中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张嵩元, 汪卫东. 2021
- Songyuan Zhang, Weidong Wang. 2021
- 基因工程微生物合成鼠李糖脂表面活性剂的研究进展
- Recent advances in the production of rhamnolipid biosurfactant by genetically engineered microorganisms
- 微生物学报, 61(10): 3059-3075
- Acta Microbiologica Sinica, 61(10): 3059-3075
-
文章历史
- 收稿日期:2020-11-24
- 修回日期:2021-03-02
- 网络出版日期:2021-03-15
2. 中石化重点实验室微生物采油研究中心, 胜利油田石油工程技术研究院, 山东 东营 257000
2. Sinopec Key Laboratory of MEOR, Petroleum Engineering Technology Research Institute, Shengli Oilfield Company, SINOPEC, Dongying 257000, Shandong Province, China
鼠李糖脂(rhamnolipid)是一类研究时间长、应用技术成熟的生物表面活性剂。其包含1−2分子鼠李糖残基组成的亲水基团和1-2个β-羟基脂肪酸单元组成的疏水基团,二者通过β-糖苷键连接。鼠李糖脂具有丰富的结构多样性(糖基数量、脂链数量、脂链长度、饱和度等),约60种鼠李糖脂同类物(congener)或同系物(homologue)已被鉴定出来[1],其分子式可表述为Rhl-Cx、Rhl-Cx-Cy、Rhl-Rhl-Cx或Rhl-Rhl-Cx-Cy (Rhl为鼠李糖;x、y通常为8-16)。
与传统的化学表面活性剂相比,鼠李糖脂不但具有更优异的表面活性和稳定性,还具有无毒、无污染、能生物降解、生物相容性好等优点,从而在医药、食品、农业、石油开采、环境污染修复等众多领域展示了其独特的应用前景[2-3]。例如,鼠李糖脂已被成功应用于微生物采油(microbial enhanced oil recovery,MEOR)中[4-5]。鼠李糖脂能够通过降低油水界面张力、乳化分散残余油、改变岩石润湿性等多种机理提高水驱采油的效率,而且能在高温、高矿化度的油藏环境中保持其性能稳定性[6-7]。在使用时,地上生产的发酵液无需进行分离纯化,直接注入地下即可,大大降低了应用成本。随着人类社会的进步发展和人们环保意识的增强,生物表面活性剂将有更加广阔的前景,有望成为传统的化学合成表面活性剂的替代品。
提升鼠李糖脂产量主要通过基因工程改造和发酵工艺优化(如培养条件、培养基成分、过程控制等)两种途径。虽然发酵优化能够显著提升鼠李糖脂的产量[8-9],但仍存在诸多限制,例如:转化效率、发酵底物选择、产物类型等重要生产指标仍受菌种遗传背景限制;发酵罐中的高产不代表在发酵罐外的复杂应用场景(如油藏、土壤等)中的高产,自然条件下的低氧[10]、高渗[11]、高温[12]胁迫均会对鼠李糖合成产生较大负面影响;主流生产菌株铜绿假单胞菌(Pseudomonas aeruginosa)具有较强致病性等。因此,利用基因工程开发更高效的鼠李糖脂合成菌株受到了越来越多的关注,与发酵工艺优化具有同等重要的地位。
随着代谢工程和合成生物学技术的发展,多种改造策略被用于优化鼠李糖脂生产(指标包括产量、生产安全性、产物类型定制化等),如代谢通路改造[13]、酶工程[14]、定向进化[15]、底盘工程[16]、实验室适应进化[17]等,未来也将有更多技术应用于其中。本文以基因工程策略为线索,对近年来基因工程微生物合成鼠李糖脂的研究进展进行综述,并对其未来发展方向作出展望。
1 鼠李糖脂的合成通路及其调控鼠李糖脂的合成通路目前研究得较为透彻,如图 1所示。其亲水基团来自dTDP-L-鼠李糖。首先,葡萄糖-6-磷酸在AlgC的作用下转化为葡萄糖-1-磷酸,随后在RmlA、RmlB、RmlC和RmlD的作用下依次经历活化、脱水、异构、还原四步合成dTDP-L-鼠李糖。β-羟基脂肪酸提供鼠李糖脂的疏水基团,但其究竟来源于脂肪酸从头合成(以β-羟脂酰ACP的形式,由RhlG/FabG合成)还是脂肪酸β-氧化(以β-羟脂酰CoA的形式,由RhlYZ合成)仍存在争议,早期研究支持前者[18],而多项后续研究倾向后者[19-21]。RhlA催化2个β-羟基脂肪酸单体发生酯化反应形成HAA (hydroxyalkanoyloxy-alkanoic acid)。RhlB和RhlC依次在HAA上引入第一、二个鼠李糖基,形成单、双鼠李糖脂。
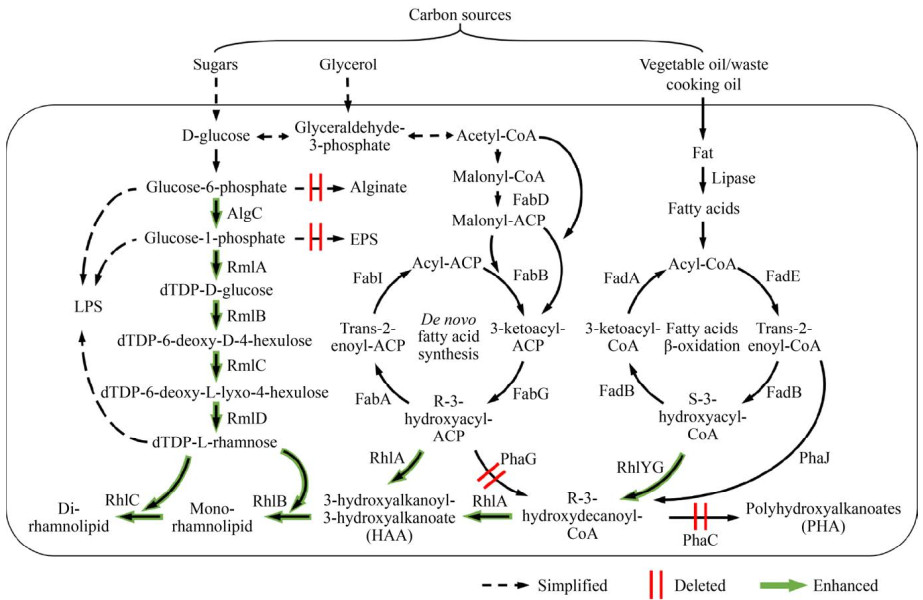
|
| 图 1 鼠李糖脂合成通路和改造靶点 Figure 1 Rhamnolipid biosynthesis pathways and genetic engineering targets. Sugar, glycerol, and vegetable oil are most frequently used as carbon resources in research and commercial production. Precursor dTDP-L-Rhamnose derives from D-glucose. 3-hydroxy fatty acid can be synthesized via de novo fatty acid synthesis (in -ACP form) or fatty acid β-oxidation (in -CoA form). RhlA condenses two 3-hydroxy fatty acids to form HAA. RhlB and RhlC sequentially incorporate two rhamnose groups to the lipid part. To increase the yield, some steps can be enhanced[16, 33, 45, 50-52] (thick arrow), while some competitive pathways can be deleted (double lines), including alginate[45], extracellular polysaccharide[13, 45] (EPS), and PHA[13, 16, 41] biosynthesis pathways. |
在天然鼠李糖脂合成菌株中,鼠李糖脂的合成通常受到群体响应(quorum sensing,QS)系统调控,即细胞达到一定密度时(例如指数生长后期) 才开始合成鼠李糖脂[22]。P. aeruginosa中,至少有4套QS系统通过复杂的相互作用协调各个鼠李糖脂合成基因的表达,但相关机制尚未被研究清楚,详细内容可参见相关综述[23-24]。虽然细胞密度依赖的鼠李糖脂合成对于微生物适应复杂环境和执行群体行为中具有重要意义,但这在工业生产中并不是一个好性状,因为人们希望工业菌株能在整个发酵过程中持续合成鼠李糖脂。群体响应使得细菌通常在指数生长后期才合成鼠李糖脂,导致了鼠李糖脂的发酵周期过长、转化率低、生产速率低,阻碍了鼠李糖脂的工业化开发。而且,群体响应背后复杂的基因表达调控也加大了基因工程改造的难度。
2 底盘微生物的选择 2.1 鼠李糖脂天然合成菌株鼠李糖脂合成酶系天然存在于多种生物中[25]。其中,人们对假单胞菌属(Pseudomonas)和博克霍尔德氏菌属(Burkholderia)的鼠李糖脂合成研究较为深入。P. aeruginosa在好氧条件下合成鼠李糖脂产量较高,是当前工业生产的主流菌株。从油藏中分离出的P. aeruginosa SG菌株能够在厌氧条件下生长并合成鼠李糖脂,在原位微生物采油(in situ-MEOR)中具有很好的应用前景[26]。然而,P. aeruginosa作为一种条件致病菌,生物安全性较差。Burkholderia属的某些菌种虽然生物安全性较好,且能够合成表面活性更优的长链鼠李糖脂,但其发酵速度慢、产量较低、且遗传背景不够清晰,很少真正用于工业生产中。
2.2 鼠李糖脂的异源合成由于天然合成菌株致病性等一些缺点,鼠李糖脂在公认为安全级(generally recognized as safe,GRAS)微生物中的异源合成受到了广泛关注。鼠李糖脂的两个重要前体dTDP-L-鼠李糖和β-羟基脂肪酸合成通路在各种微生物中普遍存在,理论上只需异源表达rhlABC基因即可实现鼠李糖脂的合成。
异源合成的首选是假单胞菌属的其他菌种。德国亚琛工业大学Lars M. Blank研究组致力于利用恶臭假单胞菌(Pseudomonas putida KT2440)合成鼠李糖脂。P. putida作为假单胞菌属模式菌株,被广泛用作细胞工厂底盘(chassis),具有较好的生物安全性[27]。研究发现,P. putida能耐受高达90 g/L的鼠李糖脂,而且人工引入的鼠李糖脂合成通路不受QS的调控,能够实现在各个阶段合成鼠李糖脂,而非仅在生长后期合成[28]。中国科学院沈阳应用生态研究所张颖研究组在利用厌氧微生物合成鼠李糖脂上进行了一系列研究工作[29]。Zhao等在施氏假单胞菌Pseudomonas stutzeri DQ1中引入rhlABRI基因,首次实现了用厌氧微生物异源合成鼠李糖脂,厌氧条件下产量可达1.61 g/L[30]。还有一些工作利用绿针假单胞菌[31] (Pseudomonas chlororaphis)、防御假单胞菌[32] (Pseudomonas protegens)等底盘合成鼠李糖脂。值得一提的是,上述两个菌种不但安全性好,还可用于生物防治,有望扩展鼠李糖脂在农业中的应用。
大肠杆菌(Escherichia coli)是应用最广泛、基因工程操作最为成熟的底盘微生物。中国科学院天津工业生物技术研究所王钦宏研究组在异源合成鼠李糖脂上取得了一系列进展[15, 33-34]。虽然大肠杆菌获得的鼠李糖脂产量较低(<1 g/L),但便于快速验证一些概念,能够为其他高产菌种的改造工作提供重要参考。
除了原核生物,在真核生物酿酒酵母(Saccharomyces cerevisiae) 中异源合成鼠李糖脂的概念也得到了初步验证[35]。
3 合成生物学策略在鼠李糖脂合成中的应用合成生物学是指在工程学思想指导下,按照特定目标理性设计、改造乃至从头合成人工生物体系[36]。细胞工厂的设计和构建是合成生物学的一个重要研究方向,目前已发展了从分子、途径到基因组各个层次的微生物细胞工厂设计和工程化构建策略,相比于传统的筛选、诱变育种等非理性设计方式,创制效率大幅提升[37]。对于代谢途径中的关键酶,合成生物学提供了多种蛋白质工程手段来优化其催化性能[38];在代谢途径层面,合成生物学提供了大量基因表达调控元件来精细调控甚至动态调控各个途径酶的表达量[39];蓬勃发展的基因编辑和DNA合成技术使大规模改造甚至从头合成宿主基因组成为可能,将为细胞工厂创造更优良的底盘生物[40]。在上述策略的指导下,产鼠李糖脂细胞工厂的构建近年来也取得了长足进步(图 2,表 1)。

|
| 图 2 基因工程微生物合成鼠李糖中应用的代谢工程策略 Figure 2 Overview of the engineering strategies used to optimize rhamnolipid production. These strategies include heterogeneous production (mainly P. putida[16, 28, 50, 51, 70-71], E. coli[15, 33-34]), pathway engineering, gene expression engineering[50-54, 56], chassis engineering[16-17, 60], and protein engineering[14-15]. Besides increasing the yield, tailor-made rhamnolipid production[69-71] also draw more and more attentions. |
| Chassis | Engineering strategies | Medium and carbon source | Maximum yield | Increased/% | References |
| P. aeruginosa SG | Blocking exopolysaccharide and PHA synthesis pathways | 60 g/L glycerol | 21.496 g/L | 69.7 | [13] |
| P. putida KT2440 | Chromosomally integrated rhl genes expressed by constitutive promoter P14ffg; Deleting flagella machinery | Delft medium with 10 g/L glucose | ~1.1 g/L | 130 | [16] |
| P. putida KT2440 | Chromosomally integrated rhl genes expressed by constitutive promoter P14ffg; Blocking PHA synthesis | Delft medium with 10 g/L glucose | ~1 g/L | 115 | [16] |
| P. putida KT2440 | Chromosomally integrated rhl genes expressed by salicylate inducible promoter (NagR/PnagAa) | LB medium with 10 g/L glucose | ~1 g/L (high genetic stability) | - | [16] |
| P. aeruginosa PA14 | Blocking PHA synthesis and over-expressing the rhlAB-R operon | PPGAS medium | ~1500 μmol/L | 59 | [41] |
| P. aeruginosa PAO1 | Overexpressing EstA | PPGAS medium (0.5% glucose) | 1.7 g/L | 325 | [42] |
| P. aeruginosa PAO1 | Overexpressing EstA | 42 g/L glycerol | 22 g/L | 290 | [43] |
| P. aeruginosa PAO1 | Overexpressing LipC | M9-minimal broth medium (4 g/L glucose) | 0.8 g/L | 100 | [44] |
| E. coli BL21 (DE3) | Co-expressing rhlABC and rfbD | LB medium with 0.4% Glucose | 0.64 g/L | 43 | [33] |
| P. aeruginosa PAO1 | Blocking exopolysaccharide, alginate, PHA synthesis and overexpressing rmlBDAC, rhlYZ, algC, fadD4, lipC/estA | MS medium (75 g/L palm oil) | 43.7 g/L | 108 | [45] |
| E. coli BL21 (DE3) | Directed evolution of RhlB (L168E) | Mineral salt or LB medium with 0.4% glucose | Surface tension: from 0.26 to 0.021 mN/m | - | [15] |
| P. aeruginosa PA14 | Semi-rational evolution of RhlA (R202K) | MSM-glycerol broth (15 g/L glycerol) | 800 mg/L·OD600 | 101 | [14] |
| P. putida KT2440 | Using synthetic promoter library to drive the expression of rhlA-rhlB-gfp operon (biofilm as the production platform) | FAB medium with 1 mmol/L sodium citrate | Yield linearly correlated with enzyme level | - | [50] |
| P. putida KT2440 | Synthetic promoter library | LB medium with 10 g/L glucose | 2.8 g/L | - | [51] |
| P. aeruginosa SG | Plasmid-encoded rhlAB fused with the strong oprL promoter | Glycerol-nitrate medium (45 g/L glycerol) | 20.98 g/L | 183 | [52] |
| B. thailandensis E264 | Deleting three QS systems (TΔbtaI1ΔbtaI2ΔbtaI3) | Nutrient broth with 4 g/L glycerol | 4.46 g/L | 374 | [53] |
| B. thailandensis E264 | Releasing QS’s repression on rhl genes by deleting scmR | Nutrient broth (NB) medium with 2 or 4% glycerol | 3 g/L | 200 | [54] |
| P. aeruginosa PAK | Mutating the 5′UTR of rhlI to release repression by sRNA P27 | M8-based agar plate with 0.2% glucose | Diameter ratio from 1.72 to 1.95 (CTAB agar test) | - | [56] |
| P. aeruginosa (NRRL B-771) | Expressing the Vitreoscilla hemoglobin gene (vgb) | MM media with 1% glucose | 8.373 g/L | 100 | [60] |
| P. putida KT2440 | Performing adaptive laboratory evolution in order to use ethanol as carbon source and defoamer | Modified M9 medium with 0.96% ethanol | 0.94 g/L | 47 | [17] |
| -: none. | |||||
3.1 代谢通路优化提升前体物质供给
删除竞争旁路或过表达通路中的关键酶是提升目标通路中代谢流的常用方法。对于鼠李糖脂合成,提升脂前体β-羟基脂肪酸和糖前体dTDP-L-鼠李糖合成通路中的代谢流都可以提高鼠李糖脂产量。
假单胞菌中,聚羟基脂肪酸酯(polyhydroxyalkanoates,PHA)合成与鼠李糖脂合成共享β-羟基脂肪酸前体。多项工作涉及删除PHA合成通路,将代谢流引向鼠李糖脂合成[13, 16, 41]。在P. aeruginosa中过表达脂肪酶EstA[42-43]和LipC[44]也可提升鼠李糖脂的产量,但机制尚不明确,猜测可能与脂肪代谢有关。
胞外多糖(exopolysaccharide,EPS)的合成消耗dTDP-L-鼠李糖合成通路中的dTDP-L-葡萄糖。Lei等敲除了P. aeruginosa SG的胞外多糖合成基因pslAB,鼠李糖脂产量较野生型提升21%,在此基础上再敲除phaC1DC2,产量较野生型提升69.7%,达到21.496 g/L[13]。
Du等在E. coli中优化rhlABC的表达量,并引入额外rfbD基因(同假单胞菌中rmlD基因)拷贝以提高前体物质dTDP-L-鼠李糖的供给,鼠李糖脂的产量从0.446 g/L提高至0.64 g/L,创造了利用大肠杆菌合成鼠李糖脂的最高产量[33]。
组合使用多种策略在一定程度上可以产生叠加效果。一项来自中国科学院微生物研究所的专利将P. aeruginosa PAO1一系列消耗前体物质的旁路基因(胞外多糖合成基因簇pslA-pslO和pelA-pelG、藻酸盐合成基因簇algD-alg8-alg44-algKEGXLIJF、PHA合成基因簇phaC1-D-C2)逐步替换为有助于提升产量的基因(rmlBDAC、rhlYZ、algC、fadD4、lipC/estA),产量从21 g/L逐步提升至43.7 g/L[45]。
3.2 蛋白质工程优化鼠李糖脂合成酶除了优化代谢通路提升目标通路上的代谢流量,也可采取蛋白质工程手段直接对酶进行改造。蛋白质工程经历了从定向进化到计算设计的发展历程,前者需要进行多轮大规模的突变和筛选从而引导蛋白质朝着人类想要的方向进化,而后者基于对蛋白质结构和机理的研究以及高性能计算方法从头设计蛋白质[46]。
由于RhlA、RhlB、RhlC均缺少实验获得的晶体学结构数据,已有工作均采用定向进化[47] (directed evolution)和半理性设计[48] (semi-rational design)两种较为初级的方式对鼠李糖脂合成酶进行改造。2014年,Han等首次将定向进化的方法引入鼠李糖脂代谢工程研究中。该研究对rhlB基因进行易错PCR,并利用鼠李糖脂的抗菌性设计了一种通量较高的筛选方案:在发酵上清液中培养枯草芽孢杆菌,OD值小说明鼠李糖脂合成量高。该方案筛选出了L168这一关键位点,将其突变为谷氨酸后,产物中长链鼠李糖脂比例高,表面活性和模拟驱油能力都有所提升[15]。2019年,Dulcey等对RhlA进行了“半理性”改造。该研究利用同源建模(homology modeling)、点突变、构建嵌合酶等方式鉴定了一系列参与催化和底物识别的重要位点,其中,针对负责底物识别的cap-domain的一些突变可以提升酶对底物的识别效率(尤其是长链底物),从而提升产量以及产物中长链鼠李糖脂的比例。其中,Y165F和R202K突变使产量提升约一倍[14]。
3.3 基因表达层面的优化在一定范围内,细胞工厂产量与关键合成酶的表达量直接相关[49]。通过采用强启动子、增加基因拷贝数等方式提高合成酶的表达量也是提高鼠李糖脂产量的一种方式。
Wigneswaran等在P. putida中构建了人工合成启动子文库(synthetic promoter library,SPL),用于驱动rhlA-rhlB-gfp多顺反子的表达。用高效液相色谱-高分辨率质谱(UHPLC-HRMS)和流式细胞仪分别对鼠李糖脂产量与鼠李糖脂合成酶表达量进行精确定量,发现二者在很大范围内具有线性相关性[50]。
Tiso等以鼠李糖脂合成为例验证了一种“需求驱动” (driven-by-demand)的代谢工程策略。该策略基于“外周代谢途径受转录调控,而中心碳代谢途径在代谢反应层面被调控”这一事实。在P. putida中,通过较强的组成型启动子提高rhlAB基因的表达量后,为鼠李糖脂合成提供前体物质的中心碳代谢途径上的代谢流也随之提升,以满足鼠李糖脂合成这一外周代谢途径的需求,解释了产量与基因表达量的相关性[51]。
除了启动子强度,基因拷贝数可以通过剂量效应(gene dosage effect)影响基因表达水平。Zhao等在能够厌氧合成鼠李糖脂的P. aeruginosa SG中引入质粒编码的rhlAB基因,并由oprL基因的组成型启动子驱动表达,鼠李糖脂产量从11 g/L提升至21 g/L[52]。
质粒虽然由于剂量效应表达量高,但稳定性较差,需要抗生素来维持以避免丢失,不利于工业生产。Tiso等利用Tn7转座子将鼠李糖脂合成基因整合入P. putida基因组中,解决了这一问题。然而,即使在基因组上,过强的组成型表达也会为宿主造成代谢压力,导致基因组不稳定,自发地将目的基因丢失。采用水杨酸诱导型启动子驱动鼠李糖脂合成基因表达不但增强了工程菌株的稳定性,还获得了更好的产量[16]。
随着人们对群体响应(QS)控制鼠李糖脂合成机制理解的加深,QS系统成为了提升鼠李糖脂产量的新靶标。在Burkholderia thailandensis中,QS系统在指数生长期抑制鼠李糖脂合成酶基因的表达。敲除B. thailandensis 3条QS途径的信号分子合成酶,鼠李糖脂产量从0.94±0.06 g/L提升至4.46±0.345 g/L[53]。后续研究发现,在指数生长期,QS系统通过激活scmR基因抑制鼠李糖脂合成基因表达,同时促进PHA合成基因的表达,因此,ΔscmR菌株能更早地开始合成鼠李糖脂,同时将更多代谢流从PHA合成引向鼠李糖脂,具有更高产量[54]。然而,不同菌种的QS系统对鼠李糖脂合成的调控模式不同。例如,在P. aeruginosa和Burkholderia glumae中,QS对鼠李糖脂合成基因起正调控作用,敲除后会降低产量[22, 55],若要提升产量,则需要上调群体响应水平。P. aeruginosa中,小RNA (small RNA,sRNA) 在群体响应调控中扮演着重要角色,sRNA P27通过与群体响应关键组分rhlI mRNA的5′UTR相互作用抑制群体响应,从而下调鼠李糖脂合成,通过点突变解除sRNA P27对rhlI mRNA的抑制作用可以提高鼠李糖脂产量[56]。值得注意的是,对于铜绿假单胞菌社会行为的基础研究表明,将鼠李糖脂合成与QS解偶联后,利用群体生产的鼠李糖脂而自身不合成鼠李糖脂的“欺骗者”的比例会逐渐提高[57],这种现象对时间跨度较长的发酵过程可能产生一定影响。
3.4 底盘工程细胞工厂的产量与宿主的生理活动密切相关[58]。在不改变合成线路的情况下对底盘生物进行优化,也能够间接提升细胞工厂的产量。
透明颤菌血红蛋白(Vitreoscilla hemoglobin,VHb)能够提升细胞对氧气的摄取能力,全局性地优化细胞生长,被广泛应用于代谢工程中[59]。在P. aeruginosa中引入VHb的同时增加供氧量,鼠李糖脂产量从不足4 g/L提升至8.373 g/L[60]。
实验室适应性进化(adaptive laboratory evolution,ALE)也是一种常用的菌种改造手段[61]。Bator等对P. putida KT2440进行实验室适应性进化,获得了能高效利用乙醇为碳源合成鼠李糖脂的菌株,同时,乙醇作为消泡剂解决了发酵过程中的泡沫问题,通过对进化而来的菌株进行基因组重测序,乙醇适应性产生的机理也得到揭示[17]。
基因组中许多基因虽然不直接与目标合成通路竞争底物,但其表达和复制消耗了大量能量,对基因组进行精简有助于提高细胞工厂产量[62]。Tiso等删除鞭毛相关基因,使更多能量能够用于鼠李糖脂合成,P. putida产量从0.47 g/L提高了130%[16]。
4 鼠李糖脂的“定制化”合成微生物合成的鼠李糖脂是由多种同系物组成的混合物,不同类型的鼠李糖脂具有不同的性质。例如,在微生物强化采油中,单、双鼠李糖脂具有不同的界面活性、润湿性能、洗油效率等性质,在总量不变的情况下,优化配比也能显著提升采油性能[63-66]。此外,单、双鼠李糖脂的比例还影响鼠李糖脂的抗菌性[67],烃链长度也和表面活性呈现正相关性[68]。基于不同应用场景对于不同类型鼠李糖脂的需求,Wittgens等提出了“定制化”合成鼠李糖脂(tailor-made rhamnolipids)的概念[69]。
目前,基因工程方法能够控制鼠李糖脂的糖基数量和脂链长度两个参数。仅表达rhlA和rhlB合成单糖鼠李糖脂,再表达rhlC则可合成双糖鼠李糖脂。脂链长度与酶的底物选择性有关。来自P. aeruginosa的合成酶能够合成中等脂链长度(C8至C12)的鼠李糖脂,以C10-C10为主,而来自Burkholderia属(如B. glumae)的合成酶能合成C12至C16的长链鼠李糖脂。在E. coli中转入来自P. aeruginosa和B. pseudomallei的RhlAB和RhlC,获得了烃链长度分布不同的单、双鼠李糖脂[33]。在P. putida中组合来自P. aeruginosa和B. glumae的rhlA和rhlB,获得了烃链长度分布不同的单鼠李糖脂[70-71]。前文中提到的利用半理性设计和定向进化手段对RhlA和RhlB酶的改造都增大了底物口袋,使得长链底物更好进入,提高了长链鼠李糖脂的比例[14-15]。
5 展望本课题组长期致力于微生物产鼠李糖脂表面活性剂在石油开采中的应用,目前已经获得比较理想的效果[63],对不同条件和性质的油藏也有较好的适应性[7],但是其大规模应用还要进一步降低生产成本。由于鼠李糖脂在油田中使用时不需要进行提取和纯化,直接注入发酵菌液即可,而且可以采用废弃油脂等低成本原料作为培养基[72-73],因此生产成本主要由产率决定。而产率的决定性因素是菌株本身,所以通过基因工程手段提升产率对于鼠李糖脂在石油开采中的应用具有重要意义。
上述利用基因工程微生物合成鼠李糖脂的工作主要集中在异源合成、产量提升、定制化合成3个方面。虽然应用了多种改造策略,且取得了一些研究进展,但多数研究工作仍围绕基因敲除与过表达等传统代谢工程手段。快速发展的合成生物学技术有望为鼠李糖脂合成带来新的工具和思路,进一步提升鼠李糖脂的产量(图 3)。
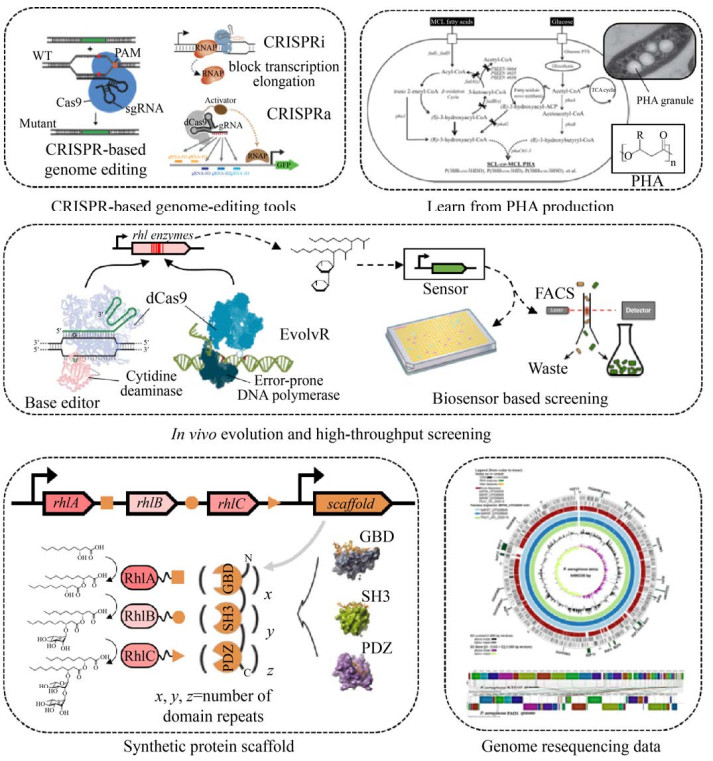
|
| 图 3 可用于提升鼠李糖脂产量的潜在策略 Figure 3 Potential strategies that can be used to optimize rhamnolipid production. Available tools include the CRISPR-based genome engineering toolkit[78-80], in vivo directed evolution platform[78, 87], synthetic protein scaffold[83-84], etc. Besides, we can learn from medium-chain PHA production[86], which shares same precursors with rhamnolipid, and genome resequencing data of rhamnolipid high-yield strains[90-91]. |
(1) 应用基于CRISPR的基因编辑工具。迄今为止,所有对产鼠李糖脂微生物进行基因改造的研究工作均采用转座子插入[74-75]和同源重组[76-77]两种方式,费时费力。近年来,基于CRISPR的基因编辑技术发展迅速:假单胞菌属的CRISPR基因编辑工具箱能够以接近100%的效率进行基因突变、删除、插入以及单碱基编辑[78];CRISPR干扰系统[79] (CRISPR interference,CRISPRi)也在假单胞菌中成功建立[80],无需对靶基因进行突变即可实现对任意基因表达的控制。上述方法将大大加速增产基因筛选及工程菌构建的速度。
(2) 采用日益丰富的基因表达调控元件。如前文所述,鼠李糖脂产量与合成酶的表达量密切相关。近年来,P. putida的常表达启动子库被成功构建[74, 81],能在很大范围内调节基因的表达量。此外,来自噬菌体的T7或类T7聚合酶表达系统具有表达强度高、可调性好、能跨物种使用等优点[82],有望在非模式微生物中实现高产鼠李糖脂。
(3) 利用合成蛋白支架(synthetic protein scaffold)。合成支架可以将一条通路中的多个酶在空间上共定位,通过提高酶和反应物的局部浓度,产生“底物通道”效应,提升反应效率[83-84]。利用该支架将鼠李糖脂合成通路中的某些酶共定位(如RmlD、RhlG、RhlYZ、RhlA、RhlB),有望提高鼠李糖脂产量。
(4) 借鉴PHA增产思路。PHA与鼠李糖脂在代谢通路上具有高度相关性[85]。作为一种性能优秀的生物聚合物生物产品,PHA的研究和产业化比鼠李糖脂要成熟得多,多种合成生物学策略都已成功应用于PHA生产中[86]。国内,清华大学陈国强研究组和蓝晶微生物科技有限公司(Bluepha Co.,Ltd.)在PHA合成领域处于世界领先地位。PHA的增产策略有望为鼠李糖脂提供思路。
(5) 构建体内定向进化及高通量筛选平台。3.2中提到的两项改造鼠李糖脂合成酶的工作[14-15]均需要人工在体外构建突变体库,费时费力。而体内进化方法能在特定区域高效引入突变。例如,一种名为EvolvR的系统可对长达250 bp的目标窗口进行高效突变[87]。单碱基编辑器可对较小的突变窗口(3-8 bp)进行突变[78]。除突变外,定向进化的另一个要素是筛选。传统的鼠李糖脂定性、定量检测方法[88]难以满足定向进化对高通量筛选的需求[14],开发一种鼠李糖脂生物传感器或许是一种可行方法[89]。
(6) 挖掘高产菌株的基因组学数据。Liu等对一株高产双鼠李糖脂的铜绿假单胞菌进行重测序,发现其特异地表达多种脂肪酶基因,猜测与该菌株能够高效利用油脂合成鼠李糖脂有关,同时还在rhlI、rhlA、rmlA、rmlC基因上发现了一些单核苷酸多态性位点,可能与酶的高催化活性有关[90]。Xu等对油田中分离的3株具有不同鼠李糖脂产量的P. aeruginosa进行重测序,发现群体响应强度对鼠李糖脂合成影响很大[91]。这些重测序数据为鼠李糖脂工程菌的改造提供了宝贵的思路。
致谢
感谢中国大学生公益行动-金牌实习生工作组的支持和资助。
| [1] | Abdel-Mawgoud AM, Lépine F, Déziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Applied Microbiology and Biotechnology, 2010, 86(5): 1323-1336. DOI:10.1007/s00253-010-2498-2 |
| [2] | Moutinho LF, Moura FR, Silvestre RC, Romão-Dumaresq AS. Microbial biosurfactants: a broad analysis of properties, applications, biosynthesis, and techno-economical assessment of rhamnolipid production. Biotechnology Progress, 2020: e3093. |
| [3] | Kumar R, Das AJ. Application of rhamnolipids in agriculture and food industry. Singapore: Springer Singapore, 2018: 97-109. |
| [4] |
Wang WD. Laboratory research and field trials of microbial oil recovery technique. Oil Drilling & Production Technology, 2012, 34(1): 107-113.
(in Chinese) 汪卫东. 微生物采油技术研究及试验. 石油钻采工艺, 2012, 34(1): 107-113. DOI:10.3969/j.issn.1000-7393.2012.01.030 |
| [5] |
Wang WD. Application of microbial enhanced oil recovery and bioreactor in oil reservoir. Chinese Journal of Bioprocess Engineering, 2017, 15(3): 74-78.
(in Chinese) 汪卫东. 微生物采油与油藏生物反应器的应用. 生物加工过程, 2017, 15(3): 74-78. DOI:10.3969/j.issn.1672-3678.2017.03.012 |
| [6] |
Liu L, Fan HF, Zhao J. Progress of biosurfactant in EOR. Oilfield Chemistry, 2018, 35(4): 738-743.
(in Chinese) 刘珑, 范洪富, 赵娟. 生物表面活性剂提高采收率的研究进展. 油田化学, 2018, 35(4): 738-743. |
| [7] | Ding MS, Wang J, Lin JZ, Sun GZ, Wang WD. Potential application of biosurfactants mixtures in high-temperature and high-salinity reservoirs. Petroleum Science and Technology, 2017, 35(12): 1189-1195. DOI:10.1080/10916466.2016.1225088 |
| [8] | Eslami P, Hajfarajollah H, Bazsefidpar S. Recent advancements in the production of rhamnolipid biosurfactants by Pseudomonas aeruginosa. RSC Advances, 2020, 10(56): 34014-34032. DOI:10.1039/D0RA04953K |
| [9] | Li QX. Rhamnolipid synthesis and production with diverse resources. Frontiers of Chemical Science and Engineering, 2017, 11(1): 27-36. DOI:10.1007/s11705-016-1607-x |
| [10] | Zhao F, Shi RJ, Ma F, Han SQ, Zhang Y. Oxygen effects on rhamnolipids production by Pseudomonas aeruginosa. Microbial Cell Factories, 2018, 17(1): 1-11. DOI:10.1186/s12934-017-0850-2 |
| [11] | Bazire A, Dheilly A, Diab F, Morin D, Jebbar M, Haras D, Dufour A. Osmotic stress and phosphate limitation alter production of cell-to-cell signal molecules and rhamnolipid biosurfactant by Pseudomonas aeruginosa. FEMS Microbiology Letters, 2005, 253(1): 125-131. DOI:10.1016/j.femsle.2005.09.029 |
| [12] | Noll P, Treinen C, Müller S, Senkalla S, Lilge L, Hausmann R, Henkel M. Evaluating temperature-induced regulation of a ROSE-like RNA-thermometer for heterologous rhamnolipid production in Pseudomonas putida KT2440. AMB Express, 2019, 9(1): 154. DOI:10.1186/s13568-019-0883-5 |
| [13] | Lei LY, Zhao F, Han SQ, Zhang Y. Enhanced rhamnolipids production in Pseudomonas aeruginosa SG by selectively blocking metabolic bypasses of glycosyl and fatty acid precursors. Biotechnology Letters, 2020, 42(6): 997-1002. DOI:10.1007/s10529-020-02838-9 |
| [14] | Dulcey CE, López de los Santos Y, Létourneau M, Déziel E, Doucet N. Semi-rational evolution of the 3-(3-hydroxyalkanoyloxy)alkanoate (HAA) synthase RhlA to improve rhamnolipid production in Pseudomonas aeruginosa and Burkholderia glumae. The FEBS Journal, 2019, 286(20): 4036-4059. DOI:10.1111/febs.14954 |
| [15] | Han L, Liu P, Peng Y, Lin J, Wang Q, Ma Y. Engineering the biosynthesis of novel rhamnolipids in Escherichia coli for enhanced oil recovery. Journal of Applied Microbiology, 2014, 117(1): 139-150. DOI:10.1111/jam.12515 |
| [16] | Tiso T, Ihling N, Kubicki S, Biselli A, Schonhoff A, Bator I, Thies S, Karmainski T, Kruth S, Willenbrink AL, Loeschcke A, Zapp P, Jupke A, Jaeger KE, Büchs J, Blank LM. Integration of genetic and process engineering for optimized rhamnolipid production using Pseudomonas putida. Frontiers in Bioengineering and Biotechnology, 2020, 8: 976. DOI:10.3389/fbioe.2020.00976 |
| [17] | Bator I, Karmainski T, Tiso T, Blank LM. Corrigendum: killing two birds with one stone-strain engineering facilitates the development of a unique rhamnolipid production process. Frontiers in Bioengineering and Biotechnology, 2020, 8: 899. DOI:10.3389/fbioe.2020.00899 |
| [18] | Zhu K, Rock CO. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. Journal of Bacteriology, 2008, 190(9): 3147-3154. DOI:10.1128/JB.00080-08 |
| [19] | Abdel-Mawgoud AM, Lépine F, Déziel E. A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chemistry & Biology, 2014, 21(1): 156-164. |
| [20] | Zhang L, Veres-Schalnat TA, Somogyi A, Pemberton JE, Maier RM. Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays. Applied and Environmental Microbiology, 2012, 78(24): 8611-8622. DOI:10.1128/AEM.02111-12 |
| [21] | Bazire A, Dufour A. The Pseudomonas aeruginosa rhlG and rhlAB genes are inversely regulated and RhlG is not required for rhamnolipid synthesis. BMC Microbiology, 2014, 14(1): 1-9. DOI:10.1186/1471-2180-14-1 |
| [22] | Van Ditmarsch D, Xavier JB. High-resolution time series of Pseudomonas aeruginosa gene expression and rhamnolipid secretion through growth curve synchronization. BMC Microbiology, 2011, 11: 140. DOI:10.1186/1471-2180-11-140 |
| [23] | Dusane DH, Zinjarde SS, Venugopalan VP, McLean RJ, Weber MM, Rahman PKSM. Quorum sensing: implications on Rhamnolipid biosurfactant production. Biotechnology and Genetic Engineering Reviews, 2010, 27(1): 159-184. DOI:10.1080/02648725.2010.10648149 |
| [24] | Reis RS, Pereira AG, Neves BC, Freire DMG. Gene regulation of rhamnolipid production in Pseudomonas aeruginosa-A review. Bioresource Technology, 2011, 102(11): 6377-6384. DOI:10.1016/j.biortech.2011.03.074 |
| [25] | Germer A, Tiso T, Müller C, Behrens B, Vosse C, Scholz K, Froning M, Hayen H, Blank LM. Exploiting the natural diversity of RhlA acyltransferases for the synthesis of the rhamnolipid precursor 3-(3-hydroxyalkanoyloxy)alkanoic acid. Applied and Environmental Microbiology, 2020, 86(6): e02317-19. DOI:10.1128/aem.02317-19 |
| [26] | Zhao F, Zhang J, Shi RJ, Han SQ, Ma F, Zhang Y. Production of biosurfactant by a Pseudomonas aeruginosa isolate and its applicability to in situ microbial enhanced oil recovery under anoxic conditions. RSC Advances, 2015, 5(45): 36044-36050. DOI:10.1039/C5RA03559G |
| [27] | Nikel PI, Martínez-García E, de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nature Reviews Microbiology, 2014, 12(5): 368-379. DOI:10.1038/nrmicro3253 |
| [28] | Wittgens A, Tiso T, Arndt TT, Wenk P, Hemmerich J, Müller C, Wichmann R, Küpper B, Zwick M, Wilhelm S, Hausmann R, Syldatk C, Rosenau F, Blank LM. Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microbial Cell Factories, 2011, 10(1): 1-18. DOI:10.1186/1475-2859-10-1 |
| [29] |
Zhao F, Zhang Y. Advances in enhanced oil recovery by biosurfactant producing microorganisms under anaerobic conditions. Biotic Resources, 2018, 40(2): 101-106.
(in Chinese) 赵峰, 张颖. 厌氧产表面活性剂微生物提高原油采收率的研究进展. 生物资源, 2018, 40(2): 101-106. |
| [30] | Zhao F, Shi R, Zhao J, Li G, Bai X, Han S, Zhang Y. Heterologous production of Pseudomonas aeruginosa rhamnolipid under anaerobic conditions for microbial enhanced oil recovery. Journal of Applied Microbiology, 2015, 118(2): 379-389. DOI:10.1111/jam.12698 |
| [31] | Solaiman DKY, Ashby RD, Gunther NW, Zerkowski JA. Dirhamnose-lipid production by recombinant nonpathogenic bacterium Pseudomonas chlororaphis. Applied Microbiology and Biotechnology, 2015, 99(10): 4333-4342. DOI:10.1007/s00253-015-6433-4 |
| [32] |
Xie ZL, Chen HN, Zhong L, Xu JY, Li Y, Pan D, Liu F, Ding XZ, Xia LQ, Zhang YM, Tu Q. Heterologous expression and activity of rhamnolipid gene regulated by three different constitutive promoters in Pseudomonas protegens. Acta Laser Biology Sinica, 2019, 28(3): 229-238.
(in Chinese) 谢芝玲, 陈汉娜, 钟林, 徐佳莹, 李演, 潘登, 刘峰, 丁学知, 夏立秋, 张友明, 涂强. 三种组成型启动子调控的鼠李糖脂基因在防御假单胞菌中的异源表达及活性研究. 激光生物学报, 2019, 28(3): 229-238. DOI:10.3969/j.issn.1007-7146.2019.03.004 |
| [33] | Du J, Zhang AJ, Hao JA, Wang J. Biosynthesis of di-rhamnolipids and variations of congeners composition in genetically-engineered Escherichia coli. Biotechnology Letters, 2017, 39(7): 1041-1048. DOI:10.1007/s10529-017-2333-2 |
| [34] |
Gong ZJ, Peng YF, Zhang YT, Song GT, Chen WJ, Jia SR, Wang QH. Construction and optimization of Escherichia coli for producing rhamnolipid biosurfactant. Chinese Journal of Biotechnology, 2015, 31(7): 1050-1062.
(in Chinese) 巩志金, 彭彦峰, 张煜婷, 宋国田, 陈五九, 贾士儒, 王钦宏. 产鼠李糖脂生物表面活性剂大肠杆菌的构建与优化. 生物工程学报, 2015, 31(7): 1050-1062. |
| [35] | Bahia FM, de Almeida GC, de Andrade LP, Campos CG, Queiroz LR, da Silva RLV, Abdelnur PV, Corrêa JR, Bettiga M, Parachin NS. Rhamnolipids production from sucrose by engineered Saccharomyces cerevisiae. Scientific Reports, 2018, 8: 2905. DOI:10.1038/s41598-018-21230-2 |
| [36] |
Ding MZ, Li BZ, Wang Y, Xie ZX, Liu D, Yuan YJ. Significant research progress in synthetic biology. Synthetic Biology Journal, 2020, 1(1): 7-28.
(in Chinese) 丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进. 合成生物学重要研究方向进展. 合成生物学, 2020, 1(1): 7-28. |
| [37] |
Yuan YM, Xing XH, Zhang C. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale. Synthetic Biology Journal, 2020, 1(6): 656-673.
(in Chinese) 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建: 从诱变育种到全基因组定制化创制. 合成生物学, 2020, 1(6): 656-673. |
| [38] | Xu N, Liu YW, Jiang HF, Liu J, Ma YH. Combining protein and metabolic engineering to construct efficient microbial cell factories. Current Opinion in Biotechnology, 2020, 66: 27-35. DOI:10.1016/j.copbio.2020.06.001 |
| [39] | Holtz WJ, Keasling JD. Engineering static and dynamic control of synthetic pathways. Cell, 2010, 140(1): 19-23. DOI:10.1016/j.cell.2009.12.029 |
| [40] | Liu JY, Wu X, Yao MD, Xiao WH, Zha J. Chassis engineering for microbial production of chemicals: from natural microbes to synthetic organisms. Current Opinion in Biotechnology, 2020, 66: 105-112. DOI:10.1016/j.copbio.2020.06.013 |
| [41] | Gutiérrez-Gómez U, Soto-Aceves MP, Servín-González L, Soberón-Chávez G. Overproduction of rhamnolipids in Pseudomonas aeruginosa PA14 by redirection of the carbon flux from polyhydroxyalkanoate synthesis and overexpression of the rhlAB-R operon. Biotechnology Letters, 2018, 40(11/12): 1561-1566. DOI:10.1007/s10529-018-2610-8 |
| [42] | Wilhelm S, Gdynia A, Tielen P, Rosenau F, Jaeger KE. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. Journal of Bacteriology, 2007, 189(18): 6695-6703. DOI:10.1128/JB.00023-07 |
| [43] | Dobler L, de Carvalho BR, Alves WDS, Neves BC, Freire DMG, Almeida RV. Enhanced rhamnolipid production by Pseudomonas aeruginosa overexpressing estA in a simple medium. PLoS ONE, 2017, 12(8): e0183857. DOI:10.1371/journal.pone.0183857 |
| [44] | Rosenau F, Isenhardt S, Gdynia A, Tielker D, Schmidt E, Tielen P, Schobert M, Jahn D, Wilhelm S, Jaeger KE. Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa. FEMS Microbiology Letters, 2010, 309(1): 25-34. |
| [45] | 朱坤, 于海英. 一种提高鼠李糖脂产量的铜绿假单胞菌及其构建方法. 中国: CN108060111A. 2018-05-22. |
| [46] |
Qu G, Zhu T, Jiang YY, Wu B, Sun ZT. Protein engineering: from directed evolution to computational design. Chinese Journal of Biotechnology, 2019, 35(10): 1843-1856.
(in Chinese) 曲戈, 朱彤, 蒋迎迎, 吴边, 孙周通. 蛋白质工程: 从定向进化到计算设计. 生物工程学报, 2019, 35(10): 1843-1856. |
| [47] | Packer MS, Liu DR. Methods for the directed evolution of proteins. Nature Reviews Genetics, 2015, 16(7): 379-394. DOI:10.1038/nrg3927 |
| [48] | Lutz S. Beyond directed evolution-semi-rational protein engineering and design. Current Opinion in Biotechnology, 2010, 21(6): 734-743. DOI:10.1016/j.copbio.2010.08.011 |
| [49] | Jeschek M, Gerngross D, Panke S. Combinatorial pathway optimization for streamlined metabolic engineering. Current Opinion in Biotechnology, 2017, 47: 142-151. DOI:10.1016/j.copbio.2017.06.014 |
| [50] | Wigneswaran V, Nielsen KF, Sternberg C, Jensen PR, Folkesson A, Jelsbak L. Biofilm as a production platform for heterologous production of rhamnolipids by the non-pathogenic strain Pseudomonas putida KT2440. Microbial Cell Factories, 2016, 15(1): 181. DOI:10.1186/s12934-016-0581-9 |
| [51] | Tiso T, Sabelhaus P, Behrens B, Wittgens A, Rosenau F, Hayen H, Blank LM. Creating metabolic demand as an engineering strategy in Pseudomonas putida-Rhamnolipid synthesis as an example. Metabolic Engineering Communications, 2016, 3: 234-244. DOI:10.1016/j.meteno.2016.08.002 |
| [52] | Zhao F, Cui QF, Han SQ, Dong HP, Zhang J, Ma F, Zhang Y. Enhanced rhamnolipid production of Pseudomonas aeruginosa SG by increasing copy number of rhlAB genes with modified promoter. RSC Advances, 2015, 5(86): 70546-70552. DOI:10.1039/C5RA13415C |
| [53] | Victor IU, Kwiencien M, Tripathi L, Cobice D, McClean S, Marchant R, Banat IM. Quorum sensing as a potential target for increased production of rhamnolipid biosurfactant in Burkholderia thailandensis E264. Applied Microbiology and Biotechnology, 2019, 103(16): 6505-6517. DOI:10.1007/s00253-019-09942-5 |
| [54] | Martinez S, Humery A, Groleau MC, Déziel E. Quorum sensing controls both rhamnolipid and polyhydroxyalkanoate production in Burkholderia thailandensis through ScmR regulation. Frontiers in Bioengineering and Biotechnology, 2020, 8: 1033. DOI:10.3389/fbioe.2020.01033 |
| [55] | Nickzad A, Lépine F, Déziel E. Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS ONE, 2015, 10(6): e0128509. DOI:10.1371/journal.pone.0128509 |
| [56] | Chen RH, Wei XY, Li ZP, Weng YD, Xia YS, Ren WR, Wang XX, Jin YX, Bai F, Cheng ZH, Jin SG, Wu WH. Identification of a small RNA that directly controls the translation of the quorum sensing signal synthase gene rhlI in Pseudomonas aeruginosa. Environmental Microbiology, 2019, 21(8): 2933-2947. DOI:10.1111/1462-2920.14686 |
| [57] | Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Molecular Microbiology, 2011, 79(1): 166-179. DOI:10.1111/j.1365-2958.2010.07436.x |
| [58] | Liao C, Blanchard AE, Lu T. An integrative circuit-host modelling framework for predicting synthetic gene network behaviours. Nature Microbiology, 2017, 2(12): 1658-1666. DOI:10.1038/s41564-017-0022-5 |
| [59] | Stark BC, Pagilla KR, Dikshit KL. Recent applications of Vitreoscilla hemoglobin technology in bioproduct synthesis and bioremediation. Applied Microbiology and Biotechnology, 2015, 99(4): 1627-1636. DOI:10.1007/s00253-014-6350-y |
| [60] | Kahraman H, Erenler SO. Rhamnolipid production by Pseudomonas aeruginosa engineered with the Vitreoscilla hemoglobin gene. Applied Biochemistry and Microbiology, 2012, 48(2): 188-193. DOI:10.1134/S000368381202007X |
| [61] | Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metabolic Engineering, 2019, 56: 1-16. DOI:10.1016/j.ymben.2019.08.004 |
| [62] | Lieder S, Nikel PI, de Lorenzo V, Takors R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microbial Cell Factories, 2015, 14: 23. DOI:10.1186/s12934-015-0207-7 |
| [63] |
Ding MS, Wang J, Lin JZ, Wang WD. Oil displacement performance of rhamnolipid fermentation broths. Journal of Southwest Petroleum University: Science & Technology Edition, 2019, 41(5): 112-119.
(in Chinese) 丁明山, 王静, 林军章, 汪卫东. 鼠李糖脂发酵液驱油性能研究. 西南石油大学学报: 自然科学版, 2019, 41(5): 112-119. |
| [64] |
Bao HX, Zhang X, Zhao F, Zhao JY, Li GQ, Shi RJ, Han SQ, Li P, Zhang Y. Effects of rhamnolipids with different structure ratios on the emulsification activity and oily sludge cleaning. Chinese Journal of Ecology, 2020, 39(1): 243-251.
(in Chinese) 包红旭, 张欣, 赵峰, 赵劲毅, 李国桥, 史荣久, 韩斯琴, 李萍, 张颖. 不同结构配比的鼠李糖脂乳化活性与油泥清洗效果. 生态学杂志, 2020, 39(1): 243-251. |
| [65] | Rocha VAL, de Castilho LVA, de Castro RPV, Teixeira DB, Magalhães AV, Gomez JGC, Freire DMG. Comparison of mono-rhamnolipids and di-rhamnolipids on microbial enhanced oil recovery (MEOR) applications. Biotechnology Progress, 2020, 36(4): e2981. |
| [66] | Li ZZ, Zhang YM, Lin JZ, Wang WD, Li S. High-yield di-rhamnolipid production by Pseudomonas aeruginosa YM4 and its potential application in MEOR. Molecules, 2019, 24(7): 1433. DOI:10.3390/molecules24071433 |
| [67] | Das P, Yang XP, Ma LZ. Analysis of biosurfactants from industrially viable Pseudomonas strain isolated from crude oil suggests how rhamnolipids congeners affect emulsification property and antimicrobial activity. Frontiers in Microbiology, 2014, 5: 696. |
| [68] |
Wu H, Wang W, Han SY. Recent progress on rhamnolipid biosurfactant. Microbiology, 2007, 34(1): 148-152.
(in Chinese) 吴虹, 汪薇, 韩双艳. 鼠李糖脂生物表面活性剂的研究进展. 微生物学通报, 2007, 34(1): 148-152. DOI:10.3969/j.issn.0253-2654.2007.01.035 |
| [69] | Wittgens A, Rosenau F. On the road towards tailor-made rhamnolipids: current state and perspectives. Applied Microbiology and Biotechnology, 2018, 102(19): 8175-8185. DOI:10.1007/s00253-018-9240-x |
| [70] | Wittgens A, Kovacic F, Müller MM, Gerlitzki M, Santiago-Schübel B, Hofmann D, Tiso T, Blank LM, Henkel M, Hausmann R, Syldatk C, Wilhelm S, Rosenau F. Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Applied Microbiology and Biotechnology, 2017, 101(7): 2865-2878. DOI:10.1007/s00253-016-8041-3 |
| [71] | Tiso T, Zauter R, Tulke H, Leuchtle B, Li WJ, Behrens B, Wittgens A, Rosenau F, Hayen H, Blank LM. Designer rhamnolipids by reduction of congener diversity: production and characterization. Microbial Cell Factories, 2017, 16(1): 1-14. DOI:10.1186/s12934-016-0616-2 |
| [72] |
Liang SK, Wang XL, Shan BT, Wang WD. Study on rhamnolipid production from refined vegetable oil wastewater by Pseudomonas sp. O-2-2. Modern Chemical Industry, 2005, 25(S1): 192-196.
(in Chinese) 梁生康, 王修林, 单宝田, 汪卫东. 假单胞菌O-2-2利用油脂废水生产鼠李糖脂研究. 现代化工, 2005, 25(S1): 192-196. |
| [73] | Shi J, Chen YC, Liu XF, Li D. Rhamnolipid production from waste cooking oil using newly isolated halotolerant Pseudomonas aeruginosa M4. Journal of Cleaner Production, 2021, 278: 123879. DOI:10.1016/j.jclepro.2020.123879 |
| [74] | Zobel S, Benedetti I, Eisenbach L, de Lorenzo V, Wierckx N, Blank LM. Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synthetic Biology, 2015, 4(12): 1341-1351. DOI:10.1021/acssynbio.5b00058 |
| [75] | Choi KH, Schweizer HP. Mini-Tn 7 insertion in bacteria with single att Tn 7 sites: example Pseudomonas aeruginosa. Nature Protocols, 2006, 1(1): 153-161. DOI:10.1038/nprot.2006.24 |
| [76] | Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin CY, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Lynne Howell P, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nature Protocols, 2015, 10(11): 1820-1841. DOI:10.1038/nprot.2015.115 |
| [77] | Liang RB, Liu JH. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiology, 2010, 10(1): 1-9. DOI:10.1186/1471-2180-10-1 |
| [78] | Chen WZ, Zhang Y, Zhang YF, Pi YS, Gu TN, Song LQ, Wang Y, Ji QJ. CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience, 2018, 6: 222-231. DOI:10.1016/j.isci.2018.07.024 |
| [79] | Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 2013, 152(5): 1173-1183. DOI:10.1016/j.cell.2013.02.022 |
| [80] | Tan SZ, Reisch CR, Prather KLJ. A robust CRISPR interference gene repression system in In. Journal of Bacteriology, 2018, 200(7): e00575-17. DOI:10.1128/jb.00575-17 |
| [81] | Elmore JR, Furches A, Wolff GN, Gorday K, Guss AM. Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metabolic Engineering Communications, 2017, 5: 1-8. DOI:10.1016/j.meteno.2017.04.001 |
| [82] | Zhao H, Zhang HM, Chen XB, Li T, Wu Q, Ouyang Q, Chen GQ. Novel T7-like expression systems used for Halomonas. Metabolic Engineering, 2017, 39: 128-140. DOI:10.1016/j.ymben.2016.11.007 |
| [83] | Whitaker WR, Dueber JE. Metabolic pathway flux enhancement by synthetic protein scaffolding. Methods in Enzymology, 2011, 497: 447-468. |
| [84] |
Yin X, Liang C, Feng Y, Zhang H, Wang Y, Li YH. Research progress on synthetic scaffold in metabolic engineering-a review. Chinese Journal of Biotechnology, 2019, 35(3): 363-374.
(in Chinese) 尹雪, 梁晨, 冯玥, 张贺, 王宇, 李玉花. 合成支架在代谢工程中的研究进展. 生物工程学报, 2019, 35(3): 363-374. |
| [85] | Nitschke M, Costa SGVAO, Contiero J. Rhamnolipids and PHAs: Recent reports on Pseudomonas-derived molecules of increasing industrial interest. Process Biochemistry, 2011, 46(3): 621-630. DOI:10.1016/j.procbio.2010.12.012 |
| [86] | Zhang X, Lin YN, Wu Q, Wang Y, Chen GQ. Synthetic biology and genome-editing tools for improving PHA metabolic engineering. Trends in Biotechnology, 2020, 38(7): 689-700. DOI:10.1016/j.tibtech.2019.10.006 |
| [87] | Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV, Dueber JE. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature, 2018, 560(7717): 248-252. DOI:10.1038/s41586-018-0384-8 |
| [88] | Jiang JJ, Jin MJ, Li XY, Meng Q, Niu J, Long XW. Recent progress and trends in the analysis and identification of rhamnolipids. Applied Microbiology and Biotechnology, 2020, 104(19): 8171-8186. DOI:10.1007/s00253-020-10841-3 |
| [89] | Williams TC, Pretorius IS, Paulsen IT. Synthetic evolution of metabolic productivity using biosensors. Trends in Biotechnology, 2016, 34(5): 371-381. DOI:10.1016/j.tibtech.2016.02.002 |
| [90] | Liu S, Xu N, Liu H, Zhou J, Xin F, Zhang W, Qian X, Jiang M, Dong W. Genome characterization of Pseudomonas aeruginosa KT1115, a high di-rhamnolipid-producing strain with strong oils metabolizing ability. Current Microbiology, 2020, 77(8): 1890-1895. DOI:10.1007/s00284-020-02009-z |
| [91] | Xu AM, Wang D, Ding YC, Zheng YQ, Wang B, Wei Q, Wang SW, Yang L, Ma LZ. Integrated comparative genomic analysis and phenotypic profiling of Pseudomonas aeruginosa isolates from crude oil. Frontiers in Microbiology, 2020, 11: 519. DOI:10.3389/fmicb.2020.00519 |
 2021, Vol. 61
2021, Vol. 61




