中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 吕倩, 马莉, 骆巧, 罗雪, 陈久兵, 罗正中, 雍康, 姚学萍, 余树民, 沈留红, 曹随忠. 2021
- Qian Lv, Li Ma, Qiao Luo, Xue Luo, Jiubing Chen, Zhengzhong Luo, Kang Yong, Xueping Yao, Shumin Yu, Liuhong Shen, Suizhong Cao. 2021
- 基于高通量测序技术分析四川两个奶牛场乳汁的菌群差异
- Analysis of the differences in the microflora of milk from two dairy farms in Sichuan province based on high-throughput sequencing technology
- 微生物学报, 61(10): 3087-3102
- Acta Microbiologica Sinica, 61(10): 3087-3102
-
文章历史
- 收稿日期:2020-11-25
- 修回日期:2021-03-11
- 网络出版日期:2021-03-28
2. 重庆三峡职业学院 动物科技学院, 重庆 404155
2. College of Animal Science and Technology, Chongqing Three Gorges Vocational College, Chongqing 404155, China
奶牛乳房炎,最常见的原因是由细菌引起的乳房内感染[1-3],是影响全球奶牛业的主要疾病之一[4-5]。众所周知,不同奶牛场的乳房炎发病率有很大差异,导致乳房炎的主要病原体也有很大差异。为了制定高效的乳房炎控制程序,确定引起乳房炎的病原体是非常重要的环节。然而,由于细菌浓度低或过高的营养和生长要求,使用传统方法在20%–40%的乳房炎病例的乳汁样本中没有检测到细菌[6]。此外,最近的研究表明,乳房炎不仅与乳房炎病原体有关,还与乳汁微生物区系失衡有关[7-8]。从另一个角度来看,乳汁微生物区系的嗜冷菌等与原奶(raw milk)的保存和质量有很大关系[9]。因此,了解乳汁中存在的细菌群落对于建立有针对性的乳房炎防控体系、提高乳制品的安全性和质量具有重要意义。
在过去二十年中,实时荧光定量PCR[10]、多重PCR[11]和变性梯度凝胶电泳PCR[12]等方法已被用于鉴定乳汁样品中的细菌DNA。近年来,越来越多的证据表明,16S rRNA基因测序几乎可以识别整个细菌群落,包括共生和致病细菌群落,因为它可以克服基于培养的细菌检测方法的局限性[13]。目前乳房炎乳汁样本中微生物区系的多样性相关研究较多,Kuehn等[8]使用16S rRNA测序技术揭示了健康乳汁和乳房炎乳汁的微生物菌群之间具有显著性差异。Oultram等[14]报道了16S rRNA基因测序可以作为临床诊断乳房炎的工具,且大多数致病菌能被检测到。然而,关于不同奶牛场健康乳汁与乳房炎乳汁样本中的微生物区系比较相关研究较少。为了比较不同奶牛场健康乳汁和乳房炎乳汁的微生物菌群,本试验采用Illumina Hiseq2500测序平台,分别测定两个奶牛场健康乳汁和乳房炎乳汁中微生物16S rRNA V4区序列,并对其群落结构和多样性进行比较分析,为两个奶牛场在乳房炎的预防工作以及原料奶的质量控制上提供理论依据。
1 材料和方法 1.1 样本采集于2019年7–9月从四川省成都市某规模化奶牛场(F1:总存栏荷斯坦奶牛1500头,泌乳牛存栏1000头)和四川省资阳市某规模化奶牛场(F2:总存栏荷斯坦奶牛2000头,泌乳牛存栏1200头)分别采集健康的奶牛乳样10份和临床型乳房炎乳样各6份,两个奶牛场均为自由卧栏式牧场,均为第二胎荷斯坦奶牛,均饲喂统一的饲料,为确保两个奶牛场温湿度一致,选择7月样本,不选择湿度有显著性差异的8月和9月样本(图 1)。按常规挤奶消毒程序对每个目标奶牛进行消毒收集试验样本50 mL后,随即置于附有冰袋的保温箱内,立即送回实验室,所有样本在超低温(–80 ℃)冰箱中保存备用。
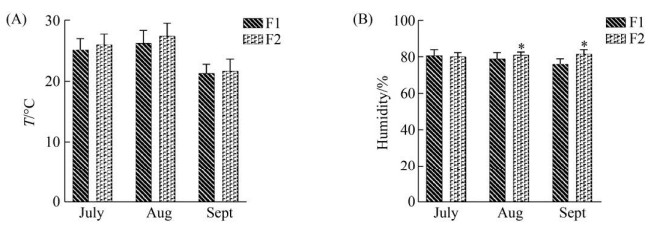
|
| 图 1 两个奶牛场的温度和湿度 Figure 1 Temperature and humidity of two dairy farms. The abscissa is the species of the month, the ordinate is the temperature or humidity, and * represents the significance of the difference test (***: P < 0.001; **: 0.001≤ P≤0.01; *: 0.01≤P≤0.05). A and B represent the temperature and humidity histograms of the two dairy farms in July, August and September, respectively. |
1.2 隐性乳房炎乳样的排除
为确保所有健康奶牛不存在隐性乳房炎,所有健康奶牛的乳汁均由美国Elitech Wescor MAS-D-TEC便携式隐性乳房炎检测仪检测,剔除隐性乳房炎样本,最终F1与F2选入6头健康奶牛的乳汁,编号分别为F1H1–F1H6、F2H1–F2H6。同时将F1与F2的6头乳房炎乳汁编号为F1M1–F1M6和F2M1–F2M6。样本共计24份。
1.3 DNA的提取和16S rRNA基因扩增及高通量测序按照Meta类样本的操作步骤提取乳汁中细菌的总DNA;将得到的总DNA为模板对细菌16S rRNA的V4区进行PCR扩增,扩增引物如下:515F (5′-GTGCCAGCMGCCGCGGTAA-3′)和806R (5′-GGACTACHVGGGTWTCTAAT-3′)。使用Illumina Hiseq2500测序平台进行高通量测序,将下机数据使用FLASH和USEARCH软件对原始数据进行拼接过滤,得到优化序列,将相似度为97%有效序列进行操作分类单元聚类(operational taxonomic uunits,OTUs),并挑选出相对丰度最高的序列作为每个OTUs的代表性序列。将OTUs代表序列与相应微生物数据库进行比对,得到各样本的物种分类信息与各水平注释信息。同时获得基于聚类结果的多样性分析与基于注释结果的各分类水平物种组成信息。
1.4 统计学方法和数据分析本研究使用多种统计学分析,具体如下:使用秩和检验进行分析比较物种组成分析、物种差异分析、多样性分析、关联分析;使用PICRUSt软件检测不同丰度的KEGG;分析作图软件为SPSS 22.0统计软件和Vegan R package软件;P < 0.05表示差异显著。
2 结果和分析 2.1 样本序列信息统计经测序,4组24个乳汁样本共获得4013234条原始序列,经过滤后获得2887024条优化序列,将相似度为97%有效序列进行操作分类单元聚类(OTUs),在F1H和F1M组中,共得到2105个OTUs (图 2-A),其中F1H样本中特有1080个OTUs,F1M样本中特有305个OTUs,两者之间共有720个OTUs;在F2H和F2M组中,共得到1803个OTUs (图 2-B),其中F2H样本中特有657个OTUs,F2M样本中特有376个OTUs,两者之间共有770个OTUs;在F1H和F2H组中,共得2246个OTUs (图 2-C),其中F1H样本中特有819个OTUs,F2H样本中特有446个OTUs,两者之间共有981个OTUs;在F1M和F2M组中,共得1509个OTUs (图 2-D),其中F1M样本中特有363个OTUs,F2M样本中特有484个OTUs,两者之间共有662个OTUs。
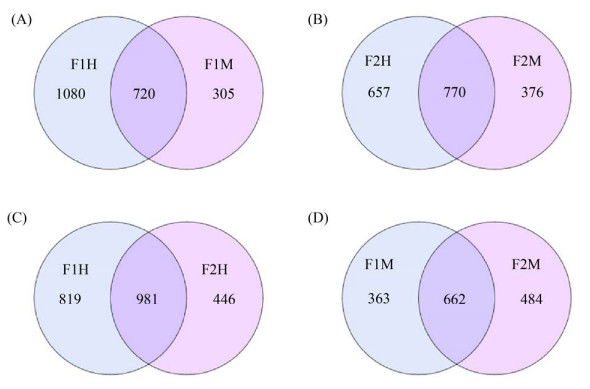
|
| 图 2 OTUs Venn图 Figure 2 OTUs Venn diagram. F1: farm1; F2: farm2; H: healthy; M: mastitis; A–D: Venn diagram showing the numbers of unique and shared bacterial OTUs according to milk sample fractions, the left part and the right part are the unique OTUs numbers of the sample, respectively, and the middle crossing part is the common number of OTUs. |
2.2 物种多样性与丰富度分析
Alpha多样性通常用来反映样本内部微生物群落的多样性和丰富度,包括多种分析手段。本试验采用Chao指数、Ace指数、Shannon指数、Simpson指数评估4组样本的Alpha多样性。其中Chao指数、Ace指数反映物种丰富度,Shannon指数、Simpson指数综合体现物种的多样性。4组样本F1H-F1M (图 3-A)、F2H-F2M (图 3-B)、F1H-F2H (图 3-C)、F1M-F2M (图 3-D)的Chao指数、Ace指数、Shannon指数、Simpson指数差异均不显著(P>0.05),说明4组样本乳汁的多样性和丰富度相似。同时,4组样本的Chao指数多样性稀释曲线图也趋于平缓,说明该样本的采样量足够,能覆盖大多数物种。
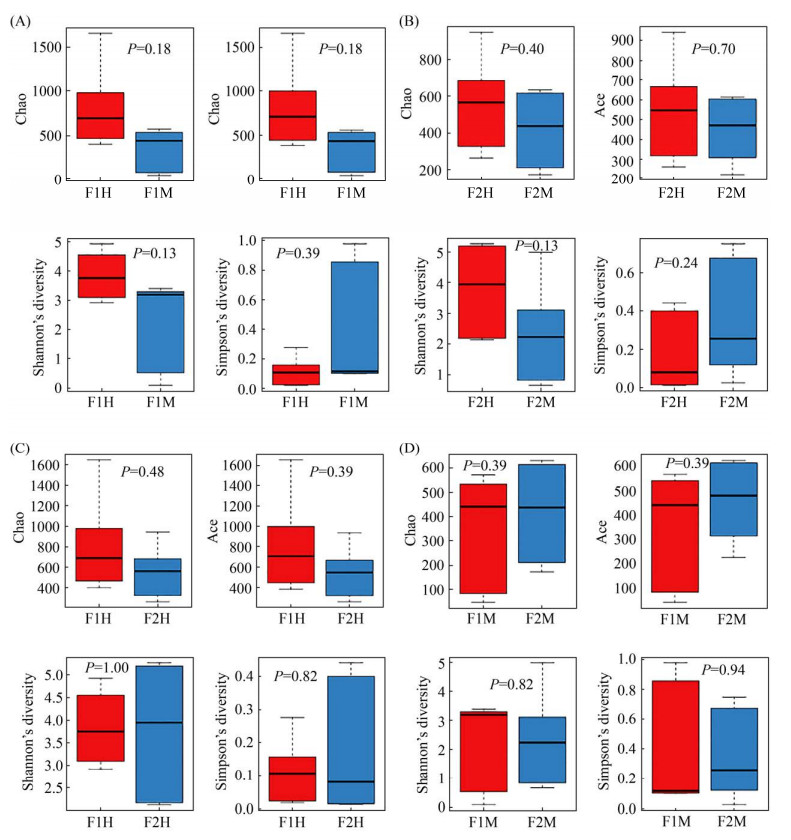
|
| 图 3 Alpha多样性箱型图 Figure 3 Alpha diversity estimates of the bacterial communities. F1: farm1; F2: farm2; H: healthy; M: mastitis. A–D: boxplots of bacterial alpha diversities evaluated by Chao index, Ace index, Shannon index and Simpson index. Different numbers in the picture denoted a P value of different alpha diversity index among groups. The five lines from bottom to top are: minimum, first quartile, median, third quartile and maximum. Outliers are represented by ".". |
Beta多样性主要分析样本与样本间微生物群落组成的相似性,PLS-DA多变量统计分析方法可将主成分分析和多元回归的功能相结合,故本试验通过PLS-DA回归模型基于OTU水平对四组样本进行Beta多样性分析(图 4),可看出F1H与F1M、F2H与F2M、F1H与F2H、F1M与F2M四组样本都分别两两聚在一起,说明分组具有意义,均具有显著性差异。

|
| 图 4 基于PLS-DA分析的细菌群落散点图 Figure 4 Scatterplot from pls-da analysis in bacterial communitise. F1: farm1; F2: farm2; H: healthy; M: mastitis. A–D: the points of different colors and shapes represent the sample groups under different conditions, and the scale of the horizontal and vertical axes is the relative distance, which has no practical significance. The closer the sample is on the graph, the more similar the composition of the sample is. |
2.3 物种组成分析及差异性分析 2.3.1 门分类学水平:
基于门分类水平研究发现(表 1),4组样本丰度>1%的优势菌门有7个,分别为变形菌门(Proteobacteria)、厚壁菌门(Firmicutes)、栖热菌门(Thermi)、拟杆菌门(Bacteroidetes)、放线菌门(Actinobacteria)、绿弯菌门(Chloroflexi)、梭杆菌门(Fusobacteria)。与场1健康乳汁(F1H)组相比,场1乳房炎乳汁(F1M)组的放线菌门(Actinobacteria)和绿弯菌门(Chloroflexi)的丰度显著下降(P<0.05)。与场2健康乳汁(F2H)组相比,场2乳房炎(F2M)组的放线菌门(Actinobacteria)的丰度也显著下降(P<0.05)。在两个奶牛场的健康乳汁中,场1 (F1H)组的放线菌门(Actinobacteria)、栖热菌门(Thermi)、绿弯菌门(Chloroflexi)的丰度显著高于场2 (F2H) (P<0.05);在场1乳房炎(F1M)和场2乳房炎(F2M)对比组中,未见显著性差异的菌门。
| Phylum | Relative abundance | P-value | |||||||
| F1H | F1M | F2H | F2M | F1H VS F1M | F2H VS F2M | F1H VS F2H | F1M VS F2M | ||
| Firmicutes | 37.55 | 32.49 | 61.38 | 39.49 | ns | ns | ns | ns | |
| Proteobacteria | 29.76 | 52.00 | 22.59 | 47.23 | ns | ns | ns | ns | |
| Actinobacteria | 21.16 | 1.80 | 4.97 | 1.21 | ** | ** | ** | ns | |
| Bacteroidetes | 7.23 | 2.98 | 9.24 | 4.46 | ns | ns | ns | ns | |
| Thermi | 0.47 | 10.05 | 0.042 | 5.72 | ns | ns | ** | ns | |
| Chloroflexi | 1.89 | 0.021 | 0.063 | 0 | ** | ns | ** | ns | |
| Fusobacteria | 0.00030 | 0.0078 | 0.26 | 1.27 | ns | ns | ** | ns | |
| * represents the significance of the difference test. ***: P<0.001; **: 0.001≤P≤0.01; *: 0.01≤P≤0.05; ns: P>0.05. The same below. | |||||||||
2.3.2 属分类学水平:
在已检测到的329种菌属中,排名前十五的菌属(表 2)分别为克雷伯氏菌属(Klebsiella)、链球菌属(Streptococcus)、埃希氏菌属(Escherichia)、假单胞菌属(Pseudomonas)、气球菌属(Aerococcus)、乳球菌属(Lactococcus)、不动杆菌属(Acinetobacter)、栖热菌属(Thermus)、耶尔森氏菌属(Yersinia)、葡萄球菌属(Staphylococcus)、土芽孢杆菌属(Geobacillus)、棒状杆菌属(Corynebacterium)、梭菌属(Clostridium)、巨球菌属(Macrococcus)、嗜冷杆菌属(Psychrobacter)。与场1健康乳汁(F1H)组相比,场1乳房炎乳汁(F1M)的气球菌属(Aerococcus)、棒状杆菌属(Corynebacterium)、梭菌属(Clostridium)、巨球菌属(Macrococcus)的丰度显著降低(P<0.05);克雷伯氏菌属(Klebsiella)、埃希氏菌属(Escherichia)、栖热菌属(Thermus)、土芽孢杆菌属(Geobacillus)的丰度显著上升(P<0.05)。与场2健康乳汁(F2H)组相比,场2乳房炎乳汁(F2M)的气球菌属(Aerococcus)、葡萄球菌属(Staphylococcus)、棒状杆菌属(Corynebacterium)的丰度显著下降(P<0.05);埃希氏菌属(Escherichia)的丰度显著上升(P<0.05)。在场1健康乳汁(F1H)和场2健康乳汁(F2H)对比组中,F2H的埃希氏菌属(Escherichia)、葡萄球菌属(Staphylococcus)的丰度显著高于F1H (P<0.05);棒状杆菌属(Corynebacterium)、巨球菌属(Macrococcus)的丰度显著低于F1H(P<0.05)。在与场1乳房炎乳汁(F1M)组相比,场2乳房炎乳汁(F2M)中的埃希氏菌属(Escherichia)、乳球菌属(Lactococcus)的丰度显著升高(P<0.05)。
| Genus | Relative abundance | P-value | |||||||
| F1H | F1M | F2H | F2M | F1H VS F1M | F2H VS F2M | F1H VS F2H | F1M VS F2M | ||
| Klebsiella | 0.50 | 29.63 | 0.65 | 8.95 | * | ns | ns | ns | |
| Streptococcus | 0.90 | 17.73 | 11.24 | 3.49 | ns | ns | ns | ns | |
| Escherichia | 0.044 | 1.31 | 3.92 | 25.09 | * | * | * | * | |
| Pseudomonas | 16.91 | 4.54 | 2.14 | 2.58 | ns | ns | ns | ns | |
| Aerococcus | 12.79 | 0.20 | 9.84 | 1.32 | ** | ** | ns | ns | |
| Lactococcus | 0.18 | 0.21 | 1.40 | 17.23 | ns | ns | ns | * | |
| Acinetobacter | 6.87 | 2.60 | 5.12 | 2.34 | ns | ns | ns | ns | |
| Thermus | 0 | 9.98 | 0 | 5.71 | * | ns | ns | ns | |
| Yersinia | 0.14 | 9.40 | 0.026 | 4.93 | ns | ns | ns | ns | |
| Staphylococcus | 0.38 | 0.34 | 11.79 | 0.36 | ns | * | * | ns | |
| Geobacillus | 0 | 7.79 | 0.0014 | 3.75 | * | ns | ns | ns | |
| Corynebacterium | 5.57 | 0.38 | 2.08 | 0.42 | ** | ** | ** | ns | |
| Clostridium | 1.50 | 0.22 | 1.83 | 2.45 | ** | ns | ns | ns | |
| Macrococcus | 5.75 | 0.020 | 0.023 | 0.013 | ** | ns | ** | ns | |
| Psychrobacter | 0.36 | 0.36 | 4.14 | 0.23 | ns | ns | ns | ns | |
2.4 关联分析与功能预测
为评估两场健康乳汁与乳房炎乳汁优势物种之间的关系,根据秩和检验得到属水平上的差异物种绘制物种之间Spearman相关性热图,筛选出30种菌属。结果如下:在场1中(图 5-A),土芽孢杆菌属(Geobacillus)与栖热菌属(Thermus)、埃希氏菌属(Escherichia)与耶尔森氏菌属(Yersinia)、藤黄单胞菌属(Luteimonas)与里氏杆菌属(Riemerella)、卟啉单胞菌属(Porphyromonas)与土芽孢杆菌属(Geobacillus) 四组菌属之间具有极显著性正相关,其相关系数均大于0.9;厌氧球菌属(Anaerococcus)与里氏杆菌属(Riemerella)、厌氧球菌属(Anaerococcus)与肠杆菌属(Marinobacter)、肠杆菌属(Marinobacter)与埃希氏菌属(Escherichia)三组菌属之间具有显著性负相关,其相关系数均大于0.7。在场2中(图 5-B),密螺旋体属(Treponema)与不动杆菌属(Acinetobacter)、Jeotgalicoccus与棒状杆菌属(Corynebacterium)两组菌属之间具有极显著性正相关,其相关系数均大于0.9;棒状杆菌属(Corynebacterium)与埃希氏菌属(Escherichia)、Jeotgalicoccus与埃希氏菌属(Escherichia)、巨球菌属(Macrococcus)与梭杆菌属(Fusobacterium) 三组菌属之间具有显著性负相关,其相关系数均大于0.7。
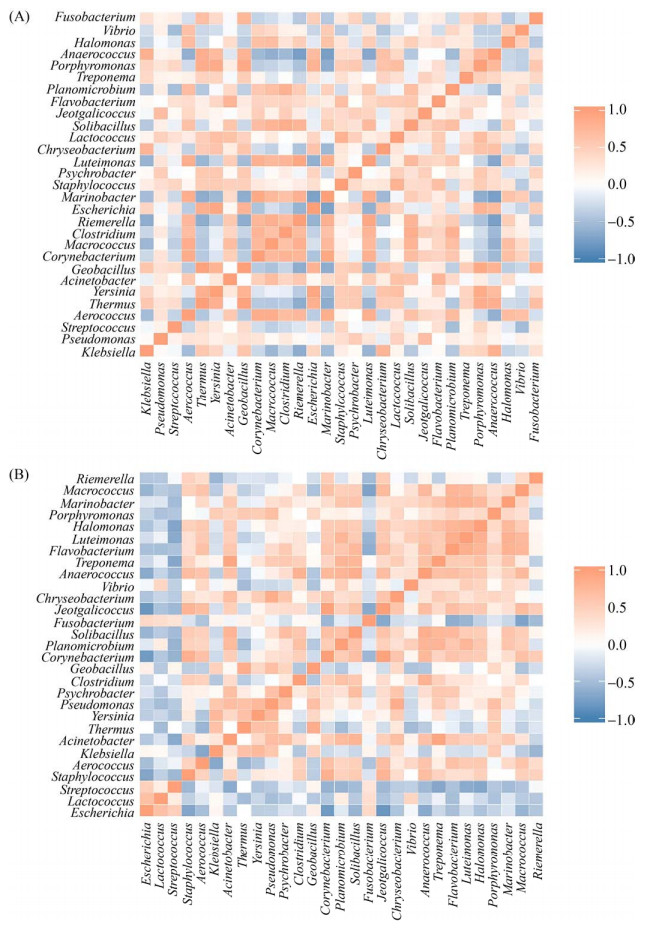
|
| 图 5 属水平Spearman相关性热图 Figure 5 Spearman correlation heat map at genus lever. The figure shows the correlation between species that differ at the taxonomic level of the genus. The darker the color, the stronger the correlation between species. A represents the correlation heat map of F1 differential bacteria, and B represents the correlation heat map of F2 differential bacteria. |
为评估两场乳房炎对奶牛乳汁代谢影响是否相同,本试验通过PICCRUSt方法分析乳汁在代谢过程中的潜在功能,分析细菌KEGG同源序列(KO)Pathway Hierarchy下的第一层级(Level 1)和第二层级(Level 2),在场1中,基于Level 1水平(图 6-A),发现有4种代谢通路具有显著性差异(P<0.05),其中有3种代谢通路在乳房炎感染后显著下调,包括细胞过程(cellular processes)、新陈代谢(metabolism)、生物体系统(organismal systems);另外一种环境信息处理(environmental information processing)代谢通路在乳房炎感染后显著上调。在Level 2水平(图 6-B),有12种代谢通路具有显著性差异(P<0.05且FDR<0.05),其中有7种代谢通路在感染乳房炎后显著下调,包括氨基酸代谢(amino acid metabolism)、能量代谢(energy metabolism)、脂质代谢(lipid metabolism)、运输和分解代谢(transport and catabolism)、折叠,分类和降解(folding, sorting and degradation)、细胞通讯(cell communication)、细胞运动(cell motility);有5种代谢通路显著上调,分别为多聚糖的生物合成与代谢(glycan biosynthesis and metabolism)、传染病(infectious diseases)、膜转运(membrane transport)、转录(transcription)、糖代谢(carbohydrate metabolism)。
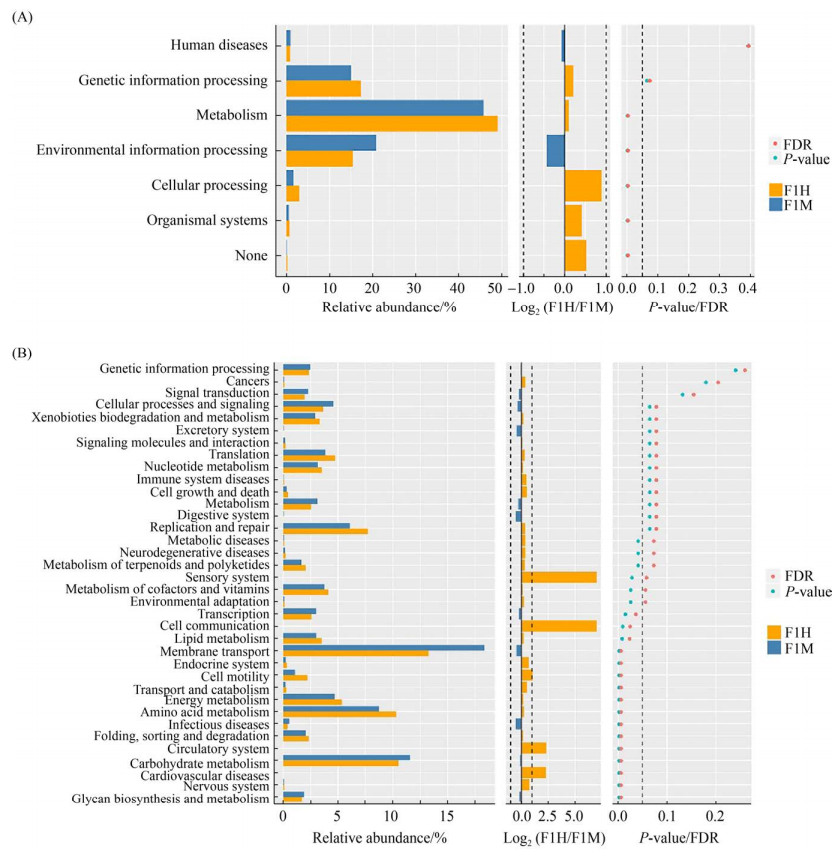
|
| 图 6 通路差异Wilcoxon结果图 Figure 6 Pathway differences Wilcoxon result diagram. On the left is a histogram of the relative abundance of pathways in each group. In the middle is the log2 value of the mean ratio of relative abundance of the same path in two groups. The figure on the right is the P-value and FDR (False Discover Rate) values obtained by Wilcox's test. If the P-value and FDR values were less than 0.05, the pathway was significantly different between the two groups. A represents the functional difference of Level 1 and B represents the functional difference of Level 2. |
在场2中,基于KEGG Pathway Hierarchy下的Level 1、Level 2水平,均未发现具有显著性差异的代谢通路(P<0.05且FDR<0.05)。
3 讨论 3.1 四组乳汁微生物菌群在不同分类水平上的差异 3.1.1 门分类学水平上的菌群结构差异:本试验结果显示:4组乳汁中最丰富的细菌门是厚壁菌门(Firmicutes)、变形菌门(Proteobacteria)、放线菌门(Actinobacteria)、拟杆菌门(Bacteroidetes),这与Paola Cremonesi等[15]研究结果一致。通过Wilcoxon rank-sum test分析发现:无论是在场1还是场2中,当奶牛发生乳房炎之后,没有发现相对丰度显著上升的菌门,仅发现相对丰度显著下降的菌门;在两场的健康乳汁对比中,发现场1中的放线菌门(Actinobacteria)、栖热菌门(Thermi)、绿弯菌门(Chloroflexi)的相对丰度显著高于场2,场2中的梭杆菌门(Fusobacteria)的相对丰度显著高于场1。在曾雪琴等[16]的研究中,发现梭杆菌门与奶牛乳房炎的发生密切相关,而梭杆菌属与多种疾病的发生具有相关性,在本实验中,场1未检测到梭杆菌属,而场2检测到梭杆菌属的相对丰度为0.016%,虽然场2梭杆菌属的相对丰度较低,但在平时的预防中,也应注意梭杆菌属的预防。在两场的乳房炎乳汁对比中,未发现有显著性差异的菌门。
3.1.2 属分类学水平上的菌群结构差异:在过去的研究中,Levison等[17]在研究加拿大地区奶牛乳房炎时发现无论是在传统奶牛场还是有机奶牛场,分离到最多的乳房炎病原体是葡萄球菌属(凝固酶阴性葡萄球菌、金黄色葡萄球菌)、埃希氏菌属(大肠杆菌)、链球菌属;Vakkamäki等[18]在研究芬兰地区奶牛乳房炎时同样发现葡萄球菌属(凝固酶阴性葡萄球菌、金黄色葡萄球菌)是奶牛乳房炎中最常见的病原体。在本试验中,两个奶牛场在奶牛感染乳房炎后,葡萄球菌属、链球菌属均未发现显著性升高,在场1中,克雷伯氏菌属(Klebsiella)、埃希氏菌属(Escherichia)、栖热菌属(Thermus)和土芽孢杆菌属(Geobacillus)的相对丰度显著升高;在场2中仅发现埃希氏菌属(Escherichia)的相对丰度显著升高。克雷伯氏菌(Klebsiella)是环境型乳房炎的条件致病菌,在乳房炎样本中,约有2%–9%的乳汁样本中分离出该菌[19];埃希氏菌属的代表菌株大肠杆菌被认为是临床型奶牛乳房炎的首要致病菌,也广泛存在于外界环境中[20-22],本研究2个奶牛场均应注意环境病原菌的预防。栖热菌属有8个有效公布的种,常在温泉、人工水热环境中被分离到[23],关于在奶牛乳汁方面的相关报道较少,在本研究中,两场健康乳汁中均未发现栖热菌属,仅在乳房炎乳汁中发现,且在场1中具有显著性差异,故栖热菌属与奶牛乳房炎的关系有待进一步的研究。据报道,在油田、火山口、温泉、土壤等高温条件下均可分离到土芽孢杆菌属(Geobacillus)的菌株[24],在本试验中,土芽孢杆菌属(Geobacillus)在2场的乳房炎乳汁中的相对丰度均升高,且在场1中具有显著性差异,而在场2中未见显著性差异,Levison等[17]曾在乳房炎乳汁中发现芽孢杆菌属也是常见病原体之一,这与本研究一致,分析两场产生差异的原因可能是由不同垫料造成的,在场1中,卧床垫料为河沙,而场2是橡胶垫,故乳房炎与卧床垫料的关系需进一步的研究。在两场健康组对比中,场2中的埃希氏菌属(Escherichia)和葡萄球菌属(Staphylococcus)的相对丰度显著高于场1,属于葡萄球菌属的金黄色葡萄球菌是奶牛乳房炎的主要传染性病原菌[25-26],故场2在平时奶牛乳房炎预防工作中除了预防环境性病原菌,还应格外注意金黄色葡萄球菌的预防。两场乳房炎对比组在门分类学水平上未发现具有显著性差异的菌门,而在属分类学水平上发现了具有显著性差异的菌属,场2中的埃希氏菌属(Escherichia)和乳球菌属(Lactococcus)的相对丰度显著高于场1。Rodrigues等[27]报道乳球菌属(Lactococcus)是一个潜在的乳房炎新发病原菌,发现乳球菌属在健康乳汁中并不丰富,反而是乳房炎乳汁中的优势菌属。这与本研究场2相似,场2乳房炎乳汁中的乳球菌属是第二优势菌属,在平时乳房炎预防中应注意乳球菌属的预防。
据报道,影响乳汁质量的细菌有3种,分别为嗜热菌、嗜温菌、嗜冷菌。嗜冷微生物影响乳汁质量已被广泛研究,在冷藏过程中,嗜冷微生物可以产生细胞外蛋白酶和脂肪酶,这些酶对巴氏消毒甚至超高温(UHT)处理具有抗性,从而导致了牛奶和乳制品的变质[28]。常见的嗜冷微生物有以下几种:革兰氏阳性菌中的假单胞菌属(Pseudomonas)、不动杆菌属(Acinetobacter)、哈夫尼菌属(Hafnia)、拉恩氏菌属(Rahnella)、产碱杆菌属(Alcaligenes)、无色杆菌属(Achromobacter)、气单胞菌属(Aeromonas)、沙雷氏菌属(Serratia)、肠杆菌属(Enterobacter)、金黄杆菌属(Chryseobacterium)、色杆菌属(Chromobacterium)、黄杆菌属(Flavobacterium);革兰氏阴性菌中的芽孢杆菌属(Bacillus)、梭菌属(Clostridium)、棒状杆菌属(Corynebacterium)、链球菌属(Streptococcus)、微球菌属(Micrococcus)、葡萄球菌属(Staphylococcus)、肠球菌属(Enterococcus)、乳杆菌属(Lactobacillus)、微杆菌属(Microbacterium)[29]。在本研究中,分离到的优势菌属中有7个嗜冷菌,分别为链球菌属(Streptococcus)、假单胞菌属(Pseudomonas)、不动杆菌属(Acinetobacter)、葡萄球菌属(Staphylococcus)、棒状杆菌属(Corynebacterium)、梭菌属(Clostridium)、嗜冷杆菌属(Psychrobacter)。嗜冷菌在两个奶牛场的健康乳汁中总丰度分别为:场1为31.87%、场2为38.72%。有研究报道,在严格按照卫生标准时,嗜冷菌的总丰度占整个微生物菌群的10%以下[30],本研究的两个奶牛场健康乳汁中的嗜冷菌总丰度均大于10%,嗜冷菌可在–10 ℃至7 ℃下生长,奶牛场乳汁储存罐的温度为4 ℃,故大量的嗜冷菌可在冷链运输过程中生长,进而造成乳汁变质。两个奶牛场均应注意冷链运输过程中的环境污染。
在本研究中,两个奶牛场乳房炎乳汁的第一优势菌属分别为克雷伯氏菌属(Klebsiella)和埃希氏菌属(Escherichia),均属于环境性病原菌,两个奶牛场均应改善奶牛场整体环境的卫生。在环境方面,需保证过道和卧床的干净和清洁,做到及时清粪,及时更换垫料;严格评估奶牛粪便,及时调整饲喂配方,避免因为踩踏、飞溅行为将稀粪溅到乳房上;保证通风,保证整体环境的干燥。在挤奶方面,应严格遵循挤奶前消毒程序,同时应避免过度挤奶。
3.2 关联分析与功能预测在2个奶牛场的健康乳汁与乳房炎乳汁的优势物种的相关性上,2个奶牛场均未发现某种菌属在感染乳房炎过程中与其他菌属均存在协同或拮抗关系,挑选相关系数最高的优势物种来看,在场1中共预测到4对正相关菌属,这与场2共预测到2对正相关菌属均不一样。同理负相关菌属在2个奶牛场也均不一样。这说明在乳房炎奶牛中,不同奶牛场其优势物种的相关性差异较大。
3.3 奶牛乳房炎发生前后乳汁菌群变化对乳汁代谢的影响在2个奶牛场的乳汁中,每种菌群存在的相对丰度不同,这表明编码每种功能的基因比例可能不同,可能反映了不同的代谢活动。通过PICCRUSt功能预测分析发现,场1的奶牛在感染乳房炎后,level 2水平上11种代谢通路相关的乳汁菌群显著下降,其中氨基酸代谢、能量代谢、脂质代谢等与席晓敏等[31]研究结果一致。而在场2中,未发现具有显著性差异的代谢通路。这表明虽然同是荷斯坦奶牛,同是乳房炎,不同奶牛场具有不同的功能差异。而造成该结果的原因可能是奶牛个体差异、奶牛场环境、奶牛场饲喂方式等不同。提示研究者在对乳房炎乳汁代谢通路进行研究时,应对具体奶牛场具体分析。
不依赖培养的高通量测序方法为我们提供了更加深入和准确的乳汁微生物多样性的评估。乳汁微生物菌群的组成在不同奶牛场之间有定性和定量的差异,影响乳汁微生物菌群的因素是多样且复杂的,奶牛场温湿度、垫料、挤奶设备等[32-33]因素均能污染乳头,从而进入乳汁中。本研究在两奶牛场温湿度一致的基础上,研究两场场内乳汁和场间乳汁的菌群差异。结果表明四组乳汁样本无论是在场内比较还是场间比较,均存在较大的差异,每个奶牛场在乳房炎的预防工作中均应依据本场菌群的分布特征制定相应的方案。本研究进一步为不同奶牛场的乳房炎预防和原料奶的质量控制上提供了理论依据。
| [1] | Bhattarai D, Worku T, Dad R, Rehman ZU, Gong XL, Zhang SJ. Mechanism of pattern recognition receptors (PRRs) and host pathogen interplay in bovine mastitis. Microbial Pathogenesis, 2018, 120: 64-70. DOI:10.1016/j.micpath.2018.04.010 |
| [2] | Miles AM, Huson HJ. Graduate Student Literature Review: Understanding the genetic mechanisms underlying mastitis. Journal of Dairy Science, 2021, 104(1): 1183-1191. DOI:10.3168/jds.2020-18297 |
| [3] | El-Ashker M, Gwida M, Monecke S, Ehricht R, Elsayed M, El-Gohary F, Reißig A, Müller E, Paul A, Igbinosa EO, Beshiru A, Maurischat S. Microarray-based detection of resistance genes in coagulase-negative staphylococci isolated from cattle and buffalo with mastitis in Egypt. Tropical Animal Health and Production, 2020, 52(6): 3855-3862. DOI:10.1007/s11250-020-02424-1 |
| [4] | Keller D, Sundrum A. Comparative effectiveness of individualised homeopathy and antibiotics in the treatment of bovine clinical mastitis: randomised controlled trial. The Veterinary Record, 2018, 182(14): 407. DOI:10.1136/vr.104555 |
| [5] | Azooz MF, El-Wakeel SA, Yousef HM. Financial and economic analyses of the impact of cattle mastitis on the profitability of Egyptian dairy farms. Veterinary World, 2020, 13(9): 1750-1759. DOI:10.14202/vetworld.2020.1750-1759 |
| [6] | Taponen S, Salmikivi L, Simojoki H, Koskinen MT, Pyörälä S. Real-time polymerase chain reaction-based identification of bacteria in milk samples from bovine clinical mastitis with no growth in conventional culturing. Journal of Dairy Science, 2009, 92(6): 2610-2617. DOI:10.3168/jds.2008-1729 |
| [7] | Oikonomou G, Machado VS, Santisteban C, Schukken YH, Bicalho RC. Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA. PLoS One, 2012, 7(10): e47671. DOI:10.1371/journal.pone.0047671 |
| [8] | Kuehn JS, Gorden PJ, Munro D, Rong RC, Dong QF, Plummer PJ, Wang C, Phillips GJ. Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLoS ONE, 2013, 8(4): e61959. DOI:10.1371/journal.pone.0061959 |
| [9] | Marchand S, Heylen K, Messens W, Coudijzer K, De Vos P, Dewettinck K, Herman L, De Block J, Heyndrickx M. Seasonal influence on heat-resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environmental Microbiology, 2009, 11(2): 467-482. DOI:10.1111/j.1462-2920.2008.01785.x |
| [10] | Katholm J, Bennedsgaard TW, Koskinen MT, Rattenborg E. Quality of bulk tank milk samples from Danish dairy herds based on real-time polymerase chain reaction identification of mastitis pathogens. Journal of Dairy Science, 2012, 95(10): 5702-5708. DOI:10.3168/jds.2011-5307 |
| [11] | Shome BR, Das Mitra S, Bhuvana M, Krithiga N, Velu D, Shome R, Isloor S, Barbuddhe SB, Rahman H. Multiplex PCR assay for species identification of bovine mastitis pathogens. Journal of Applied Microbiology, 2011, 111(6): 1349-1356. DOI:10.1111/j.1365-2672.2011.05169.x |
| [12] | Kuang Y, Tani KR, Synnott AJ, Ohshima K, Higuchi H, Nagahata H, Tanji Y. Characterization of bacterial population of raw milk from bovine mastitis by culture-independent PCR-DGGE method. Biochemical Engineering Journal, 2009, 45(1): 76-81. DOI:10.1016/j.bej.2009.02.010 |
| [13] | Kennedy R, Lappin DF, Dixon PM, Buijs MJ, Zaura E, Crielaard W, O'Donnell L, Bennett D, Brandt BW, Riggio MP. The microbiome associated with equine periodontitis and oral health. Veterinary Research, 2016, 47: 49. DOI:10.1186/s13567-016-0333-1 |
| [14] | Oultram JWH, Ganda EK, Boulding SC, Bicalho RC, Oikonomou G. A metataxonomic approach could be considered for cattle clinical mastitis diagnostics. Frontiers in Veterinary Science, 2017, 4: 36. |
| [15] | Cremonesi P, Ceccarani C, Curone G, Severgnini M, Pollera C, Bronzo V, Riva F, Addis MF, Filipe J, Amadori M, Trevisi E, Vigo D, Moroni P, Castiglioni B. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE, 2018, 13(10): e0205054. DOI:10.1371/journal.pone.0205054 |
| [16] |
Zeng XQ, Liu CJ, Yang X, Li XR. Microbial community structure and diversity of mastitis cows by 16S rRNA high-throughput sequencing. Acta Agriculturae Zhejiangensis, 2019, 31(9): 1437-1445.
(in Chinese) 曾学琴, 柳陈坚, 杨雪, 李晓然. 高通量测序法检测奶牛乳房炎关联微生物群落结构及多样性. 浙江农业学报, 2019, 31(9): 1437-1445. DOI:10.3969/j.issn.1004-1524.2019.09.06 |
| [17] | Levison LJ, Miller-Cushon EK, Tucker AL, Bergeron R, Leslie KE, Barkema HW, DeVries TJ. Incidence rate of pathogen-specific clinical mastitis on conventional and organic Canadian dairy farms. Journal of Dairy Science, 2016, 99(2): 1341-1350. DOI:10.3168/jds.2015-9809 |
| [18] | Vakkamäki J, Taponen S, Heikkilä AM, Pyörälä S. Bacteriological etiology and treatment of mastitis in Finnish dairy herds. Acta Veterinaria Scandinavica, 2017, 59(1): 33. DOI:10.1186/s13028-017-0301-4 |
| [19] | Massé J, Dufour S, Archambault M. Characterization of Klebsiella isolates obtained from clinical mastitis cases in dairy cattle. Journal of Dairy Science, 2020, 103(4): 3392-3400. DOI:10.3168/jds.2019-17324 |
| [20] | Kerro Dego O, Oliver SP, Almeida RA. Host-pathogen gene expression profiles during infection of primary bovine mammary epithelial cells with Escherichia coli strains associated with acute or persistent bovine mastitis. Veterinary Microbiology, 2012, 155(2/3/4): 291-297. |
| [21] | Zhang J, Jiang Y, Xia X, Wu J, Almeida R, Eda S, Qi HC. An on-site, highly specific immunosensor for Escherichia coli detection in field milk samples from mastitis-affected dairy cattle. Biosensors and Bioelectronics, 2020, 165: 112366. DOI:10.1016/j.bios.2020.112366 |
| [22] | Vasquez AK, Ganda EK, Capel MB, Eicker S, Virkler PD, Bicalho RC, Nydam DV. The microbiome of Escherichia coli and culture-negative nonsevere clinical mastitis: Characterization and associations with linear score and milk production. Journal of Dairy Science, 2019, 102(1): 578-594. DOI:10.3168/jds.2018-15062 |
| [23] | 韩天赐. 西藏曲才热泉的细菌多样性及栖热菌属比较基因组学. 云南大学硕士学位论文, 2018. |
| [24] |
Li CF, Li Y, Wu XY, Cao YB, Wang WD, Bao MT. Study on the activation Geobacillus species in Zhan 3 blocks of Shengli Oilfield. Journal of China University of Petroleum: Edition of Natural Science, 2016, 40(1): 163-167.
(in Chinese) 李彩风, 李阳, 吴昕宇, 曹嫣镔, 汪卫东, 包木太. 胜利油田沾3区块油藏中Geobacillus菌的激活研究. 中国石油大学学报: 自然科学版, 2016, 40(1): 163-167. |
| [25] | Hoekstra J, Zomer AL, Rutten VPMG, Benedictus L, Stegeman A, Spaninks MP, Bennedsgaard TW, Biggs A, De Vliegher S, Mateo DH, Huber-Schlenstedt R, Katholm J, Kovács P, Krömker V, Lequeux G, Moroni P, Pinho L, Smulski S, Supré K, Swinkels JM, Holmes MA, Lam TJGM, Koop G. Genomic analysis of European bovine Staphylococcus aureus from clinical versus subclinical mastitis. Scientific Reports, 2020, 10(1): 18172. DOI:10.1038/s41598-020-75179-2 |
| [26] | Pérez VKC, Custódio DAC, Silva EMM, Oliveira J, Guimarães AS, Brito MAVP, Souza-Filho AF, Heinemann MB, Lage AP, Dorneles EMS. Virulence factors and antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis in Brazil. Brazilian Journal of Microbiology, 2020, 51(4): 2111-2122. DOI:10.1007/s42770-020-00363-5 |
| [27] | Rodrigues MX, Lima SF, Higgins CH, Canniatti-Brazaca SG, Bicalho RC. The Lactococcus genus as a potential emerging mastitis pathogen group: a report on an outbreak investigation. Journal of Dairy Science, 2016, 99(12): 9864-9874. DOI:10.3168/jds.2016-11143 |
| [28] | de Oliveira GB, Favarin L, Luchese RH, McIntosh D. Psychrotrophic bacteria in milk: How much do we really know?. Brazilian Journal of Microbiology, 2015, 46(2): 313-321. DOI:10.1590/S1517-838246220130963 |
| [29] | Vithanage NR, Dissanayake M, Bolge G, Palombo EA, Yeager TR, Datta N. Biodiversity of culturable psychrotrophic microbiota in raw milk attributable to refrigeration conditions, seasonality and their spoilage potential. International Dairy Journal, 2016, 57: 80-90. DOI:10.1016/j.idairyj.2016.02.042 |
| [30] | Vithanage NR, Dissanayake M, Bolge G, Palombo EA, Yeager TR, Datta N. Microbiological quality of raw milk attributable to prolonged refrigeration conditions. The Journal of Dairy Research, 2017, 84(1): 92-101. DOI:10.1017/S0022029916000728 |
| [31] | 席晓敏. 乳房炎牛乳中微生物多样性及代谢组学研究. 呼和浩特: 内蒙古农业大学硕士学位论文, 2016. |
| [32] | Santman-Berends IMGA, Swinkels JM, Lam TJGM, Keurentjes J, van Schaik G. Evaluation of udder health parameters and risk factors for clinical mastitis in Dutch dairy herds in the context of a restricted antimicrobial usage policy. Journal of Dairy Science, 2016, 99(4): 2930-2939. DOI:10.3168/jds.2015-10398 |
| [33] | Fukushima Y, Kino E, Furutani A, Minamino T, Mikurino Y, Horii Y, Honkawa K, Sasaki Y. Epidemiological study to investigate the incidence and prevalence of clinical mastitis, peracute mastitis, metabolic disorders and peripartum disorders, on a dairy farm in a temperate zone in Japan. BMC Veterinary Research, 2020, 16(1): 389. DOI:10.1186/s12917-020-02613-y |
 2021, Vol. 61
2021, Vol. 61




