重悬在1 mL binding buffer中,离心并去上清,重复1−2次。将结合有蛋白的DNA探针沉淀用100 μL的elution buffer (10% V/V甘油,20 mmol/L Tris-HCl,1 mol/L NaCl,1 mol/L EDTA,pH 6.8)重悬,离心,上清液即为与DNA探针相结合的蛋白。蛋白质谱检测由上海厚基生物科技有限公司完成,采用Label-free定量蛋白质组学分析技术,通过LC-MS/ MS技术对蛋白质酶解肽段进行质谱分析。
1.3 突变株构建 1.3.1 基因敲除:以ACPL_1889基因(本实验调控蛋白基因序列均可在NCBI数据库中获得,GenBank:CP003170.1)敲除菌株的构建为例。以游动放线菌SE50/110基因组DNA为模板,利用ACPL_1889-LF/R和ACPL_1889-RF/R两对引物,分别扩增出带有酶切位点的上游和下游同源臂。将测序正确的片段经酶切后连接至游离型载体pLQ752中,获得同源重组质粒pLQ1461。将重组质粒转入大肠杆菌ET12567(pUZ8002)中,采用菌丝体接合转移的方式[20]将质粒导入游动放线菌QQ-2中,于SFM培养基30 ℃培养5−7 d后,挑出接合子扩大培养,经转接2轮无抗性SM培养基松弛培养和双交换筛选,获得基因缺失突变候选株。提取基因组DNA,以ACPL_1889-YZ-F/R为引物进行PCR验证,得到突变株WXM-12。
1.3.2 基因强化表达:以ACPL_1889基因强化表达菌株的构建为例。以pDR4-K*质粒DNA为模板,利用引物kasOp*-F/R,通过PCR扩增得到126 bp的kasOp*启动子片段。以游动放线菌SE50/110基因组DNA为模板,利用引物ACPL_1889GB-F/R,通过PCR扩增得到228 bp的ACPL_1889基因片段。以获得的kasOp*启动子片段和ACPL_1889基因片段为模板,利用引物kasOp*-F/ACPL_1889GB-R,通过重叠PCR扩增得到354 bp的kasOp*-ACPL_1889基因片段。将测序正确的基因片段经酶切后连接至载体pSET152中,得到整合型质粒pLQ1450。将质粒转入大肠杆菌ET12567(pUZ8002)中,采用菌丝体接合转移的方式[20]将质粒导入QQ-2中。于SFM培养基30 ℃培养5−7 d后,挑出接合子扩大培养,经过抗性验证及提取基因组DNA和PCR验证后,得到基因强化表达突变株WXM-01。
1.4 突变株发酵将菌液涂布至STY平板30 ℃培养2−3 d后,将菌体接种至SM培养基,30 ℃、220 r/min培养30−36 h,转接4 mL至40 mL种子培养基中,30 ℃、220 r/min培养30−36 h,然后转接7.5 mL种子液至42.5 mL发酵培养基中,根据实验需求于30 ℃、220 r/min培养2−4 d.
1.5 RT-qPCR分析阿卡波糖生物合成基因的转录水平收取1 mL发酵48 h的菌体,离心弃去上清加入1 mL带有玻璃珠的Redzol溶液,在细胞破碎仪中破碎细胞。加入300 μL酚-氯仿,混合均匀于4 ℃、12000 r/min离心10 min,吸取上清,重复此步骤3–4直到蛋白几乎除净。加入200 μL无水乙醇,充分混匀后加入离心柱中,于4 ℃、12000 r/min离心2 min,弃去收集管内液体,向离心柱中心加入50 μL DEPC处理过的水,静置片刻后于4 ℃、12000 r/min离心2 min,获得总mRNA,反转录成cDNA。以cDNA为模板、编码RNA聚合酶Sigma因子的hrdB基因(GenBank No. AEV82165.1)作为内参进行RT-qPCR,使用SYBR Green为信号检测染料,具体计算分析方法参照文献[24]。
1.6 体外蛋白表达及纯化 1.6.1 体外蛋白菌株的构建:以ACPL_1889蛋白表达菌株的构建为例。以游动放线菌SE50/110基因组DNA为模板,利用引物ACPL_1889-EX-F/R,通过PCR扩增得到228 bp带有EcoR I/Hind Ⅲ的ACPL_1889基因片段。将测序正确的基因片段经酶切后连接至载体pET30a中,得到蛋白表达质粒pLQ1466。将蛋白表达质粒转入大肠杆菌BL21(DE3)中,获得蛋白表达菌株。
1.6.2 蛋白表达与纯化:将构建好的蛋白表达菌株在含50 mg/L卡那霉素的LB培养基中于37 ℃、220 r/min条件过夜培养,然后按2%的转接量转接至1 L含50 mg/L卡那霉素的LB培养基继续培养至OD600达到0.6−0.8时,加入终浓度为0.8 mmol/L的IPTG,在16 ℃条件下继续诱导培养18−20 h。然后离心收集菌体,用buffer A重悬菌体。利用超声破碎仪进行破碎,离心收集上清,经镍柱亲和层析后,以20 mL浓度为50 mmol/L的咪唑洗去杂蛋白,再以10 mL浓度为250 mmol/L的咪唑洗脱目的蛋白。经Millipore超滤管脱盐浓缩后,保存于10%甘油中。
1.7 凝胶阻滞实验以游动放线菌SE50/110基因组DNA为模板,利用FAM荧光标记的引物PW-FAM-F/PW-R,通过PCR扩增得到100 ng荧光标记的DNA探针。将获取的目的蛋白依次稀释到不同浓度(0−4000 nmol),配制EMSA反应体系,同时以1 μg非标记探针作为特异性竞争对照、1 μg poly(dI: dC)作为非特异性竞争对照,25 ℃反应30 min。在4 ℃、100 V下进行非变性聚丙烯酰胺凝胶电泳60 min,在多功能激光扫描仪中观察结合情况。
1.8 阿卡波糖产量分析取1 mL发酵液离心,收集上清,用ddH2O将上清稀释5倍后加入等体积氯仿,离心除去蛋白质杂质。取上清,经滤膜过滤,利用HPLC进行含量检测。使用Agilent phenomenex NH2柱(4.6 mm×250 mm,5 μm),流动相为65%乙腈-35%磷酸盐(0.6 g/L KH2PO4,0.704 g/L Na2HPO4·12H2O,100 mL乙腈),检测波长为210 nm。
2 结果和分析 2.1 钓取阿卡波糖生物合成基因簇的转录调控蛋白在SE50/110中,负责阿卡波糖生物合成的基因成簇分布,组成acb基因簇。其中PWV、PAB两个双向启动子控制了阿卡波糖生物合成基因的转录,PAB控制acbA与acbB的转录,PWV控制acbVUSRPIJKMLNOC及转运基因acbWXY的转录。但是在acb基因簇中未发现调控基因的存在,因此我们推测acb基因簇的转录很可能受到基因组中其他位置的调控因子的调控。基于上述假设,本研究利用PWV、PAB两个双向启动子区域的DNA作为探针(图 5-A),以亲和层析的方式钓取与启动子特异结合的蛋白,并结合蛋白组质谱分析,从中筛选出可能的转录调控因子。亲和层析后将洗脱液经SDS聚丙烯酰胺凝胶电泳和银染检测,初步判断是否存在蛋白及洗脱特异性。
将洗脱2次与3次的蛋白洗脱液分别进行蛋白组质谱检测,质谱鉴定结果显示,与PAB结合的蛋白共有228个,与PWV结合的蛋白共有220个。通过与KEGG数据库进行比对分析,在PAB结合蛋白中有23个、在PWV结合蛋白中有16个被注释为转录调控相关的蛋白;被直接注释为调控因子的蛋白共有14个,其中ACPL_566、ACPL_1896、ACPL_6780、ACPL_3083只与PAB启动子区域结合,ACPL_151只与PWV启动子区域结合,ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104、ACPL_3989、ACPL_5445、ACPL_7617、ACPL_8270与PAB、PWV两个启动子区域均结合,最终确定这9个蛋白(表 2)的编码基因为候选基因,进行后续的体内、体外功能验证。
| Proteins | Score | Putative gene products |
| ACPL_6479 | 79 | XRE-family transcriptional regulator |
| ACPL_5445 | 72 | HTH-type transcriptional repressor PurR |
| ACPL_1889 | 54 | Transcriptional regulator |
| ACPL_8104 | 51 | Regulatory protein HlyX |
| ACPL_7303 | 47 | Nitrogen regulatory protein P-Ⅱ |
| ACPL_8270 | 39 | GntR-family transcriptional regulator |
| ACPL_4236 | 35 | YezE-like HTH-type transcriptional regulator |
| ACPL_3989 | 23 | HTH-type transcriptional regulator MalT |
| ACPL_7617 | 16 | BadM/Rrf2-family transcriptional regulator |
| *score: The score of the results of mass spectrometry and the comparison results of the corresponding proteins in the database provided. | ||
2.2 候选基因体内功能分析 2.2.1 候选基因强化表达菌株构建及产量分析:
将ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104、ACPL_7617、ACPL_8270、ACPL_5445、ACPL_3989经PCR扩增后,引入强启动子kasOp*并克隆至载体pSET152中获得整合型质粒pLQ1450 (图 2-A)、pLQ1451、pLQ1452、pLQ1453、pLQ1454、pLQ1457、pLQ1458、pLQ1459、pLQ1460,采用菌丝体接合转移的方式将质粒导入游动放线菌QQ-2中,分别获得基因强化表达突变株WXM-01 (图 2-B)、WXM-02、WXM-03、WXM-04、WXM-05、WXM-08、WXM-09、WXM-10、WXM-11。将质粒整合菌株与突变株进行发酵,利用HPLC检测发酵液中阿卡波糖产量。结果显示(图 2-C),强化表达ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104,突变株的产量比整合有载体pSET152的对照菌株分别提高了25%、26%、18%、22%、15%;强化表达ACPL_5445、ACPL_7617、ACPL_3989、ACPL_8270,突变株的产量比对照菌株分别降低了12%、23%、39%、9%。
2.2.2 候选基因敲除菌株构建及产量分析:通过PCR扩增,分别获得ACPL_5445、ACPL_3989、ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8270、ACPL_8104、ACPL_7617的左右同源臂,酶切连接至pLQ752中构建同源重组质粒pLQ1455、pLQ1456、pLQ1461、pLQ1462、pLQ1463、pLQ1464、pLQ1465、pLQ1466、pLQ1467。采用菌丝体接合转移的方式将质粒导入游动放线菌QQ-2中,获得基因缺失突变株WXM-06、WXM-02、WXM-12、WXM-13、WXM-14、WXM-15、WXM-16。未能获得ACPL_8270、ACPL_8104这2个基因的敲除突变菌株,推测它们可能为必需基因。将出发菌株与突变株进行发酵,利用HPLC检测发酵液中阿卡波糖含量。结果显示(图 3-C),相较于出发菌株,ACPL_5445和ACPL_3989突变株的产量分别提高了13%、8%,ACPL_1889突变株的产量降低了22%,ACPL_7617、ACPL_7303、ACPL_4236、ACPL_6479突变株的产量无明显变化。
2.2.3 阿卡波糖生物合成基因转录水平分析:基因强化表达和敲除突变株产量结果显示,ACPL_1889可能是正调控因子,ACPL_5445和ACPL_3989可能是负调控因子,因此,利用qPCR分析这3个基因的突变株中阿卡波糖生物合成基因acbW、acbV、acbA和acbB的转录水平。收集发酵第2天的菌体进行RNA提取和qPCR分析。结果显示(图 4),与对照菌株相比,强化表达ACPL_3989使acbW、acbV、acbB的表达水平分别下调约4倍、2倍和5倍,敲除ACPL_3989使acbW和acbA的表达水平分别提高了约90倍和40倍;强化表达ACPL_1889使acbW、acbV、acbA的表达水平均提高了约2倍、acbB的表达水平提高了约6倍,敲除ACPL_1889使acbW、acbA、acbB的表达水平下调约2倍,acbV表达水平下调约9倍;强化表达ACPL_5445后acbW、acbV、acbA、acbB表达水平变化不大,而敲除ACPL_5445使acbW、acbV、、acbA、acbB的表达水平分别提高了约3倍、2倍、5.6倍和4.7倍。这些结果进一步表明,ACPL_1889对阿卡波糖的生物合成起到正调控作用,ACPL_5445和ACPL_3989对阿卡波糖的生物合成起到负调控作用。
2.3 候选调控蛋白的体外功能分析为实现候选调控蛋白的异源表达,首先利用pET30a载体构建ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104、ACPL_5445、ACPL_3989、ACPL_7617、ACPL_8270的表达质粒pLQ1468、pLQ1469、pLQ1470、pLQ1471、pLQ1472、pLQ1473、pLQ1474、pLQ1475、pLQ1476。将这些质粒导入BL21(DE3),经诱导表达和镍柱纯化得到可溶性蛋白。
按照1.6的实验方法,分别以FAM荧光标记的PWV和PAB中4个启动子区域DNA PW、PV、PA、PB (图 5-A)为探针,利用凝胶阻滞实验(EMSA)检测候选蛋白与启动子区域的结合特异性,以10倍浓度非标记启动子探针作为特异性竞争对照、poly (dI: dC)作为非特异性竞争对照。结果显示,ACPL_1889蛋白与PAB和PWV两个启动子DNA均能结合,发生迁移阻滞,随着蛋白浓度的增加阻滞越明显,加入特异性竞争性探针的阻滞明显减弱,加入非特异性竞争性探针的阻滞也有不同程度的减弱,说明ACPL_1889能特异性地结合PAB和PWV两个启动子DNA,但结合能力不强;ACPL_3989蛋白能与PAB启动子DNA结合,发生迁移阻滞,随着蛋白浓度的增加阻滞越强,加入特异性竞争性探针的阻滞消失,加入非特异性竞争性探针的阻滞无明显变化,说明ACPL_3989蛋白与PAB启动子DNA有特异性结合。
2.4 组合突变提高阿卡波糖产量候选调控蛋白的体内验证结果显示,强化表达ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104或者敲除基因ACPL_5445、ACPL_3989,突变株的产量提高了8%−26%。因此,为获取更高产的阿卡波糖突变株,首先将ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104这5个基因在pLQ752中串联,构建重组质粒PLQ1477 (图 6-A)。采用菌丝体接合转移的方式将质粒导入游动放线菌QQ-2中,获得突变株WXM-17。经HPLC检测,阿卡波糖产量相较于整合空载质粒的QQ-2提高了27%。在此基础上进一步敲除ACPL_5445、ACPL_3989,获得突变株WXM-18,其阿卡波糖产量相较于整合空载质粒的QQ-2提高了32% (图 6-B)。

|
| 图 6 阿卡波糖高产菌株的构建及产量 Figure 6 Construction of acarbose high-yield mutants and yields. A: recombinant plasmid pLQ1477 for the overexpression of 5 positive regulatory genes; B: yields of high-yield mutants. *: significant difference at P < 0.05; **: significant difference at P < 0.01; ***: significant difference at P < 0.001. Error bars, mean±SD (n=3 biological replicates). |
3 讨论
随着acb基因簇中各基因功能的解析,为阿卡波糖高产菌株的工程改造提供了基础与目标。然而,迄今为止,涉及阿卡波糖生物合成基因的转录调控因子的研究仍旧很少。虽然有研究发现在SE50/110中,蛋白AcrC能与acb基因簇中acbD和acbE之间的区域相结合,对acbD和acbE的转录有抑制作用,从而对阿卡波糖的早期合成有一定的调控作用,但acbD和acbE并不是阿卡波糖生物合成的关键基因,对阿卡波糖产量并未产生明显影响[25]。因此,本研究以acb基因簇中2个双向启动子区域PWV和PAB为探针,通过DNA亲和层析结合蛋白组质谱分析,挖掘基因簇外转录调控因子,并结合体内体外实验,以验证其对阿卡波糖生物合成的调控作用。
DNA亲和层析技术目前已在放线菌中被有效应用。曲爽等在吸水链霉井冈变种5008 (S. hygroscopicus var. jinggangensis 5008)中以启动子valK-valA-int为探针,钓取到全局性调控因子GlnR[14]。Ajith等也利用DNA亲和层析在S. peucetius 29050中寻找到一种具有二氢脂酰胺脱
氢酶(DLDH)活性的序列特异性DNA结合蛋白,该蛋白可能参与了柔红霉素生物合成的调控[26]。本实验中,通过DNA亲和层析和蛋白质组质谱分析,成功钓取到9个与阿卡波糖生物合成簇的2个双向启动子区域PWV、PAB均结合的调控因子ACPL_1889、ACPL_4236、ACPL_7303、ACPL_6479、ACPL_8104、ACPL_8270、ACPL_5445、ACPL_3989、ACPL_7617。
随后,在体内验证中,通过引入强启动子kasOp*和同源重组的方式分别构建基因强化表达和敲除突变株,对于ACPL_8270和ACPL_8104这2个基因,我们未能成功在QQ-2中敲除。近一步分析发现ACPL_8270与ACPL_8104这2个基因在基因组中的位置距离复制起点较近,且通过NCBI数据库比对发现,ACPL_8104同源调控因子Crp在S. coelicolor中缺失,会导致菌体形态学发育缺陷,还会导致次级代谢物生物合成的改变[27]。ACPL_8270属于GntR家族调控因子,这类调控因子在放线菌的初级和次级代谢中都发挥重要作用,有研究发现在S. coelicolor M145中缺失GntR家族调控因子dasR基因,突变株无法正常产生气生菌丝和孢子[28]。因此,我们判断ACPL_8270与ACPL_8104可能为游动放线菌SE50/110基因组中的必需基因。
综合体内体外实验验证,我们推断ACPL_1889可能为阿卡波糖生物合成正调控因子,ACPL_3989和ACPL_5445可能为2个负调控因子。目前,对于ACPL_1889蛋白家族及其同源蛋白的有关报道非常少。在本实验中,ACPL_1889能与acb基因簇中2个双向启动子PWV和PAB均结合,并对阿卡波糖生物合成基因转录及阿卡波糖产量具有一定的促进,强化表达基因ACPL_1889,阿卡波糖生物合成相关基因acbW、acbV、acbA、acbB的表达水平均提高,阿卡波糖产量提升25%,敲除该基因后,阿卡波糖的产量下降了22%,表现出正调控作用,其结构和作用机制有待进一步探究。ACPL_3989是HTH型调控因子MalT,其属于LuxR家族调控因子,不同的LuxR家族蛋白表现出复杂的调控机制,大部分为正调控因子,少部分具有负调控作用。在S. hygrospinosus var. beijingensis中,AniF能激活aniR-G基因的转录并参与茴香霉素生物合成[29]。本实验中,蛋白ACPL_3989能与启动子PAB区域结合并调控阿卡波糖生物合成基因的转录,强化表达基因ACPL_3989,阿卡波糖产量降低39%,敲除该基因后,阿卡波糖产量提高8%,表明其起到负调控作用。ACPL_5445属HTH型抑制因子PurR,PurR调控因子主要负责调控嘌呤生物合成操纵子基因purCSQLF和purDEK以及核苷ABC转运蛋白基因的转录[30−31]。本实验中,ACPL_5445虽未直接与阿卡波糖生物合成基因簇启动子区域结合,但强化表达基因ACPL_5445,阿卡波糖产量降低12%,敲除该基因后,阿卡波糖生物合成基因转录水平有所提高,阿卡波糖产量提高13%,表明其可能通过间接作用而发挥抑制功能。
本研究采用自下而上的研究方法,以acb基因簇的2个双向启动子为探针,利用DNA亲和层析技术钓取到与启动子区域结合的调控蛋白,并结合体内体外验证实验,寻找到阿卡波糖生物合成的调控因子,不但为揭示阿卡波糖生物合成的转录调控机制奠定了基础,而且这些调控基因的改造显著提升了阿卡波糖的产量。
| [1] |
Diabetes Society of Chinese Medical Association. Guidelines for the prevention and treatment of type 2 diabetes in China (2013 Edition). Chinese Journal of Diabetes Mellitus, 2014, 6(7): 447-498.
(in Chinese) 中华医学会糖尿病学分会. 中国2型糖尿病防治指南(2013年版). 中国糖尿病杂志, 2014, 6(7): 447-498. |
| [2] | Petersmann A, Nauck M, Müller-Wieland D, Kerner W, Müller UA, Landgraf R, Freckmann G, Heinemann L. Definition, classification and diagnosis of diabetes mellitus. Experimental and Clinical Endocrinology & Diabetes, 2018, 126(7): 406-410. |
| [3] | Hu FB, Manson JE, Stampfer MJ, Colditz G, Liu S, Solomon CG, Willett WC. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. The New England Journal of Medicine, 2001, 345(11): 790-797. DOI:10.1056/NEJMoa010492 |
| [4] |
Liu YG, Tian H, Xie XX, Shen XY, Chen CQ. Research progress in non-insulin drugs used for treating type 2 diabetes. Drugs & Clinic, 2013, 28(2): 108-113.
(in Chinese) 刘永贵, 田红, 解学星, 沈雪砚, 陈常青. 治疗2型糖尿病的非胰岛素类药物的研究进展. 现代药物与临床, 2013, 28(2): 108-113. |
| [5] |
Feng ZH, Wang YS, Zheng YG. Progress in biosynthesis pathway of acarbose. Biotechnology Bulletin, 2011(8): 60-67.
(in Chinese) 冯志华, 王远山, 郑裕国. 阿卡波糖的生物合成途径研究进展. 生物技术通报, 2011(8): 60-67. |
| [6] |
He ST, Xu JY, Chen DJ. Antidiabetic drugs with α-glucosidase inhibition activities. Industrial Microbiology, 2003, 33(1): 43-49.
(in Chinese) 何素婷, 许激扬, 陈代杰. 具有α-葡糖苷酶抑制作用的抗糖尿病药物. 工业微生物, 2003, 33(1): 43-49. DOI:10.3969/j.issn.1001-6678.2003.01.011 |
| [7] | Wehmeier UF, Piepersberg W. Biotechnology and molecular biology of the α-glucosidase inhibitor acarbose. Applied Microbiology and Biotechnology, 2004, 63(6): 613-625. DOI:10.1007/s00253-003-1477-2 |
| [8] | Zhang CS, Stratmann A, Block O, Brückner R, Podeschwa M, Altenbach HJ, Wehmeier UF, Piepersberg W. Biosynthesis of the C7-cyclitol moiety of acarbose in Actinoplanes species SE50/110:7-O-phosphorylation of the initial cyclitol precursor leads to proposal of a new biosynthetic pathway. Journal of Biological Chemistry, 2002, 277(25): 22853-22862. |
| [9] |
Zhang Y, Zhou CL. Application of metabolic regulation in the antibiotic biosynthesis of actinomycetes. Journal of China Pharmaceutical University, 2015, 46(4): 393-399.
(in Chinese) 张亚, 周长林. 代谢调控技术在放线菌生物合成抗生素的应用进展. 中国药科大学学报, 2015, 46(4): 393-399. |
| [10] | Gao C, Hindra, Mulder D, Yin C, Elliot MA. Crp is a global regulator of antibiotic production in streptomyces. mBio, 2012, 3(6): e00407-e00412. |
| [11] | Kang SH, Huang JQ, Lee HN, Hur YA, Cohen SN, Kim ES. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. Journal of Bacteriology, 2007, 189(11): 4315-4319. DOI:10.1128/JB.01789-06 |
| [12] | Wang J, Zhao GP. GlnR positively regulates NasA transcription in Streptomyces coelicolor. Biochemical and Biophysical Research Communications, 2009, 386(1): 77-81. DOI:10.1016/j.bbrc.2009.05.147 |
| [13] | 曲爽. 井冈霉素生物合成的调控机制. 上海交通大学博士学位论文, 2015. |
| [14] | Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Molecular Microbiology, 2008, 67(4): 861-880. DOI:10.1111/j.1365-2958.2007.06092.x |
| [15] | Zhang YY, Zou ZZ, Niu GQ, Tan HR. jadR* and jadR2 act synergistically to repress jadomycin biosynthesis. Science China Life Sciences, 2013, 56(7): 584-590. DOI:10.1007/s11427-013-4508-y |
| [16] | Schaffert L, Schneiker-Bekel S, Dymek S, Droste J, Persicke M, Busche T, Brandt D, Pühler A, Kalinowski J. Essentiality of the maltase AmlE in maltose utilization and its transcriptional regulation by the repressor AmlR in the acarbose-producing bacterium Actinoplanes sp. SE50/110. Frontiers in Microbiology, 2019, 10: 2448. |
| [17] | Zhu YP, Xu WH, Zhang J, Zhang PP, Zhao ZL, Sheng DH, Ma W, Zhang YZ, Bai LQ, Pang XH. A hierarchical network of four regulatory genes controlling production of the polyene antibiotic candicidin in Streptomyces sp. strain FR-008. Applied and Environmental Microbiology, 2020, 86(9): e00055-20. |
| [18] | Mao XM, Sun N, Zheng Y, Li YQ. Development of series of affinity tags in streptomyces. Scientific Reports, 2017, 7(1): 6854. |
| [19] | Yu P, Liu SP, Bu QT, Zhou ZX, Zhu ZH, Huang FL, Li YQ. WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Applied and Environmental Microbiology, 2014, 80(22): 6879-6887. DOI:10.1128/AEM.01849-14 |
| [20] | Zhao QQ, Xie HX, Peng Y, Wang XR, Bai LQ. Improving acarbose production and eliminating the by-product component C with an efficient genetic manipulation system of Actinoplanes sp. SE50/110. Synthetic and Systems Biotechnology, 2017, 2(4): 302-309. DOI:10.1016/j.synbio.2017.11.005 |
| [21] | Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). Journal of Bacteriology, 1999, 181(1): 204-211. |
| [22] | Wang WS, Li X, Wang J, Xiang SH, Feng XZ, Yang KQ. An engineered strong promoter for streptomycetes. Applied and Environmental Microbiology, 2013, 79(14): 4484-4492. |
| [23] | Bierman M, Logan R, O'Brien K, Seno ET, Nagaraja Rao R, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp.. Gene, 1992, 116(1): 43-49. |
| [24] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25(4): 402-408. |
| [25] | Wolf T, Droste J, Gren T, Ortseifen V, Schneiker-Bekel S, Zemke T, Pühler A, Kalinowski J. The MalR type regulator AcrC is a transcriptional repressor of acarbose biosynthetic genes in Actinoplanes sp. SE50/110. BMC Genomics, 2017, 18(1): 562. |
| [26] | Ajith VK, Prasad R. A novel protein that binds to dnrN-dnrO intergenic region of Streptomyces peucetius purified by DNA affinity capture has dihydrolipoamide dehydrogenase activity. Protein Expression and Purification, 2009, 67(2): 132-138. |
| [27] | Derouaux A, Halici S, Nothaft H, Neutelings T, Moutzourelis G, Dusart J, Titgemeyer F, Rigali S. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. Journal of Bacteriology, 2004, 186(6): 1893-1897. |
| [28] | Rigali S, Nothaft H, Noens EEE, Schlicht M, Colson S, Müller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Molecular Microbiology, 2006, 61(5): 1237-1251. |
| [29] | Shen JF, Kong LX, Li Y, Zheng XQ, Wang Q, Yang WN, Deng ZX, You DL. A LuxR family transcriptional regulator AniF promotes the production of anisomycin and its derivatives in Streptomyces hygrospinosus var. beijingensis. Synthetic and Systems Biotechnology, 2019, 4(1): 40-48. |
| [30] | Beyer NH, Roepstorff P, Hammer K, Kilstrup M. Proteome analysis of the purine stimulon from Lactococcus lactis. PROTEOMICS, 2003, 3(5): 786-797. |
| [31] | Martinussen J, Sørensen C, Jendresen CB, Kilstrup M. Two nucleoside transporters in Lactococcus lactis with different substrate specificities. Microbiology: Reading, England, 2010, 156(Pt 10): 3148-3157. |
 2021, Vol. 61
2021, Vol. 61




