中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 靳海洋, 王慧, 张燕辉, 胡天龙, 林志斌, 刘本娟, 蔺兴武, 谢祖彬. 2021
- Haiyang Jin, Hui Wang, Yanhui Zhang, Tianlong Hu, Zhibin Lin, Benjuan Liu, Xingwu Lin, Zubin Xie. 2021
- 基于基因组的一株土壤固氮菌分离菌株鉴定及其促生作用
- Genome-based identification and plant growth promotion of a nitrogen-fixing strain isolated from soil
- 微生物学报, 61(10): 3249-3263
- Acta Microbiologica Sinica, 61(10): 3249-3263
-
文章历史
- 收稿日期:2021-01-10
- 修回日期:2021-02-15
- 网络出版日期:2021-08-05
2. 河南省农业科学院小麦研究所, 河南 郑州 450002;
3. 中国科学院大学, 北京 100049
2. Wheat Research Institute, Henan Academy of Agricultural Sciences, Zhengzhou 450002, Henan Province, China;
3. University of Chinese Academy of Sciences, Beijing 100049, China
生物固氮(Biological nitrogen fixation,BNF)是固氮微生物将氮气转化为生物可利用活性氮的过程,作为氮循环的重要组成部分,是氮素可利用转化的主要自然途径[1-4]。与化学氮肥相比,固氮微生物固定的氮素是农业生态系统中环境友好的活性氮来源。最大化地开发利用生物固氮作用,从而减少化学氮肥的施用量,有利于农业的可持续发展。开展固氮菌资源的收集及特性研究对于实现固氮菌资源有效利用、提高农田生物固氮能力具有重要的现实意义。
非共生固氮(asymbiotic nitrogen fixation)是不依靠微生物与植物形成共生关系的生物固氮形式,在陆地生态系统中广泛分布[3]。除固氮作用外,前人研究中分离获得的非共生固氮菌株大多可在体外检测到生长素的生成,部分菌株还具有铁载体生成和溶磷活性等促进植物生长潜力[5-6]。前人研究表明,接种固氮促生菌能够对甘蔗、水稻、玉米、小麦等作物的生长产生多方面有益作用[7-10]。在巴西、阿根廷等南美洲地区,商业化的固氮促生菌(固氮螺菌Azospirillum spp.)接种剂应用较为普遍,约有350万hm2的小麦、玉米和高粱等作物已被接种,且使用量还在不断增加[11-12]。在阿根廷,Azospirillum brasilense Az39用于玉米、小麦和其他非豆类作物的接种在全国范围内推广,在研究和应用推广中,增产效果得到了充分证实[13-14]。相比之下,国内的非共生固氮微生物接种剂产品较少且推广应用效果不够理想。
微生物菌株的分离培养工作是微生物学研究的基础,分离获得微生物菌株是微生物系统分类学和微生物资源利用的重要基础[15-16],纯培养物挖掘越来越受到国内外微生物研究学者的重视。近年来,国内研究学者从农田土壤、作物体内分离筛选得到了越来越多的非共生固氮菌种资源[17-19]。固氮酶基因的定量及无氮培养基培养计数的结果均表明,土壤中的固氮菌显著多于根系[20]。水稻土的淹水环境为固氮微生物提供了良好的生存环境[21-22],是固氮菌株分离的重要来源。课题组前期研究[23]表明,不同类型水稻土的生物固氮量差异很大,紫色土发育水稻土的固氮能力显著高于其他水稻土类型(红壤发育水稻土、黑土发育水稻土和下位砂姜土发育水稻土)。因此,本研究选用固氮能力较高的紫色土发育水稻土作为分离来源,通过富集纯化法分离固氮微生物菌株,采用基于基因组的方法对其进行物种水平鉴定,并对其固氮能力和促生作用进行研究,以评价其开发应用潜力,为研究利用生物固氮提供微生物资源。
1 材料和方法 1.1 土壤样品本研究分离土壤固氮菌的来源为紫色土发育水稻土,采自四川省盐亭县水稻田耕层土壤(31°16′14″N,105°28′27″E),根据课题组前期研究结果[23],紫色土发育水稻土的固氮能力较高,约为红壤发育水稻土的9.14倍,约为黑土发育水稻土的3.00倍,约为下位砂姜土发育水稻土的2.14倍。
1.2 培养基Brown无氮培养基[24] (g/L):葡萄糖5,无水CaCl2 0.15,MgSO4·7H2O 0.2,FeSO4·7H2O 0.04,Na2MoO4 0.005,K2HPO4 0.8,pH 6.8-7.0。固体培养基另加琼脂糖10-15 g/L。
1.3 菌株的分离采用富集纯化法分离高效固氮菌株。取10 g土壤样品加入90 mL液体无氮培养基中,28 ℃振摇培养4-5 d (180 r/min),取1%的培养物接种到100 mL液体无氮培养基中进一步培养,之后每2-3 d转接1次。重复转接4次后,在固体无氮培养基上采用稀释平板法分离,挑取单菌落划线纯化。
1.4 菌株基因组DNA提取挑取纯化的单菌落接种于液体无氮培养基中,28 ℃、180 r/min振摇培养至OD600≈1,吸取1 mL菌液于1.5 mL离心管,离心收集菌体(10000×g,10 min),用细菌基因组DNA提取试剂盒(TaKaRa,Code No. 9763)提取菌株基因组DNA,ddH2O溶解洗脱DNA。
1.5 菌株16S rRNA基因序列分析采用细菌通用引物27F (5′-AGAGTTTGATC MTGGCTCAG-3′)和1492R (5′-GGTTACCTTGTT ACGACTT-3′)[25-26]对16S rRNA基因全长序列进行PCR扩增。PCR反应中的DNA Polymerase、Buffer、dNTPs Mixture等使用TaKaRa Premix Taq (Ex Taq Version 2.0 plus dye) (TaKaRa,Code No. RR902A),其中包含:TaKaRa Ex Taq 0.05 U/μL、dNTPs Mixture 0.4 mmol/L、Mg2+ 4 mmol/L。PCR反应体系包含:25 μL TaKaRa Premix Taq、50-100 ng基因组DNA、1 μL正向引物(20 μmol/L)、1 μL反向引物(20 μmol/L)、ddH2O补充至50 μL。PCR反应条件为:94 ℃ 5 min;94 ℃ 1 min,55 ℃ 1 min,72 ℃ 1.5 min,30个循环;72 ℃ 5 min。
PCR扩增后,以DL2000 (TaKaRa,Code No. 3427Q)为DNA Marker,对扩增产物进行1%琼脂糖凝胶电泳。目的DNA片段回收纯化后,通过3730×l DNA Analyzer (Applied Biosystems)进行测序,测序结果使用SeqMan (DNASTAR Lasergene)拼接。使用EzBioCloud Identify service (https://www.ezbiocloud.net/identify)[27]将所得序列与数据库进行比对。
从数据库中收集相似度较高或相近科属的模式菌株16S rRNA基因序列以重建系统发育树,在MEGA 7中采用Neighbor-Joining方法进行系统发育树构建[28-29]。Neighbor-Joining建树模式为Tamura 3-parameter model,使用Bootstrap进行检验评估,设置复制值为1000次。
1.6 菌株全基因组测序与相关指数比较使用NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs)进行文库构建。利用Illumina HiSeq测序平台进行2×150 bp测序,对测序数据通过FastQC进行质量评估,通过Trimmomatic 0.36进行质量剪切[30]。使用SPAdes 3.5.0进行基因组拼接[31],采用NCBI prokaryotic genome annotation pipeline (PGAP)进行基因组注释[32]。
从公共数据库中收集了相近模式菌株的基因组序列,使用Up-to-date bacterial core gene set and pipeline (UBCG)重建基于基因组92个核心基因的系统发育树[33],通过基因支持指数(gene support index,GSI)评估系统发育分支。
测序获得新分离菌株P208的全基因组序列(GCA_005144545.1),从公共数据库收集密切相关的模式菌株Azotobacter chroococcum ATCC 9043T (GCA_004327905.1)和A. beijerinckii DSM 378T (GCA_900110885.1)基因组序列作为对比,计算菌株之间的全基因组相关指数[34]。使用JSpeciesWS Online Service (http://jspecies.ribohost.com/jspeciesws/)计算平均核苷酸一致性(average nucleotide identity,ANI)和四核苷酸频率相关系数(tetranucleotide frequency correlation coefficient,TETRA)[35]。使用Genome-to-Genome Distance Calculator 2.1 (http://ggdc.dsmz.de/ggdc.php)计算数字DNA-DNA杂交值(digital DNA- DNA hybridization,dDDH)与G+C含量差异[36]。利用在线工具(http://enve-omics.ce.gatech.edu/aai/)计算平均氨基酸一致性(average amino acid identity,AAI)[37]。利用INRA-MUMi在线工具(http://genome.jouy.inra.fr/mumi/)计算最大唯一匹配指数(maximal unique matches index,MUMi)[38]。
1.7 菌株固氮能力的定量测定采用乙炔还原法和15N2示踪法测定对数生长中期(OD600≈0.4-0.8)菌液的固氮能力,菌液蛋白量采用Bradford法测定[39],牛血清蛋白(BSA)作为蛋白标准。选择固氮菌属模式菌株A. chroococcum ATCC 9043T作为对照。
对于乙炔还原法,将对数生长中期菌液转移到无菌玻璃瓶中。置换顶空10%的空气为高纯乙炔。黑暗条件下28 ℃、180 r/min振摇培养2 h,采集瓶内顶空气体,气相色谱法测定顶空乙烯产生情况[40],同时做无菌培养基对照和无乙炔置换对照。
对于15N2示踪法,将对数生长中期菌液转移到无菌玻璃瓶中。置换顶空10%的空气为高纯15N2。黑暗条件下28 ℃、180 r/min振摇培养2 h,通过Whatman玻璃微纤维过滤器(GF/F) (GE Healthcare,CAT No.1825-047)过滤收集细胞,烘干称重研磨后采用元素分析仪-同位素比质谱仪(EA-IRMS) (Flash 2000-Delta V advantage,Thermo Fisher Scientific)测定氮含量与15N丰度[41]。
1.8 菌株促生潜力体外测定对新分离菌株的IAA生成、溶磷活性和铁载体生成进行定量分析,并与固氮菌属模式菌株A. chroococcum ATCC 9043T进行对比。
1.8.1 IAA生成测定:菌株振摇培养于含有500 mg/L L-色氨酸的液体Brown培养基中(28 ℃、180 r/min),48 h后取菌液离心(10000×g,10 min),取上清液1 mL,加入显色液(10.8 mol/L H2SO4,4.5 g/L FeCl3) 1 mL混合均匀,室温避光反应30 min后测定其OD540。同时以吲哚-3-乙酸标准液绘制标准曲线[42]。
1.8.2 溶磷活性测定:菌株振摇培养于溶磷NBRIP培养基[43]中(28 ℃、180 r/min),7 d后取菌液离心(10000×g,10 min),取上清液用钼蓝比色法测定菌液中的可溶性磷含量[44-45]。
1.8.3 铁载体生成测定:菌株振摇培养于无铁的液体Brown培养基中(28 ℃、180 r/min),72 h后取菌液离心(10000×g,10 min),取上清液1 mL,加入CAS检测液[46]1 mL混合均匀,室温反应1 h后测定OD630,以未接菌培养基作为参比对照。
1.9 菌液接种对作物生长的影响接种液准备:将无氮培养基中28 ℃、180 r/min振摇培养24 h的菌液离心(5000×g,5 min)、无菌水漂洗后重悬,调节菌浓度约为108 CFU/mL。
水稻(武运粳23)、小麦(宁麦14)和玉米(伟科966)种子表面灭菌处理(70%乙醇1 min,2%次氯酸钠溶液5 min,无菌水冲洗5次)后在湿润滤纸上28 ℃黑暗条件下预发芽。2-3 d萌动后,在接种液(对照处理为无菌水)中浸泡3 h[47-48]。挑选萌发状态一致的萌发种子种植在Hoagland[48] 1%琼脂平板上,每个平板另加接种液(对照处理为无菌水) 1 mL,在平板上加一层无菌水防止琼脂干裂,28 ℃恒温光照培养箱中光暗交替(14:10)培养,每个处理3次重复,2周后取样测定根长、株高、根干重和茎叶干重。
2 结果和分析 2.1 分离菌株的形态与生长从固氮能力较强的紫色土发育水稻土富集培养物中分离得到菌株P208。在固体无氮培养基上,菌株P208菌落呈黏液状、表面光滑,培养5 d后菌落中产生棕黑色非水溶性色素。菌株P208细胞形态呈卵圆形,革兰氏阴性,约(2-3) μm× (4-5) μm,常成对排列(图 1)。菌株P208在固体无氮培养基上生长较快,在28 ℃条件下划线培养2 d后即可长出稳定的单菌落,1%接种于液体无氮培养基中培养18-20 h后达到对数生长中期(28 ℃、180 r/min)。
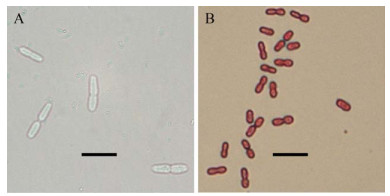
|
| 图 1 菌株P208的细胞形态(Bar=10 μm) Figure 1 Cellular morphology of strain P208 (Bar=10 μm). A: phase-contrast microscopy; B: Gram staining. |
2.2 16S rRNA基因序列相似性及系统发育分析
菌株P208的16S rRNA基因序列(MK841645)与A. chroococcum IAM 12666T(=ATCC 9043T) (AB175653,相似度为99.79%)、A. beijerinckii ATCC 19360T (=DSM 378T) (AJ308319,相似度为98.66%)具有相对较高相似度,高于物种分类的阈值(> 98.65%)[49]。与其他模式菌株16S rRNA基因序列相似度较低(< 97.1%)。
基于16S rRNA基因的系统发育分析,菌株P208与固氮菌属(Azotobacter) 7个物种的模式菌株聚集形成1个独立的簇,表明菌株P208应归于固氮菌属(Azotobacter) (图 2)。另外,菌株P208与A. chroococcum IAM 12666T (=ATCC 9043T)系统发育距离最近,其次为A. beijerinckii ATCC 19360T (=DSM 378T)。

|
| 图 2 基于16S rRNA基因的系统发育树 Figure 2 Phylogenetic tree based on 16S rRNA gene sequences. |
2.3 基因组核心基因系统发育分析
基于基因组92个核心基因的系统发育分析结果与16S rRNA基因具有一致性,证实了菌株P208与A. chroococcum ATCC 9043T的系统发育距离最近,且形成了1个区别于A. beijerinckii DSM 378T的独立分枝(GSI=90,最大值为92) (图 3)。
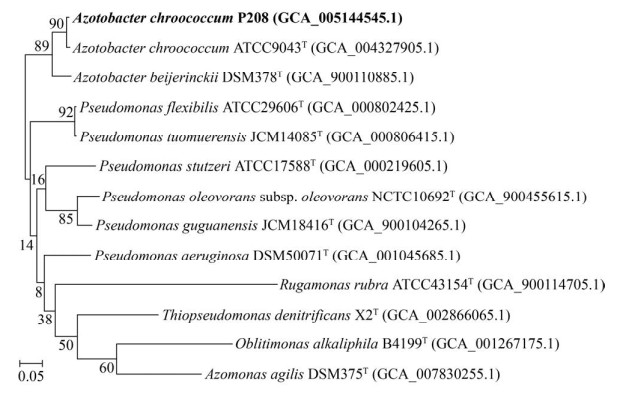
|
| 图 3 基于基因组92个核心基因的系统发育树 Figure 3 Phylogenetic tree based on UBCGs (concatenated alignment of 92 core genes). Gene support indices (GSIs) values (maximal value is 92) are given next to the branches. |
2.4 基因组特征与相关指数比较
菌株P208全基因组项目GenBank/EMBL/DDBJ登录号为SWKB00000000 (表 1)。测序组装后得到的基因组大小为5188530 bp,G+C含量为66.0 mol%。测序所得Contig 247个、N50长度49173 bp、测序深度452×,采用NCBI PGAP注释共预测出5067个基因,其中蛋白质编码基因4714个,RNA基因64个,伪基因289个。其中,64个RNA基因包括rRNA基因(5S/16S/23S) 6个、tRNA基因54个和ncRNA基因4个。通过对基因组注释结果中的固氮相关基因进行统计,新分离菌株预测得到的固氮酶基因、固氮酶结构和功能相关基因有32个,测序的基因组将为进一步研究功能基因、代谢途径和比较基因组学提供基础。
| Attributes | Values |
| GenBank accession | SWKB00000000 |
| Genome size/bp | 5188530 |
| G+C content/(mol%) | 66.0 |
| Contigs | 247 |
| N50/bp | 49173 |
| Genome coverage/× | 452 |
| Total genes | 5067 |
| Protein-coding genes | 4714 |
| RNA genes | 64 |
| rRNA genes | 6 |
| tRNA genes | 54 |
| ncRNA genes | 4 |
| Pseudo genes | 289 |
为进一步明确菌株P208在物种水平的系统分类地位,对其和密切相关的模式菌株A. chroococcum ATCC 9043T (GCA_004327905.1)、A. beijerinckii DSM 378T (GCA_900110885.1)进行基因组相关指数比较(表 2)。结果表明,菌株P208与A. chroococcum ATCC 9043T的平均核苷酸一致性(ANI)、平均氨基酸一致性(AAI)和数字DNA-DNA杂交值(dDDH)均高于物种分类的阈值(ANI > 95%-96%,AAI > 95%-96%,dDDH > 70%)[34, 50-51],而与A. beijerinckii DSM 378T的ANI、AAI、dDDH均低于物种分类的阈值。菌株P208与A. chroococcum ATCC 9043T的最大唯一匹配指数(MUMi)低于物种分类的阈值(< 0.33)[38],而与A. beijerinckii DSM 378T的MUMi值高于物种分类的阈值。同时,基因组G+C含量差异和四核苷酸频率相关系数(TETRA)均在同一物种的变异范围之内(分别为 < 1%、> 0.99)[50, 52]。因此,结合其形态特征,菌株P208可鉴定为褐球固氮菌(A. chroococcum)。
| Genomic metrics | Strains | |
| Azotobacter chroococcum ATCC 9043T | A. beijerimckii DSM 378T | |
| ANI/% | 96.82 | 89.87 |
| AAI/% | 96.05 | 89.19 |
| dDDH/% | 73.1-78.9 | 39.6-44.6 |
| MUMi | 0.268 | 0.583 |
| Difference in G+C content/% | 0.16 | 0.37 |
| TETRA | 0.9995 | 0.9971 |
| Species circumscription thresholds: ANI (> 95%-96%), AAI (> 95%-96%), dDDH (> 70%), MUMi (< 0.33), difference in G+C content (< 1%), TETRA (> 0.99). | ||
2.5 菌株的固氮能力
对数生长中期菌液在10%乙炔中培养2 h后,即可检测到较高的乙烯峰。定量结果显示,A. chroococcum P208的乙烯生成速率显著高于模式菌株A. chroococcum ATCC 9043T (P < 0.01) (图 4-A)。15N2示踪法测定固氮活性的结果与乙炔还原法测定结果一致,菌株P208的固氮活性为136.38 nmol N2/(mg protein·min),约为模式菌株A. chroococcum ATCC 9043T的2.61倍(图 4-B)。

|
| 图 4 新分离菌株与模式菌株的固氮能力 Figure 4 Nitrogen fixation activity of newly isolated strain and type strain. A: acetylene reduction assay; B: 15N2 tracer method. Error bars represent standard deviations (n=3). The different letters above bars indicate significant differences at P < 0.01 level according to LSD. |
2.6 体外促生潜力
通过定量分析,A. chroococcum P208具有IAA生成、溶磷活性和铁载体生成等促进植物生长潜力的培养特性(表 3)。A. chroococcum P208的IAA和铁载体生成能力显著高于模式菌株A. chroococcum ATCC 9043T (P < 0.05)。
| Strains | IAA/(μg/mL) | Siderophore/% | Phosphate solubilization/(μg/mL) |
| Azotobacter chroococcum ATCC 9043T | 12.47±0.28 b | 48.91±7.26 b | 1.29±0.14 a |
| A. chroococcum P208 | 18.48±0.48 a | 88.21±4.61 a | 1.32±0.16 a |
| Values are means±standard deviations (n=3). Data followed by the same letters in the same column were not significantly different at P < 0.05 level according to LSD. | |||
2.7 菌液接种对作物幼苗生长的影响
如表 4所示,与未接种对照处理相比,接种A. chroococcum P208显著促进了水稻、小麦幼苗根系的生长(P < 0.05),小麦根长增加41.3%、根干重增加42.3%,水稻幼苗根系生长受接种的影响更加显著,水稻根长增加134.5%、根干重增加43.5%。接种A. chroococcum P208对小麦、水稻幼苗的影响未体现在株高和茎叶干重的变化上。另外,玉米幼苗的生长指标在未接种对照和接种A. chroococcum P208处理间差异未达显著水平。
| Crops | Treatments | Root length/cm | Shoot length/cm | Root dry weight/(mg/plant) | Shoot dry weight/(mg/plant) |
| Rice | Control | 2.9±0.6 b | 12.4±0.5 a | 4.6±0.4 b | 11.9±0.2 a |
| Inoculation with P208 | 6.8±0.9 a | 12.2±1.2 a | 6.6±0.5 a | 11.0±1.1 a | |
| Wheat | Control | 10.9±1.6 b | 17.7±2.0 a | 9.7±0.8 b | 29.3±5.4 a |
| Inoculation with P208 | 15.4±1.0 a | 19.2±0.7 a | 13.8±0.9 a | 31.8±0.7 a | |
| Maize | Control | 18.7±1.7 a | 22.3±1.9 a | 67.1±8.2 a | 72.9±3.0 a |
| Inoculation with P208 | 18.2±2.0 a | 22.0±1.1 a | 67.3±9.3 a | 71.9±11.5 a | |
| Values are means±standard deviations (n=3). Data followed by the same letters in the same column for each crop were not significantly different at P < 0.05 level according to LSD. | |||||
3 讨论
选择特定的生境和样品作为分离来源,有利于分离出特定功能的微生物菌株。在本研究中,从固氮能力较高的土壤样品中分离获得了固氮能力较高的A. chroococcum P208,不同的培养基和培养条件能分离出不同的微生物菌株[53-54],耐酸、耐盐、耐氨、耐氧压和碳源需求广的固氮微生物菌株能够增加菌株的接种成活率和预期效果,更多培养基和培养条件下的分离筛选工作还有待进一步进行。
16S rRNA基因是编码原核生物一种rRNA的基因序列,基于其高度保守性和特异性,常被应用于原核生物的物种鉴定中[55]。随着测序技术的不断进步,基因组测序成本逐渐降低,在微生物分类鉴定中普遍采用基因组测序数据成为可能[56]。以平均核苷酸一致性(ANI)和数字DNA-DNA杂交值(dDDH)为代表的全基因组相关指数(OGRI)逐渐成为原核生物物种划分的金标准[50]。实际操作中,甚至没必要测定鉴定菌株的完整基因组序列,随机测序20%以上基因组进行分析就可以完成鉴定[50]。尽管16S rRNA基因保守性强,无法区分亲缘关系极近的菌株,但仍然是原核生物快速鉴定的重要方法,目前主要结合16S rRNA基因相似性比对和全基因组序列比较进行原核生物的物种鉴定,16S rRNA基因序列相似性97% (98.65%)[49, 57]被广泛用作物种分类的阈值。固氮菌属(Azotobacter)物种之间16S rRNA基因的相似度分析结果(图 5)显示,固氮菌属物种的16S rRNA基因具有较高的保守性,A. paspali ATCC 23833T和A. vinelandii IAM 15004T之间的16S rRNA基因相似度高达98.97%。因此,16S rRNA基因相似度97% (甚至98.65%)作为物种鉴定的阈值不适用于固氮菌属。在本研究中,菌株P208和A. beijerinckii ATCC 19360T之间的16S rRNA基因相似度高于物种分类的阈值,但它们最终被归类为不同的物种。前人研究中,16S rRNA基因序列相似度 > 99%的菌株也有分属于不同物种的可能[58-59]。该结果再次证明了16S rRNA基因相似度 > 97% (甚至98.65%)进行物种水平鉴定的不确定性,以及全基因组序列比较(如ANI、dDDH)对物种鉴定的必要性。

|
| 图 5 固氮菌属物种之间16S rRNA基因的相似度与系统发育分析 Figure 5 Pairwise sequence similarities and phylogenetic analyses based on 16S rRNA gene of Azotobacter spp.. |
乙炔还原法是简单快速测定固氮能力的方法[60],在使用中常通过系数R值(R=乙烯生成速率/固氮速率)将乙炔还原活性换算为固氮活性,前人研究中多使用R=3作为理论换算系数[61]。本研究同时采用乙炔还原法和15N2示踪法测定菌株的固氮能力(图 4),两种方法测定结果的换算系数R值为4.90-5.74,高于理论R值。前人研究中,不同条件下测定的R值与理论R值也存在明显偏差,变化幅度从小于1到大于30,大多数报道中R值为2-7[40, 62]。在大多数情况下,两种方法测定结果具备较好的一致性,而固氮速率较低时,测定结果差异较大[62]。因此,通过乙炔还原法定量测定固氮能力时,应同时结合15N2示踪法确定换算系数R值。
固氮促生菌在农业中的应用有助于减少农用化学品的使用,降低生产成本,增加经济效益和环境效益,利于生态农业的发展[63-65]。植物促生菌促进植物生长的机制主要包括:生物固氮、磷等元素的增溶和矿化、生长素等植物激素的生成、ACC脱氨酶的生成、铁载体的生成和抗生素的生成等[66]。同一物种的不同菌株其促生潜力等特征存在显著差异[66],而不同菌株的比较应在同一培养条件下进行,本研究选用模式菌株A. chroococcum ATCC 9043T作为参比,在同一条件下测定了新分离菌株P208的固氮能力及促生潜力(图 4,表 3),固氮活性为模式菌株的2.61倍,IAA生成能力为模式菌株的1.48倍,铁载体生成能力为模式菌株的1.80倍,具有潜在的开发应用价值。前人研究表明,与溶磷作用等促生特征相比,IAA和铁载体生成是植物促生菌筛选的优选特征,IAA和铁载体生成有利于增强促生菌的环境适应性,促进菌株在植物根系的定殖[67]。菌株生成的铁载体能够促进植物对铁的吸收,对植物病原微生物具有拮抗作用[68-69];菌株IAA的生成与植株根茎生物量、氮磷吸收量显著正相关[70],不同水平的IAA生成对植株根系伸长具有不同的效应,较低浓度的IAA生成与植株根系的伸长具有显著的正相关关系,而较高浓度的IAA生成抑制主根的伸长,促进侧根的生长,增加根系的生物量[67, 70-71]。
为了解分离菌株的促生应用潜力,在室内培养条件下进一步研究了A. chroococcum P208接种对作物幼苗生长的影响,结果显示可显著促进水稻和小麦幼苗的根系生长(表 4)。更长、更丰富的根系可能促进植物对水分和养分的吸收[48]。然而,结果中未检测到接种对茎叶生长的显著影响,这可能归因于较短的培养时间和Hoagland植物生长培养基中丰富的营养。在相同的试验条件下,玉米幼苗根系和茎叶的生长均对接种没有明显的响应,这表明A. chroococcum P208接种对不同作物幼苗生长的影响不同。前人研究表明,不同类型的农作物品种对细菌接种的反应不同[7, 66]。A. chroococcum P208接种对玉米幼苗生长的影响较小也可能归因于供试材料的基因型。因此,涉及更多作物和品种的接种试验研究有待进一步开展。
综上,本研究从固氮能力较高的紫色土发育水稻土中分离到一株具有较强固氮促生能力的固氮菌,利用基于基因组的方法鉴定为褐球固氮菌(Azotobacter chroococcum),A. chroococcum P208能够为进一步的生物技术提供新的固氮酶、固氮酶基因和代谢途径。此外,作为一株高效固氮促生菌株资源,A. chroococcum P208的应用前景值得进一步探索。
| [1] | Reed SC, Cleveland CC, Townsend AR. Relationships among phosphorus, molybdenum and free-living nitrogen fixation in tropical rain forests: results from observational and experimental analyses. Biogeochemistry, 2013, 114: 135-147. DOI:10.1007/s10533-013-9835-3 |
| [2] | Keuter A, Veldkamp E, Corre MD. Asymbiotic biological nitrogen fixation in a temperate grassland as affected by management practices. Soil Biology and Biochemistry, 2014, 70: 38-46. DOI:10.1016/j.soilbio.2013.12.009 |
| [3] | Reed SC, Cleveland CC, Townsend AR. Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annual Review of Ecology, Evolution, and Systematics, 2011, 42(1): 489-512. DOI:10.1146/annurev-ecolsys-102710-145034 |
| [4] | Mus F, Alleman AB, Pence N, Seefeldt LC, Peters JW. Exploring the alternatives of biological nitrogen fixation. Metallomics, 2018, 10(4): 523-538. DOI:10.1039/C8MT00038G |
| [5] | Habibi S, Djedidi S, Ohkama-Ohtsu N, Sarhadi WA, Kojima K, Rallos RV, Ramirez MDA, Yamaya H, Sekimoto H, Yokoyama T. Isolation and screening of indigenous plant growth-promoting rhizobacteria from different rice cultivars in Afghanistan soils. Microbes and Environments, 2019, 34(4): 347-355. DOI:10.1264/jsme2.ME18168 |
| [6] | Silva JF, Silva TR, Escobar IEC, Fraiz ACR, Santos JWM, Nascimento TR, Santos JMR, Peters SJW, Melo RF, Signor D, Fernandes-Júnior PI. Screening of plant growth promotion ability among bacteria isolated from field-grown Sorghum under different managements in Brazilian drylands. World Journal of Microbiology and Biotechnology, 2018, 34(12): 1-10. DOI:10.1007/s11274-018-2568-7 |
| [7] | Dos Santos SG, Chaves VA, Da Silva Ribeiro F, Alves GC, Reis VM. Rooting and growth of pre-germinated sugarcane seedlings inoculated with diazotrophic bacteria. Applied Soil Ecology, 2019, 133: 12-23. DOI:10.1016/j.apsoil.2018.08.015 |
| [8] | Banik A, Dash GK, Swain P, Kumar U, Mukhopadhyay SK, Dangar TK. Application of rice (Oryza sativa L.) root endophytic diazotrophic Azotobacter sp. strain Avi2(MCC 3432) can increase rice yield under green house and field condition. Microbiological Research, 2019, 219: 56-65. DOI:10.1016/j.micres.2018.11.004 |
| [9] | Shirinbayan S, Khosravi H, Malakouti MJ. Alleviation of drought stress in maize (Zea mays) by inoculation with Azotobacter strains isolated from semi-arid regions. Applied Soil Ecology, 2019, 133: 138-145. DOI:10.1016/j.apsoil.2018.09.015 |
| [10] | Piccinin GG, Braccini AL, Dan LGM, Scapim CA, Ricci TT, Bazo GL. Efficiency of seed inoculation with Azospirillum brasilense on agronomic characteristics and yield of wheat. Industrial Crops and Products, 2013, 43: 393-397. DOI:10.1016/j.indcrop.2012.07.052 |
| [11] | Cassán F, Diaz-Zorita M. Azospirillum sp. in current agriculture: From the laboratory to the field. Soil Biology and Biochemistry, 2016, 103: 117-130. DOI:10.1016/j.soilbio.2016.08.020 |
| [12] | Cassán FD, Okon Y, Creus CM. Handbook for Azospirillum. Cham: Springer International Publishing, 2015. |
| [13] | Perrig D, Boiero ML, Masciarelli OA, Penna C, Ruiz OA, Cassán FD, Luna MV. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Applied Microbiology and Biotechnology, 2007, 75(5): 1143-1150. DOI:10.1007/s00253-007-0909-9 |
| [14] | Cassán F, Perrig D, Sgroy V, Masciarelli O, Penna C, Luna V. Azospirillum brasilense Az39 and Bradyrhizobium japonicum E109, inoculated singly or in combination, promote seed germination and early seedling growth in corn (Zea mays L.) and soybean (Glycine max L.). European Journal of Soil Biology, 2009, 45(1): 28-35. DOI:10.1016/j.ejsobi.2008.08.005 |
| [15] | Overmann J. Significance and future role of microbial resource centers. Systematic and Applied Microbiology, 2015, 38(4): 258-265. DOI:10.1016/j.syapm.2015.02.008 |
| [16] | Viver T, Cifuentes A, Díaz S, Rodríguez-Valdecantos G, González B, Antón J, Rosselló-Móra R. Diversity of extremely halophilic cultivable prokaryotes in Mediterranean, Atlantic and Pacific solar salterns: Evidence that unexplored sites constitute sources of cultivable novelty. Systematic and Applied Microbiology, 2015, 38(4): 266-275. DOI:10.1016/j.syapm.2015.02.002 |
| [17] |
Li YX, Guo PY, Sun JG. Isolation, identification, phylogeny and growth promoting characteristics of endophytic diazotrophs from Tuber and root crops. Scientia Agricultura Sinica, 2017, 50(1): 104-122.
(in Chinese) 李艳星, 郭平毅, 孙建光. 块根块茎类作物内生固氮菌分离鉴定、系统发育与促生特性. 中国农业科学, 2017, 50(1): 104-122. |
| [18] |
Sun JG, Luo Q, Gao M, Hu HY, Xu J, Zhou YQ. Isolation and phylogeny of nitrogen-fixing endophytic bacteria in wheat, rice, maize, Chinese cabbage and celery. Scientia Agricultura Sinica, 2012, 45(7): 1303-1317.
(in Chinese) 孙建光, 罗琼, 高淼, 胡海燕, 徐晶, 周义清. 小麦、水稻、玉米、白菜、芹菜内生固氮菌及其系统发育. 中国农业科学, 2012, 45(7): 1303-1317. |
| [19] |
Sun JG, Xu J, Hu HY, Zhang YC, Liu J, Wang WB, Sun YH. Collection and investigation on asymbiotic nitrogen-fixing microbial resources from 13 provinces over China. Plant Nutrition and Fertilizer Science, 2009, 15(6): 1450-1465.
(in Chinese) 孙建光, 徐晶, 胡海燕, 张燕春, 刘君, 王文博, 孙燕华. 中国十三省市土壤中非共生固氮微生物菌种资源研究. 植物营养与肥料学报, 2009, 15(6): 1450-1465. DOI:10.3321/j.issn:1008-505X.2009.06.030 |
| [20] | Rilling JI, Acuña JJ, Sadowsky MJ, Jorquera MA. Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an andisol from southern Chile. Frontiers in Microbiology, 2018, 9: 2710. DOI:10.3389/fmicb.2018.02710 |
| [21] | Rinaudo G, Balandreau J, Dommergues Y. Algal and bacterial non-symbiotic nitrogen fixation in paddy soils. Plant and Soil, 1971, 35(1): 471-479. DOI:10.1007/BF02661872 |
| [22] | Rao VR, Kalininskaia TA, Miller IM. Study of the activity of nonsymbiotic nitrogen fixation in rice field soils using 15N2. Mikrobiologiya, 1973, 42(4): 729-734. |
| [23] | Wang XJ, Liu BJ, Ma J, Zhang YH, Hu TL, Zhang H, Feng YC, Pan HL, Xu ZW, Liu G, Lin XW, Zhu JG, Bei QC, Xie ZB. Soil aluminum oxides determine biological nitrogen fixation and diazotrophic communities across major types of paddy soils in China. Soil Biology and Biochemistry, 2019, 131: 81-89. DOI:10.1016/j.soilbio.2018.12.028 |
| [24] | Brown ME, Burlingham SK, Jackson RM. Studies on Azotobacter species in soil. Plant and Soil, 1962, 17(3): 309-319. DOI:10.1007/BF01377670 |
| [25] | Lane DJ. 16S/23S rRNA sequencing//Stackebrandt E, Goodfellow M. Nucleic acid techniques in bacterial systematics. New York: Wiley, 1991: 115-175. |
| [26] | Gauri SS, Mandal SM, Mondal KC, Dey S, Pati BR. Enhanced production and partial characterization of an extracellular polysaccharide from newly isolated Azotobacter sp. SSB81. Bioresource Technology, 2009, 100(18): 4240-4243. DOI:10.1016/j.biortech.2009.03.064 |
| [27] | Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(5): 1613-1617. DOI:10.1099/ijsem.0.001755 |
| [28] | Kumar S, Stecher G, Tamura K. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016, 33(7): 1870-1874. DOI:10.1093/molbev/msw054 |
| [29] | Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 1987, 4(4): 406-425. |
| [30] | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014, 30(15): 2114-2120. DOI:10.1093/bioinformatics/btu170 |
| [31] | Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology, 2012, 19(5): 455-477. DOI:10.1089/cmb.2012.0021 |
| [32] | Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Research, 2016, 44(14): 6614-6624. DOI:10.1093/nar/gkw569 |
| [33] | Kim J, Na SI, Kim D, Chun J. UBCG2:Up-to-date bacterial core genes and pipeline for phylogenomic analysis. Journal of Microbiology, 2021, 59(6): 609-615. DOI:10.1007/s12275-021-1231-4 |
| [34] | Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi HN, Xu XW, de Meyer S, Trujillo ME. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 2018, 68(1): 461-466. DOI:10.1099/ijsem.0.002516 |
| [35] | Richter M, Rosselló-Móra R, Oliver Gl öckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics, 2016, 32(6): 929-931. DOI:10.1093/bioinformatics/btv681 |
| [36] | Meier-Kolthoff JP, Auch AF, Klenk HP, G öker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics, 2013, 14(1): 1-14. DOI:10.1186/1471-2105-14-1 |
| [37] | Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints, 2016, 4: e1900v1901. |
| [38] | Deloger M, El Karoui M, Petit MA. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. Journal of Bacteriology, 2009, 191(1): 91-99. DOI:10.1128/JB.01202-08 |
| [39] | Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 1976, 72(1/2): 248-254. |
| [40] | Bellenger JP, Xu Y, Zhang X, Morel FMM, Kraepiel AML. Possible contribution of alternative nitrogenases to nitrogen fixation by asymbiotic N2-fixing bacteria in soils. Soil Biology and Biochemistry, 2014, 69: 413-420. DOI:10.1016/j.soilbio.2013.11.015 |
| [41] | Bellenger JP, Wichard T, Xu Y, Kraepiel AML. Essential metals for nitrogen fixation in a free-living N2-fixing bacterium: chelation, homeostasis and high use efficiency. Environmental Microbiology, 2011, 13(6): 1395-1411. DOI:10.1111/j.1462-2920.2011.02440.x |
| [42] | Glickmann E, Dessaux Y. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Applied and Environmental Microbiology, 1995, 61(2): 793-796. DOI:10.1128/aem.61.2.793-796.1995 |
| [43] | Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiology Letters, 1999, 170(1): 265-270. DOI:10.1111/j.1574-6968.1999.tb13383.x |
| [44] | Murphy J, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Analytica Chimica Acta, 1962, 27: 31-36. DOI:10.1016/S0003-2670(00)88444-5 |
| [45] | Collavino MM, Sansberro PA, Mroginski LA, Aguilar OM. Comparison of in vitro solubilization activity of diverse phosphate-solubilizing bacteria native to acid soil and their ability to promote Phaseolus vulgaris growth. Biology and Fertility of Soils, 2010, 46(7): 727-738. DOI:10.1007/s00374-010-0480-x |
| [46] | Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 1987, 160(1): 47-56. DOI:10.1016/0003-2697(87)90612-9 |
| [47] | Pereyra MA, Ballesteros FM, Creus CM, Sueldo RJ, Barassi CA. Seedlings growth promotion by Azospirillum brasilense under normal and drought conditions remains unaltered in Tebuconazole-treated wheat seeds. European Journal of Soil Biology, 2009, 45(1): 20-27. DOI:10.1016/j.ejsobi.2008.09.015 |
| [48] | Knoth JL, Kim SH, Ettl GJ, Doty SL. Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. GCB Bioenergy, 2013, 5(4): 408-418. DOI:10.1111/gcbb.12006 |
| [49] | Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 2014, 64. |
| [50] | Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(45): 19126-19131. DOI:10.1073/pnas.0906412106 |
| [51] | Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. Journal of Bacteriology, 2005, 187(18): 6258-6264. DOI:10.1128/JB.187.18.6258-6264.2005 |
| [52] | Meier-Kolthoff JP, Klenk HP, G öker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. International Journal of Systematic and Evolutionary Microbiology, 2014, 64(Pt 2): 352-356. |
| [53] | Li HB, Singh RK, Singh P, Song QQ, Xing YX, Yang LT, Li YR. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Frontiers in Microbiology, 2017, 8: 1268. DOI:10.3389/fmicb.2017.01268 |
| [54] | Kirchhof G, Reis VM, Baldani JI, Eckert B, D öbereiner J, Hartmann A. Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant and Soil, 1997, 194(1/2): 45-55. DOI:10.1023/A:1004217904546 |
| [55] | Chun J, Rainey FA. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. International Journal of Systematic and Evolutionary Microbiology, 2014, 64(Pt 2): 316-324. |
| [56] | Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek, 2017, 110(10): 1281-1286. DOI:10.1007/s10482-017-0844-4 |
| [57] | Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology, 1994, 44(4): 846-849. DOI:10.1099/00207713-44-4-846 |
| [58] | Hahn MW, Jezberová J, Koll U, Saueressig-Beck T, Schmidt J. Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. The ISME Journal, 2016, 10(7): 1642-1655. DOI:10.1038/ismej.2015.237 |
| [59] | Fox GE, Wisotzkey JD, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. International Journal of Systematic Bacteriology, 1992, 42(1): 166-170. DOI:10.1099/00207713-42-1-166 |
| [60] | Hardy RWF, Holsten RD, Jackson EK, Burns RC. The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiology, 1968, 43(8): 1185-1207. DOI:10.1104/pp.43.8.1185 |
| [61] | Saiz E, Sgouridis F, Drijfhout FP, Ullah S. Biological nitrogen fixation in peatlands: Comparison between acetylene reduction assay and 15N2 assimilation methods. Soil Biology and Biochemistry, 2019, 131: 157-165. DOI:10.1016/j.soilbio.2019.01.011 |
| [62] | Montoya JP, Voss M, Kahler P, Capone DG. A simple, high-precision, high-sensitivity tracer assay for N(inf2) fixation. Applied and Environmental Microbiology, 1996, 62(3): 986-993. DOI:10.1128/aem.62.3.986-993.1996 |
| [63] | Hayat R, Ahmed I, Sheirdil RA. An overview of plant growth promoting rhizobacteria (PGPR) for sustainable agriculture. Crop Production for Agricultural Improvement. Dordrecht: Springer Netherlands, 2012: 557-579. |
| [64] | Zahir ZA, Arshad M, Frankenberger WT Jr. Plant growth promoting rhizobacteria: applications and perspectives in agriculture. Advances in Agronomy. Amsterdam: Elsevier, 2003: 97-168. |
| [65] | Kennedy IR, Choudhury ATMA, Kecskés ML. Non-symbiotic bacterial diazotrophs in crop-farming systems: can their potential for plant growth promotion be better exploited?. Soil Biology and Biochemistry, 2004, 36(8): 1229-1244. DOI:10.1016/j.soilbio.2004.04.006 |
| [66] | Montañez A, Blanco AR, Barlocco C, Beracochea M, Sicardi M. Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (Zea mays L.) and their inoculation effects in vitro. Applied Soil Ecology, 2012, 58: 21-28. DOI:10.1016/j.apsoil.2012.02.009 |
| [67] | Etesami H, Hosseini HM, Alikhani HA, Mohammadi L. Bacterial biosynthesis of 1-aminocyclopropane-1-carboxylate (ACC) deaminase and indole-3-acetic acid (IAA) as endophytic preferential selection traits by rice plant seedlings. Journal of Plant Growth Regulation, 2014, 33(3): 654-670. DOI:10.1007/s00344-014-9415-3 |
| [68] | Khare E, Arora NK. Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea. Current Microbiology, 2010, 61(1): 64-68. DOI:10.1007/s00284-009-9577-6 |
| [69] | Khan A, Singh P, Srivastava A. Synthesis, nature and utility of universal iron Chelator-Siderophore: a review. Microbiological Research, 2018, 212/213: 103-111. DOI:10.1016/j.micres.2017.10.012 |
| [70] | Marques APGC, Pires C, Moreira H, Rangel AOSS, Castro PML. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biology and Biochemistry, 2010, 42(8): 1229-1235. DOI:10.1016/j.soilbio.2010.04.014 |
| [71] | Xie H, Pasternak JJ, Glick BR. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Current Microbiology, 1996, 32(2): 67-71. DOI:10.1007/s002849900012 |
 2021, Vol. 61
2021, Vol. 61




