中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Jinjin Li, Zhanjing Li, Jiajia Ni. 2021
- 李金金, 李占景, 倪加加. 2021
- Role and mechanism of intestinal microorganisms in the development of hepatocellular carcinoma and its feasibility as a diagnostic index
- 肠道微生物在肝细胞癌发生发展中的作用机制及其作为诊断指标的可行性
- Acta Microbiologica Sinica, 61(11): 3401-3412
- 微生物学报, 61(11): 3401-3412
-
文章历史
- 收稿日期:2021-02-03
- 修回日期:2021-04-12
- 网络出版日期:2021-08-18
2. Research and Development Center, Guangdong Meilikang Bio-Science Ltd., Dongguan 523000, Guangdong Province, China;
3. Dongguan Key Laboratory of Medical Bioactive Molecular Developmental and Translational Research, Guangdong Medical University, Dongguan 523808, Guangdong Province, China
2. 广东美立康生物科技有限公司研究与开发中心, 广东 东莞 523000;
3. 广东医科大学东莞市医学生物活性分子开发与转化研究重点实验室, 广东 东莞 523808
Numerous reports have assessed the relationship between intestinal microbiota (IM) and carcinogenesis in its various forms, including gastric, colorectal and breast cancer[1-4]. Due to the anatomical and functional connections between the gut and liver, the latter is heavily influenced by intestinal microorganisms and their components[5]. Moreover, the relationship between hepatocellular carcinoma (HCC) and IM has been discussed extensively in previous reports[6-8]. Furthermore, IM has been suggested to act as a molecular marker for early non-invasive diagnosis and serves as a therapeutic target in HCC[8-9].
While the relationship between IM and HCC and the molecular mechanism of IM-induced HCC are well understood, these findings have not been applied to the clinical diagnosis of HCC. In this review, the available information concerning the relationship between IM and HCC and the obstacles to the application of the former to the clinical diagnosis of HCC have been discussed. Moreover, we elucidate the role of IM as a feasible and useful indicator when diagnosing HCC.
1 Relationship between IM and HCC: phenomena and mechanismsChronic viral infection and liver cirrhosis are major causes of HCC, and both are closely related to the composition of the gut microbiota[5, 7, 10]. Ren et al.[8] analyzed 419 samples using high-throughput sequencing of the 16S rRNA gene and found that the fecal microbiota was richer in the phylum Actinobacteria as well as 13 genera, including Gemmiger, Parabacteroides and Paraprevotella, in early HCC as compared with those with liver cirrhosis. Meanwhile, the phylum Verrucomicrobia and 12 genera, including Alistipes, Phascolarctobacterium and Ruminococcus, were significantly decreased in early HCC compared to controls; however, 6 genera, including Klebsiella and Haemophilus, increased. It is worth noting that, although there were significant differences, the intra-group errors were very large[8]. Ponziani et al.[11] reported that the fecal microbiota of cirrhotic patients with HCC contained a higher abundance of Enterobacteriaceae and Streptococcus as well as a reduced proportion of Akkermansia compared to controls. Moreover, while Bacteroides and Ruminococcaceae were increased in the former, Bifidobacterium was found to be less abundant. Our previous study comparing HCC patients and healthy controls revealed that Desulfococcus, Enterobacter, Prevotella, Veillonella and many unidentified genera were significantly increased in all stages of HCC, while Cetobacterium was significantly reduced[12]. These results suggest that the association between the relative abundance of species in IM and HCC remains inconsistent.
Although Toffanin et al.[13] argued that IM and Toll-like receptor 4 (TLR4) do not contribute significantly to tumor initiation, IM plays a vital role in promoting the development of HCC. Increased intestinal permeability, bacterial overgrowth and the impaired ability of Kupffer cells to remove microbial products, can all lead to increased transport of IM and the metabolites thereof into the liver via portal vein circulation[14]. Increased intestinal permeability is found in many stages of chronic liver disease and cancer and can lead to increased lipopolysaccharide (LPS) levels in the circulatory system[15-18]. The translocation of bacterial components (such as LPS, peptidoglycan, and flagellin), otherwise known as pathogen-associated molecular patterns (PAMPs), triggers inflammatory responses via TLRs, the latter of which are found in most chronic liver diseases, including HCC[5]. TLRs are a class of proteins with the ability to recognize structurally conserved molecules derived from microbiota[19]. TLR4, a receptor of TLRs, is expressed by Kupffer and hepatic stellate cells (HSCs) as well as hepatocytes within the liver and induces inflammatory responses[20]. These responses ultimately lead to liver fibrosis which, in turn, can develop into HCC[10, 21-22]. Animal model-based studies have shown that an increase in the levels of bacterial LPS in cirrhotic livers activates HSCs and TLR4 in hepatocytes, thereby leading to fibrosis and regulation of epidermal growth factor secretion which, in turn, triggers tumor proliferation[7]. Moreover, liver inflammation, fibrosis, and HCC formation were inhibited by blocking TLR4 signal transduction in mice[15, 23-24]. Treatment with the TLR4 agonist LPS has been found to promote HCC development in mice, whereas germ-free status and non-absorbable antibiotics reduced hepatic inflammation, fibrosis, and HCC development[15]. Moreover, Toffanin et al.[13] found that continuous administration of low doses of LPS increased tumor number and size in conventional wild-type mice; meanwhile, systemic LPS levels and tumor overgrowth were reduced in sterile mice. Dapito et al.[23] found that the degree of hepatocarcinogenesis in chronically injured livers depended on the composition of the gut microbiota as well as the extent of TLR4 activation in non-bone- marrow-derived resident liver cells, including both hepatocytes and HSCs. These results were different from those of Yu et al.[15], who found that Kupffer cells were the main targets of LPS/TLR4 signals in the liver, playing a vital role in the induction of tumor necrosis factor α (TNFα) and IL-6. Significant reductions in cytokine production and complementary proliferation in response to diethylnitrosamine have been reported following the inactivation of Kupffer cells. Regarding HCC promotion, another key mechanism of the LPS-TLR4 axis is to prevent NF-κB B-mediated hepatocyte apoptosis. A negative correlation has been reported between tumor occurrence and the apoptosis marker caspase3 in sterile or TLR4-deficient mice[23]. Activation of the LPS-TLR4 signaling pathway in Kupffer cells has been found to cause TNF- and IL-6-dependent compensatory hepatocyte proliferation, oxidative stress, and a reduction in apoptosis[15]. Moreover, LPS appeared to activate TLR4 in HCC cell lines, enhancing their invasiveness and inducing epithelial-mesenchymal transition[25]. Other LTRs, such as TLR2 and TLR9, have also been associated with IM-induced liver injury. TLR2, TLR4, TLR9 and NLP3 receptors were prevented in experimental hepatic fibrosis in knockout mice[26]. The increase in TLR4 and TLR9 in portal vein circulation promoted the expression of liver TNFα, which has been found to drive the progression of non-alcoholic steatohepatitis[27]. Overall, intestinal dysbiosis leads to increased intestinal permeability due to the destruction of the intestinal barrier. This increase in intestinal permeability can promote the occurrence of HCC through the PAMP-TLR- mediated signaling pathway. Liver damage reduces the degradation of endotoxin and leads to endotoxin accumulation, thereby causing liver injury. Therefore, intestinal dysbiosis and HCC form a vicious circle: intestinal dysbiosis induces HCC, while HCC further aggravates intestinal dysbiosis (Figure 1).
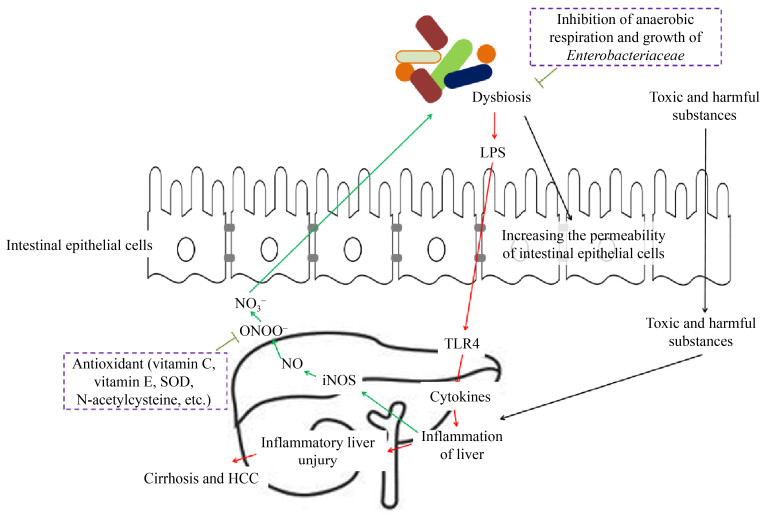
|
| Figure 1 Relationship between dysbiosis of gut microbiota and hepatocarcinogenesis. HCC: hepatocellular carcinoma; TLR4: Toll-like receptor 4; LPS: lipopolysaccharide. |
Yoshimoto et al.[28] found that mice consuming a high-fat diet experienced altered composition of the gut microbiota, resulting in increased production of deoxycholic acid, secondary bile acid, and metabolic byproduct of the gut microbiota shown to cause DNA damage. The enterohepatic circulation of deoxycholic acid provoked a senescence-associated secretory phenotype in HSCs. This phenotype led to the production of proinflammatory cytokines and tumor-promoting factors that have been found to promote HCC in the liver upon exposure to chemical carcinogens. Additionally, Ma et al.[29] reported that gut microbiome-mediated bile acid metabolism appeared to regulate liver cancer through hepatic CXCR6+ natural killer T cells.
2 The difficulties faced during clinical diagnosis of HCC using IMOwing to the distribution heterogeneity of IM as well as its sensitivity to external environmental factors, analysis is challenging, and the results are often inconsistent. Although there are several hundred species of bacteria living in the intestinal tract, their distribution is not uniform. The 50 most abundant bacterial cells often account for more than 70% of the total number of bacterial cells[12]. This heterogeneity makes it difficult to analyze changes in the composition of intestinal bacteria, especially in the species with low abundance. IM is affected by many factors, including diet[30-33], sex[34-36], geographical location[37], health status[38-39], and stochasticity[40]. Huang et al.[36] found that the IM of liver-specific tuberous sclerosis complex 1 knockout mice differed during the development of HCC based on sex. Specifically, female mice developed IM disorder earlier than did male mice. Lam et al.[41] suggested that diet could affect the composition of the IM, holding the potential to create a stable, healthy environment for the microbiota in the long term. He et al.[37] reported that regional differences in IM profoundly limited its use in clinical disease diagnosis. While factors such as sex can easily be excluded in clinical diagnosis, this is not true of all factors. For this reason, the patterns observed in animal models are not completely consistent with clinical results, making it difficult to apply such findings to the clinical auxiliary diagnosis of HCC.
Rodent models have previously been used to study the associations between gut microbiota and various host diseases[36, 42]. Usually, interfering factors, such as diet[2, 30, 32, 43], lifestyle[44-45] and genetic background[46-48], can be eliminated using animal models. Therefore, a positive result is more easily obtained in animal studies. However, caution is warranted when applying associations between gut microbiota and host diseases in animal models to clinical diagnoses. Clinically, it is very difficult to eliminate the interference of uncontrolled factors. The significant difference between the gut microbiota of rodent models and humans could be attributed to experimental errors[49].
3 A potential way to use IM for diagnosis of HCCUsing inclusive, comprehensive indicators instead of a single index may be a way to resolve the issues regarding the use of IM in the auxiliary diagnosis of HCC. Previous studies have used alpha diversity indices, the Firmicutes/Bacteroidetes and Bifidobacterium/Enterobacteriaceae ratios, and other indicators to analyze the association between IM and liver diseases. Wong et al.[50] suggested that the Firmicutes/Bacteroides ratio in feces could be used as a diagnostic indicator of nonalcoholic steatohepatitis. Lu et al.[51] proposed that the Bifidobacteria/Enterobacteriaceae ratio was indicative of the degree of biological imbalance present in the development of liver diseases. While positive results are indeed possible to obtain, it appears to be difficult to do so consistently. To circumvent this issue, we used a novel comprehensive indicator, namely the degree of dysbiosis, to measure the extent of the IM disorder. We then analyzed the correlation between the degree of dysbiosis and the extent of the deterioration caused by HCC. The results showed that the index was superior to the aforementioned indicators[12]. However, the indicator requires further refinement using subjects with differing IM bacterial compositions and HCC locations from which representative bacterial taxa can be obtained for the calculation of the degree of dysbiosis.
To enable the establishment of a comprehensive indicator, we offer the following suggestions.
(1) To ensure the universality of the final results, the samples used for the calculation should be selected from as broad a selection of locations as possible.
(2) Because many bacteria in the IM are difficult to culture and may not even have an official scientific name, it is preferable to use DNA sequences instead of species names as molecular markers. While this approach would likely be more troublesome, it is more convenient for the subsequent use of gene chips and other technical methods when investigating the relative abundances of each species.
(3) Although this review focused on the use of IM in the diagnosis of HCC, it is preferable to investigate the molecular markers of IM that possess the ability to distinguish between different cancers during screening to develop a comprehensive indicator, as the patterns of change in IM may vary based on the type of cancer[52]. The advantage of this approach is that the results obtained can also be used in the auxiliary diagnosis of other cancers.
(4) The formula warrants further refinement. We initially calculated the formula for the degree of dysbiosis as the difference between the sums of the logarithms of the relative abundances of 13 potentially harmful and 7 inherently pathogenic bacterial genera, respectively[12]. However, the results may be more pronounced when their respective ratios are used. It is generally believed that other metabolic diseases, such as obesity, type 2 diabetes, hypertension, irritable bowel syndrome, and steatohepatitis, are important factors that lead to chronic liver diseases and eventually develop into HCC[53]. Therefore, we should consider adding the intestinal microorganisms related to these diseases (as we showed in Table 1) into the calculation formula of the comprehensive indicator. This may be more helpful for the early diagnosis of HCC. Among these bacteria, Bifidobacterium and Lactobacilli protect against pathogen-induced NF-κB activation[54], thus inhibiting the local or systemic inflammatory response induced by endotoxin produced by Gram-negative bacteria, such as Bacteroides, Enterococcus and Escherichia, through the NF-κB pathway[55-57]. Besides, considering the heterogeneity of relative abundance of bacteria in intestinal microbiota, different molecular markers can be weighted to different extents.
| Taxa | Change of relative abundance | Related disease | Attribute | Metabolic characteristic | References |
| Actinobacteria | Decrease | IBS | - | - | [58] |
| Akkermansia | Decrease | Early HCC | - | Using host-derived mucins as carbon and nitrogen source and promotes barrier function partly by enhancing mucus production | [8] |
| Akkermansia muciniphila | Decrease | Obesity, diabetes and ameliorates, alcoholic liver | - | - | [59-61] |
| Bacteroides | Increase | IBS | Probiotic | Pro-inflammatory | [58, 62] |
| Bacteroides fragilis | Increase | CRC | Inherently pathogenic bacterium | - | [62-63] |
| Bacteroides fragilis | Decrease | Autism spectrum disorders | Probiotic | - | [64] |
| Bacteroides thetaiotaomicron | - | - | - | Promoting intestinal mucosal immune barrier and regulating balance of immune system | [65] |
| Bacteroidetes | Decrease | IBS | - | - | [58] |
| Bifidobacteria | Decrease | IBS | Probiotic | - | [58, 62] |
| Bifidobacterium | Decrease | IBS, HCC and liver cirrhosis | Probiotic | - | [58] |
| Bifidobacterium bifidum | - | - | Probiotic | - | [58] |
| Bifidobacterium breve Bb99 | - | - | Probiotic | - | [58] |
| Bifidobacterium infantis | - | - | Probiotic | - | [58] |
| Bifidobacterium longum | - | - | Probiotic | - | [58] |
| Citrobacter | - | - | Probiotic | - | [62] |
| Clostridiales | - | - | Probiotic | - | [62] |
| Clostridium | Increase | Liver tumor metastases | Inherently pathogenic bacterium | Increasing levels of secondary bile acids in the liver, which in turn promotes liver cancer | [29, 62, 66] |
| Clostridium IV | Decrease | Early HCC | - | Butyrate-producing bacterium | [8] |
| Collinsella | Decrease | IBS | - | - | [58] |
| Coprococcus | Decrease | IBS and early HCC | Probiotic | Butyrate-producing bacterium | [8, 58] |
| Dorea | Increase | IBS | - | - | [58] |
| Enterobacteriaceae | Increase | IBS | - | - | [58] |
| Enterococcus faecalis OG1RF | - | - | - | Causative effect of CRC | [1] |
| Escherichia | Increase | CRC | Inherently pathogenic bacterium | - | [62-63] |
| Escherichia coli NC101 | Increase | CRC | - | Causative effect of CRC | [1] |
| Eubacterium | - | - | Probiotic | - | [62] |
| Faecalibacterium | Decrease | IBS, early HCC | - | Butyrate-producing bacterium | [8, 58] |
| Firmicutes | Increase | IBS | - | - | [58] |
| Fusobacterium nucleatum | Increase | CRC | - | - | [62] |
| Haemophilus | Increase | Early HCC | - | LPS-producing bacterium | [8] |
| Klebsiella | Increase | Early HCC | - | LPS-producing bacterium | [8] |
| Lactobacilli | Decrease | IBS | Probiotic | - | [58, 62] |
| Lactobacillus | Decrease | IBS, HCC, and liver cirrhosis | Probiotic | - | [58, 62] |
| Lactobacillus acidophilus | - | - | Probiotic | - | [58] |
| Lactobacillus bulgaricus | - | - | Probiotic | - | [58] |
| Lactobacillus rhamnosus GG | - | - | Probiotic | - | [58] |
| Lactobacillus rhamnosus Lc705 | - | - | Probiotic | - | [58] |
| Leuconostoc | - | - | Probiotic | - | [62] |
| Methanobacteriales | Decrease | IBS | - | - | [58] |
| Oscillibacter | Decrease | Early HCC | - | Butyrate-producing bacterium | [8] |
| Prevotella | Decrease | IBS, and autism spectrum disorders | Inherently pathogenic bacterium | - | [58, 62, 64] |
| Propionibacterium freudenreichii spp. Shermanii JS | - | - | Probiotic | - | [58] |
| Proteobacteria | Increase | IBS | - | - | [58] |
| Ruminococcus | Increase | IBS | - | - | [58] |
| Ruminococcus | Decrease | Early HCC | Probiotic | Butyrate-producing bacterium | [8, 62] |
| Ruminococcus gnavus | Increase | IBS | - | Mucin degraders, and pro-inflammatory | [58] |
| Ruminococcus torques | Increase | IBS | - | Mucin degraders | [58] |
| Salmonella | - | - | Inherently pathogenic bacterium | - | [62] |
| Staphylococcus | - | - | Inherently pathogenic bacterium | - | [62] |
| Streptococci | Increase | IBS | - | - | [58] |
| Streptococcus gallolyticus | Increase | CRC | Inherently pathogenic bacterium | - | [62-63] |
| Streptococcus thermophilus | - | - | Probiotic | - | [58] |
| Veillonella | Increase | IBS | - | - | [58] |
| γ-proteobacteria | Increase | IBS | - | - | [58] |
| CRC: colorectal cancer; IBS: irritable bowel syndrome; LPS: lipopolysaccharide; HCC: hepatocellular carcinoma. | |||||
4 Conclusion
There is a close relationship between IM and HCC. The use of a comprehensive indicator of IM will enable the establishment of a unified index for clinical auxiliary diagnosis. However, this indicator offers scope for further refinement in future studies.
| [1] | Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science, 2012, 338(6103): 120-123. DOI:10.1126/science.1224820 |
| [2] | Feng Q, Liang SS, Jia HJ, Stadlmayr A, Tang LQ, Lan Z, Zhang DY, Xia HH, Xu XY, Jie ZY, Su LL, Li XP, Li X, Li JH, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang HM, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nature Communications, 2015, 6: 6528. DOI:10.1038/ncomms7528 |
| [3] | Qi YF, Sun JN, Ren LF, Cao XL, Dong JH, Tao K, Guan XM, Cui YN, Su W. Intestinal microbiota is altered in patients with gastric cancer from Shanxi Province, China. Digestive Diseases and Sciences, 2019, 64(5): 1193-1203. DOI:10.1007/s10620-018-5411-y |
| [4] | Alizadehmohajer N, Shojaeifar S, Nedaeinia R, Esparvarinha M, Mohammadi F, Ferns GA, Ghayour-Mobarhan M, Manian M, Balouchi A. Association between the microbiota and women's cancers-Cause or consequences?. Biomedicine & Pharmacotherapy, 2020, 127: 110203. |
| [5] | Tao XM, Wang N, Qin WX. Gut microbiota and hepatocellular carcinoma. Gastrointestinal Tumors, 2015, 2(1): 33-40. DOI:10.1159/000380895 |
| [6] | Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes, 2014, 5(4): 441-445. DOI:10.4161/gmic.29599 |
| [7] | Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nature Reviews Gastroenterology & Hepatology, 2017, 14(9): 527-539. |
| [8] | Ren ZG, Li A, Jiang JW, Zhou L, Yu ZJ, Lu HF, Xie HY, Chen XL, Shao L, Zhang RQ, Xu SY, Zhang H, Cui GY, Chen XH, Sun RR, Wen H, Lerut JP, Kan QC, Li LJ, Zheng SS. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut, 2019, 68(6): 1014-1023. DOI:10.1136/gutjnl-2017-315084 |
| [9] | Darnaud M, Faivre J, Moniaux N. Targeting gut flora to prevent progression of hepatocellular carcinoma. Journal of Hepatology, 2013, 58(2): 385-387. DOI:10.1016/j.jhep.2012.08.019 |
| [10] | Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nature Reviews Gastroenterology & Hepatology, 2018, 15(7): 397-411. |
| [11] | Ponziani FR, Bhoori S, Castelli C, Putignani L, Rivoltini L, Del Chierico F, Sanguinetti M, Morelli D, Paroni Sterbini F, Petito V, Reddel S, Calvani R, Camisaschi C, Picca A, Tuccitto A, Gasbarrini A, Pompili M, Mazzaferro V. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology, 2019, 69(1): 107-120. DOI:10.1002/hep.30036 |
| [12] | Ni JJ, Huang R, Zhou HF, Xu XP, Li Y, Cao PH, Zhong KB, Ge M, Chen XX, Hou BH, Yu M, Peng BG, Li Q, Zhang P, Gao Y. Analysis of the relationship between the degree of dysbiosis in gut microbiota and prognosis at different stages of primary hepatocellular carcinoma. Frontiers in Microbiology, 2019, 10: 1458. DOI:10.3389/fmicb.2019.01458 |
| [13] | Toffanin S, Cornella H, Harrington A, Llovet JM. HCC Is promoted by bacterial translocation and TLR-4 signaling: a new paradigm for chemoprevention and management. Hepatology, 2012, 56(5): 1998-2000. DOI:10.1002/hep.26080 |
| [14] | Sandler NG, Koh C, Roque A, Eccleston JL, Siegel RB, Demino M, Kleiner DE, Deeks SG, Liang TJ, Heller T, Douek DC. Host response to translocated microbial products predicts outcomes of patients with HBV or HCV infection. Gastroenterology, 2011, 141(4): 1220-1230. DOI:10.1053/j.gastro.2011.06.063 |
| [15] | Yu LX, Yan HX, Liu Q, Yang W, Wu HP, Dong W, Tang L, Lin Y, He YQ, Zou SS, Wang C, Zhang HL, Cao GW, Wu MC, Wang HY. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology, 2010, 52(4): 1322-1333. DOI:10.1002/hep.23845 |
| [16] | Lin RS, Lee FY, Lee SD, Tsai YT, Lin HC, Rei-Hwa L, Wan-Ching H, Huang CC, Wang SS, Kwang-Juei L. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophaegeal varices, and hyperdynamic circulation. Journal of Hepatology, 1995, 22(2): 165-172. DOI:10.1016/0168-8278(95)80424-2 |
| [17] | Zhang HL, Yu LX, Yang W, Tang L, Lin Y, Wu H, Zhai B, Tan YX, Shan L, Liu Q, Chen HY, Dai RY, Qiu BJ, He YQ, Wang C, Zheng LY, Li YQ, Wu FQ, Wang HY. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. Journal of Hepatology, 2012, 57(4): 803-812. DOI:10.1016/j.jhep.2012.06.011 |
| [18] | Nolan JP. The role of intestinal endotoxin in liver injury: a long and evolving history. Hepatology, 2010, 52(5): 1829-1835. DOI:10.1002/hep.23917 |
| [19] | Li DY, Yang M, Edwards S, Ye SQ. Nonalcoholic fatty liver disease. Journal of Parenteral and Enteral Nutrition, 2013, 37(6): 787-793. DOI:10.1177/0148607113481623 |
| [20] | Schuppan D, Afdhal NH. Liver cirrhosis. The Lancet, 2008, 371(9615): 838-851. DOI:10.1016/S0140-6736(08)60383-9 |
| [21] | Anand G, Zarrinpar A, Loomba R. Targeting dysbiosis for the treatment of liver disease. Seminars in Liver Disease, 2016, 36(1): 37-47. DOI:10.1055/s-0035-1571276 |
| [22] | Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. The Journal of Physiology, 2012, 590(3): 447-458. DOI:10.1113/jphysiol.2011.219691 |
| [23] | Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell, 2012, 21(4): 504-516. DOI:10.1016/j.ccr.2012.02.007 |
| [24] | Seki E, De Minicis S, Österreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nature Medicine, 2007, 13(11): 1324-1332. DOI:10.1038/nm1663 |
| [25] | Jing YY, Han ZP, Sun K, Zhang SS, Hou J, Liu Y, Li R, Gao L, Zhao X, Zhao QD, Wu MC, Wei LX. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Medicine, 2012, 10: 98. DOI:10.1186/1741-7015-10-98 |
| [26] | Fouts DE, Torralba M, Nelson KE, Brenner DA, Schnabl B. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. Journal of Hepatology, 2012, 56(6): 1283-1292. DOI:10.1016/j.jhep.2012.01.019 |
| [27] | Henao-Mejia J, Elinav E, Jin CC, Hao LM, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature, 2012, 482(7384): 179-185. DOI:10.1038/nature10809 |
| [28] | Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature, 2013, 499(7456): 97-101. DOI:10.1038/nature12347 |
| [29] | Ma C, Han MJ, Heinrich B, Fu Q, Zhang QF, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan WX, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science: New York, N Y, 2018, 360(6391): eaan5931. DOI:10.1126/science.aan5931 |
| [30] | Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science, 2011, 333(6038): 101-104. DOI:10.1126/science.1206025 |
| [31] | A Parnell J, A Reimer R. Prebiotic fiber modulation of the gut microbiota improves risk factors for obesity and the metabolic syndrome. Gut Microbes, 2012, 3(1): 29-34. DOI:10.4161/gmic.19246 |
| [32] | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature, 2014, 505(7484): 559-563. DOI:10.1038/nature12820 |
| [33] | Zhao LP, Zhang F, Ding XY, Wu GJ, Lam YY, Wang XJ, Fu HQ, Xue XH, Lu CH, Ma JL, Yu LH, Xu CM, Ren ZY, Xu Y, Xu SM, Shen HL, Zhu XL, Shi Y, Shen QY, Dong WP, Liu R, Ling YX, Zeng Y, Wang XP, Zhang QP, Wang J, Wang LH, Wu YQ, Zeng BH, Wei H, Zhang MH, Peng YD, Zhang CH. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science, 2018, 359(6380): 1151-1156. DOI:10.1126/science.aao5774 |
| [34] | Bridgewater LC, Zhang CH, Wu YQ, Hu WW, Zhang QP, Wang J, Li ST, Zhao LP. Gender-based differences in host behavior and gut microbiota composition in response to high fat diet and stress in a mouse model. Scientific Reports, 2017, 7: 10776. DOI:10.1038/s41598-017-11069-4 |
| [35] | Santos-Marcos JA, Rangel-Zuñiga OA, Jimenez-Lucena R, Quintana-Navarro GM, Garcia-Carpintero S, Malagon MM, Landa BB, Tena-Sempere M, Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F, Camargo A. Influence of gender and menopausal status on gut microbiota. Maturitas, 2018, 116: 43-53. DOI:10.1016/j.maturitas.2018.07.008 |
| [36] | Huang R, Li T, Ni J, Bai X, Gao Y, Li Y, Zhang P, Gong Y. Different sex-based responses of gut microbiota during the development of hepatocellular carcinoma in liver-specific Tsc1-knockout mice. Frontiers in Microbiology, 2018, 9: 1008. DOI:10.3389/fmicb.2018.01008 |
| [37] | He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, Chen MX, Chen ZH, Ji GY, Zheng ZDX, Mujagond P, Chen XJ, Rong ZH, Chen P, Lyu LY, Wang X, Wu CB, Yu N, Xu YJ, Yin J, Raes J, Knight R, Ma WJ, Zhou HW. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nature Medicine, 2018, 24(10): 1532-1535. DOI:10.1038/s41591-018-0164-x |
| [38] | Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiological Reviews, 2010, 90(3): 859-904. DOI:10.1152/physrev.00045.2009 |
| [39] | Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Medicine, 2011, 3(3): 1-12. |
| [40] | Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(11): E1326-E1332. DOI:10.1073/pnas.1414261112 |
| [41] | Lam YY, Zhang CH, Zhao LP. Causality in dietary interventions-building a case for gut microbiota. Genome Medicine, 2018, 10(1): 62. DOI:10.1186/s13073-018-0573-y |
| [42] | Xie GX, Wang XN, Liu P, Wei RM, Chen WL, Rajani C, Hernandez BY, Alegado R, Dong B, Li DF, Jia W. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget, 2016, 7(15): 19355-19366. DOI:10.18632/oncotarget.8466 |
| [43] | Moschen AR, Wieser V, Tilg H. Dietary factors: major regulators of the gut's microbiota. Gut and Liver, 2012, 6(4): 411-416. DOI:10.5009/gnl.2012.6.4.411 |
| [44] | Houghton D, Stewart CJ, Day CP, Trenell M. Gut microbiota and lifestyle interventions in NAFLD. International Journal of Molecular Sciences, 2016, 17(4): 447. DOI:10.3390/ijms17040447 |
| [45] | Shin JH, Sim M, Lee JY, Shin DM. Lifestyle and geographic insights into the distinct gut microbiota in elderly women from two different geographic locations. Journal of Physiological Anthropology, 2016, 35(1): 31. DOI:10.1186/s40101-016-0121-7 |
| [46] | Esworthy RS, Smith DD, Chu FF. A strong impact of genetic background on gut microflora in mice. International Journal of Inflammation, 2010, 2010(2010): 986046. |
| [47] | Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microbial Ecology, 2011, 61(2): 423-428. DOI:10.1007/s00248-010-9787-2 |
| [48] | Zhao LL, Wang G, Siegel P, He C, Wang HZ, Zhao WJ, Zhai ZX, Tian FW, Zhao JX, Zhang H, Sun ZK, Chen W, Zhang Y, Meng H. Quantitative genetic background of the host influences gut microbiomes in chickens. Scientific Reports, 2013, 3: 1163. DOI:10.1038/srep01163 |
| [49] | Arrieta MC, Walter J, Finlay BB. Human microbiota-associated mice: a model with challenges. Cell Host & Microbe, 2016, 19(5): 575-578. |
| [50] | Wong VWS, Tse CH, Lam TTY, Wong GLH, Chim AML, Chu WCW, Yeung DKW, Law PTW, Kwan HS, Yu J, Sung JJY, Chan HLY. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis-a longitudinal study. PLoS ONE, 2013, 8(4): e62885. DOI:10.1371/journal.pone.0062885 |
| [51] | Lu HF, Wu ZW, Xu W, Yang JZ, Chen YB, Li LJ. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Microbial Ecology, 2011, 61(3): 693-703. DOI:10.1007/s00248-010-9801-8 |
| [52] | Jia W, Rajani C, Xu HX, Zheng XJ. Gut microbiota alterations are distinct for primary colorectal cancer and hepatocellular carcinoma. Protein & Cell, 2021, 12(5): 374-393. |
| [53] | Ni J, Fu C, Huang R, Li Z, Li S, Cao P, Zhong K, Ge M, Gao Y. Metabolic syndrome cannot mask the changes of faecal microbiota compositions caused by primary hepatocellular carcinoma. Letters in Applied Microbiology, 2021, 73(1): 73-80. DOI:10.1111/lam.13477 |
| [54] | O'Mahony D, Murphy S, Boileau T, Park J, O'Brien F, Groeger D, Konieczna P, Ziegler M, Scully P, Shanahan F, Kiely B, O'Mahony L. Bifidobacterium animalis AHC7 protects against pathogen-induced NF-κB activation in vivo. BMC Immunology, 2010, 11: 63. DOI:10.1186/1471-2172-11-63 |
| [55] | Chen ZT, Li SL, Cai EQ, Wu WL, Jin JS, Zhu B. LPS induces pulmonary intravascular macrophages producing inflammatory mediators via activating NF-κB. Journal of Cellular Biochemistry, 2003, 89(6): 1206-1214. DOI:10.1002/jcb.10590 |
| [56] | Dong JS, Li JJ, Cui LY, Wang YF, Lin JQ, Qu Y, Wang H. Cortisol modulates inflammatory responses in LPS-stimulated RAW264.7 cells via the NF-κB and MAPK pathways. BMC Veterinary Research, 218, 14(1): 30. |
| [57] | Pinho-Ribeiro FA, Zarpelon AC, Mizokami SS, Borghi SM, Bordignon J, Silva RL, Cunha TM, Alves-Filho JC, Cunha FQ, Casagrande R, Verri WA Jr. The Citrus flavonone naringenin reduces lipopolysaccharide-induced inflammatory pain and leukocyte recruitment by inhibiting NF-κB activation. The Journal of Nutritional Biochemistry, 2016, 33: 8-14. DOI:10.1016/j.jnutbio.2016.03.013 |
| [58] | Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder?. American Journal of Physiology Gastrointestinal and Liver Physiology, 2017, 312(1): G52-G62. DOI:10.1152/ajpgi.00338.2016 |
| [59] | Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(22): 9066-9071. DOI:10.1073/pnas.1219451110 |
| [60] | Le Chatelier E, Nielsen T, Qin JJ, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li JH, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature, 2013, 500(7464): 541-546. DOI:10.1038/nature12506 |
| [61] | Grander C, Adolph TE, Wieser V, Lowe P, Wrzosek L, Gyongyosi B, Ward DV, Grabherr F, Gerner RR, Pfister A, Enrich B, Ciocan D, Macheiner S, Mayr L, Drach M, Moser P, Moschen AR, Perlemuter G, Szabo G, Cassard AM, Tilg H. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut, 2018, 67(5): 891-901. DOI:10.1136/gutjnl-2016-313432 |
| [62] | Oh CR. Correlation between dysbiosis of gut microbiota and human colorectal cancer. International Research Journal of Advanced Engineering and Science, 2018, 3(3): 226-231. |
| [63] | Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE, 2011, 6(1): e16393. DOI:10.1371/journal.pone.0016393 |
| [64] | Gilbert JA, Krajmalnik-Brown R, Porazinska DL, Weiss SJ, Knight R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell, 2013, 155(7): 1446-1448. DOI:10.1016/j.cell.2013.11.035 |
| [65] | Zocco MA, Ainora ME, Gasbarrini G, Gasbarrini A. Bacteroides thetaiotaomicron in the gut: molecular aspects of their interaction. Digestive and Liver Disease, 2007, 39(8): 707-712. DOI:10.1016/j.dld.2007.04.003 |
| [66] | Hartmann N, Kronenberg M. Cancer immunity thwarted by the microbiome. Science, 2018, 360(6391): 858-859. DOI:10.1126/science.aat8289 |
 2021, Vol. 61
2021, Vol. 61




