中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 彭倩楠, 林璐. 2020
- Qiannan Peng, Lu Lin. 2020
- 木质素在海洋中的生物转化及其对海洋碳循环的影响
- Biotransformation of lignin in the ocean and its impact on marine carbon cycle
- 微生物学报, 60(9): 1959-1971
- Acta Microbiologica Sinica, 60(9): 1959-1971
-
文章历史
- 收稿日期:2020-04-29
- 修回日期:2020-07-08
- 网络出版日期:2020-07-21
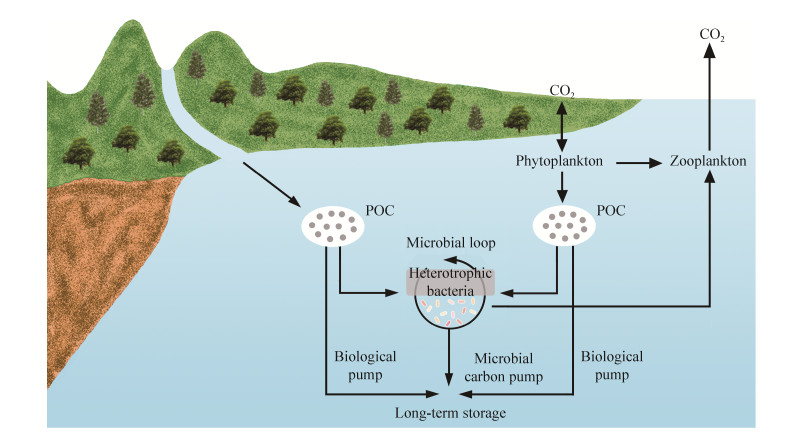
海洋是地球上最大的碳库,可吸收人为碳排放,调节全球气候变化[1]。据统计,全球每年约有0.43 Gt的颗粒有机碳(particulate organic carbon,POC)通过河川径流汇入海洋[2]。随后,POC可经由生物泵(biological pump,BP)的垂直沉降作用输送至深海,也可经由微型生物碳泵(microbial carbon pump,MCP)将其转化为惰性溶解有机碳(refractory dissolved organic carbon,RDOC)在海洋中存储上千年(图 1)[3]。故陆源POC的生物地球化学过程对于海洋碳汇乃至全球碳循环影响深远[4]。木质素是地球上丰富的陆源有机碳,以不同化学键交联的氧代苯丙醇为单位,由不同的醚键和碳-碳键连接而成的高分子芳香族杂聚物。它主要来源于陆地维管植物细胞壁[5],具有较高的化学稳定性和较强的抗降解能力。一直作为陆源有机碳生物标志物,用于研究陆源有机碳在海洋中的输入、迁移和埋藏过程[5]。但近来研究指出,木质素在海洋中的稳定性和低降解性可能被过分夸大[4, 6],除去光降解和热降解等非生物因素外,异养微生物的代谢活动是其生物降解的主要途径[7]。微生物所参与的MCP可将木质素转化为潜在RDOC,进入数千年甚至上万年周转的地球化学循环,长期封存在海洋中,故微生物驱动的木质素代谢活动是海洋碳汇的重要组成部分。然而目前对于海洋生态系统中微生物代谢木质素过程的研究极为有限,为克服上述困难,本文综述了目前参与木质素代谢的关键微生物菌群及其代谢机理,尤其是海洋微生物代谢途径,为后续海洋生态系统中微生物驱动木质素的生物地球化学过程以及海洋碳汇的研究提供参考。
1 参与木质素代谢的海洋微生物
国内外研究者已经陆续从不同海洋环境样品中筛选和分离到参与木质素代谢的海洋微生物群落。与陆地生态系统中真菌主导的木质素生物降解有所不同[8],这些群落以细菌和古菌为主[9-10],可能是由海洋特殊生境(寡营养、高盐)所造成。
迄今发现的海洋木质素降解细菌主要可分为三大类:放线菌门、变形菌门和厚壁菌门(图 2)[11]。日本海洋科技中心Yukari Ohta教授团队[12]基于纯培养和质谱技术对深海沉积物和沉木样品中的菌株进行分离和功能筛选,发现其中208个菌株能够代谢木质素衍生的芳香化合物,主要分属于厚壁菌门、放线菌门和变形菌门。Ohta据此推测海洋环境中可能富含参与木质素代谢的微生物,能够通过氧化和非氧化途径对木质素进行代谢。

|
| 图 2 参与木质素代谢的海洋细菌系统发育进化树 Figure 2 Phylogenetic tree of marine lignin-metabolizing bacteria. |
随着测序技术和数据分析方法的飞速发展,基于宏基因组和单细胞测序技术来探索环境中微生物的群落结构、组成及功能,正逐步揭开海洋中木质素代谢菌群的神秘面纱。美国田纳西大学Terry Hazen教授团队[13]结合鸟枪法宏基因组学和16S rRNA基因序列分析技术,发现γ-变形菌纲和α-变形菌纲是东地中海海平面下50 m处海水环境中参与木质素代谢的主要菌群,并指出其中参与苯乙酰CoA、香草酸和阿魏酸分解代谢途径的相关基因丰度较高,尤其是来自于假单胞菌属(Pseudomonas)和海单胞菌属(Marinomonas)的苯乙酰CoA代谢途径相关基因。日本理化学研究所Keiji团队[16]从被尿囊素污染的海域中分离、筛选到能够以木质素或其衍生物为唯一碳源直接合成聚羟基脂肪酸酯(PHA)的Oceanimonas doudoroffii菌株。笔者所在团队[17-19]也发现能代谢木质素的Pseudomonas putida菌株,且该菌株能够以木质素为碳源合成PHA。结合系统生物学方法,团队进一步揭示了该菌株木质素代谢及转化至PHA的分子机理。PHA作为微生物储碳的一种有效方式,木质素至PHA的生物转化可减少异养微生物通过呼吸作用所排放的CO2,是海洋增汇的理想途径[20]。
除上述海洋细菌外,深海古菌也积极参与了海洋木质素代谢。上海交通大学王风平教授团队[9]通过宏基因组测序和纯培养技术相结合,从海洋沉积物中发现了一个“深古菌”新门,命名为Bathyarchaeota。通过宏基因组数据分析,该团队推测Bathyarchaeota具有多种代谢潜能,并通过木质素富集实验证实其中Bathy-8类群能够在厌氧环境下以木质素为能源,以无机碳为碳源进行生长[9]。
2 微生物代谢木质素机制过去几十年里,以陆地生态系统中真菌为主导的木质素降解已得到广泛研究。真菌能够分泌多种胞外酶对木质素进行解聚[21],如漆酶(Lac)、木质素过氧化物酶(LiP)、锰过氧化物酶(MnP)、多功能过氧化物酶(VP)、染料脱色过氧化物酶(DyP)等,通过胞外催化生成化学不稳定性的木质素自由基中间体,进而引发高分子木质素内部的Cα-Cβ裂解、烷基-芳基裂解、交联、去甲氧基化及环裂解等,产生低分子量芳香族化合物[22](表 1)。与真菌类似,细菌也拥有自己的胞外氧化酶系统,能够对木质素进行解聚,如细胞色素P450单加氧酶(P450s)、染料过氧化物酶(DyP)、漆酶(Lac)、锰过氧化物酶(MnP)、双加氧酶、非血红素铁酶、超氧化物歧化酶(SOD)、β-醚酶等[19, 36-43](表 1)。其中,染料过氧化物酶(DyP)普遍存在于各种细菌中。笔者所在团队解析出Pseudomonas putida中2个B型DyP,具有独特的木质素氧化活性,能与细菌高效协同,从而极大促进细菌生长和木质素降解[19]。总之,细菌的木质素降解能力虽弱于真菌,但通常具有简单的蛋白表达系统和较强的环境适应能力,如更好的热稳定性、更广的pH和温度耐受范围等[11]。此外,细菌还能生产高价值产物,因而细菌介导的木质素代谢近年来逐渐受到广泛关注[17-18, 44]。
| Taxonomy | Lignin-degrading microorganisms | Enzymes | Pathway |
| Fungi | White-rot fungi Coriolus[23], Phanerochaete[24], Trametes[25], Bjerkandera[26], Pleurotus[27], Phlebia[28] Brown-rot fungi Gloeophyllum[29], Poria[29], Polyporus[30] Soft-rot fungi Chaetomium[31], Paecilomyces[32], Fusarium[33], Aspergillus[34] | Major enzymes such as laccases (Lac), manganese peroxidases (MnP), lignin peroxidases (LiP), dye-decolorizing peroxidases (D-type DyP), versatile peroxidases (VP). Auxiliary enzymes such as aryl-alcohol oxidases (AAO), glyoxal oxidases (GLOX), glucose oxidases (GIO). | Diaryl propane catabolic pathway, pinoresinol pathway[35], etc. |
| Bacteria | Actinobacteria Streptomyces[45], Micrococcus[46], Micromonospor[46], Arthrobacter[47], Rhodococcu[48], Microbispora[46], Nocardia[49], Thermoactinomyces[50] Proteobacteria Acinetobacter[51], Comamonas[52], Pseudomonas[47], Xanthomonas[53], Mycoplana[54], Sphingomonas[55] Firmicutes Clostridium[46], Bacillus[56] | Major enzymes such as laccases (Lac), dye-decolorizing peroxidases (DyP), manganese peroxidases (MnP). Auxiliary enzymes such as cytochrome P450, dioxygenase, non-heme enzyme, superoxide dismutase, β-etherase enzyme. | β-Aryl ether cleavage pathway, biphenyl catabolic pathway, diaryl propane catabolic pathway, biphenyl degradation pathway, vanillic acid pathway, ferulic acid degradation pathway, 3-methyl-gallic acid pathway, benzoic acid pathway, oxidative cleavage of protocatechuic acid, β-ketoadipate pathway, phenylacetic acid pathway, phenol pathway, gentisate pathway, etc. |
目前,对细菌木质素衍生物代谢途径的研究主要集中于陆地环境中所筛选到的菌株,如来自造纸厂废水环境中的Sphingomonas paucimobilis SYK-6菌株以及来自土壤环境中的Streptomyces viridosporus T7A菌株等[57-59]。而对海洋微生物代谢木质素的研究较为缺乏,故本文综述了陆地微生物木质素代谢途径,并与已知的海洋微生物代谢途径相比较,为后续海洋微生物代谢木质素途径的研究提供参考。
木质素经胞外解聚酶解聚后可得到3种不同结构的基本单元,含有2个甲氧基的S型单元,含有1个甲氧基的G型单元,以及不含甲氧基的H型单元。这些木质素衍生物输送到胞内后,经过多种外围和中心代谢途径进行开环代谢,最后进入三羧酸(TCA)循环为细胞供能[60](图 3)。

|
| 图 3 木质素三种基本结构单元代谢途径示意图 Figure 3 Metabolic pathways of three basic structural units of lignin. |
对香豆酸作为H单元的代表,其降解途径主要可分为CoA依赖型β氧化、CoA依赖型非β氧化和不依赖于CoA的代谢途径(图 3)。(1) CoA依赖型β氧化途径首先通过羟基肉桂酰CoA合成酶的催化,将对香豆酸转化为羟基肉桂酰CoA,而后通过水合、氧化和裂解生成乙酰CoA和对羟基苯甲酰CoA,并进一步转化为对羟基苯甲酸[61-62]。(2) CoA依赖型非β氧化前两步与CoA依赖型β氧化途径相同,随后发生Cα-Cβ断裂,生成乙酰CoA和对羟基苯甲醛,对羟基苯甲醛进一步氧化为对羟基苯甲酸[63-66]。(3)不依赖于CoA的代谢途径与CoA依赖型非β氧化途径相似,将对香豆酸转化为对羟基苯甲醛,进而氧化生成对羟基苯甲酸,但不形成CoA硫酯[67]。生成的羟基苯甲酸在对羟基苯甲酸-3-羟化酶的催化下可转化为原儿茶酸。原儿茶酸是芳香族化合物降解的重要中间产物,可在原儿茶酸4, 5双加氧酶、原儿茶酸3, 4双加氧酶及原儿茶酸2, 3双加氧酶的催化下分别在苯环的4, 5位、3, 4位或2, 3位间进行开环[68-70]。其中,原儿茶酸3, 4双加氧酶广泛存在于细菌中,原儿茶酸经3, 4开环裂解后能够进入β-酮己二酸芳香族分解代谢途径,最终进入TCA循环为细胞供能[71]。
G单元木质素衍生体以β-芳基醚、联苯、二芳基丙烷等为代表(图 3)。(1) β-芳基醚首先通过脱氢酶氧化生成相应的酮,而后经以谷胱甘肽为辅因子的β-醚酶的还原性醚裂解反应,转化为中间体香草酸,经甲基化进一步转化为原儿茶酸进入中心代谢途径[72-74]。(2)联苯在木质素中的含量仅次于芳基醚寡聚体。它首先在脱甲基酶的催化下进行脱甲基化,随后由双加氧酶催化苯环断裂,裂解产物在水解酶作用下发生C-C键断裂,形成5-羧基香草酸,经脱羧酶催化脱羧为香草酸后进一步转化,生成原儿茶酸,进入中心代谢途径[75-76]。(3)二芳基丙烷被某种未知酶催化生成甲醛和Lignostilbene,随后在双氧酶的作用下进一步氧化为香草醛,进入中心代谢途径[77-78]。
丁香酸是S型木质素的单体模型化合物。已知能够有效降解丁香酸的微生物较少,可能是由于S型木质素单元的芳香环含有2个甲氧基,包围了酚羟基,使得甲氧基在降解竞争中占据优势地位,其与苯环连接键最先发生断裂,然而甲氧基与苯环连接键的键能较高,较难发生断裂,因此S型比H型和G型木质素降解更为困难[79-80]。有关细菌S型木质素降解途径的研究主要集中于Sphingomonas sp. SYK-6。丁香酸首先被依赖于四氢叶酸依赖型脱甲基酶脱甲基化为3-甲基没食子酸,3-甲基没食子酸可以直接开环裂解进入原儿茶酸4, 5-裂解途径;也可以以没食子酸或4-羧基-2-羟基-6-甲氧基-6-氧-2, 4-己二烯酸(CHMOD)为中间体进行代谢,最终进入原儿茶酸的4, 5-裂解途径[81-83](图 3)。
此外,有限的研究显示,海洋微生物存在其独特的代谢途径,可能是由于其独特的生境所造成(表 2和图 3)。对于来自东地中海的海水样品进行宏基因组分析显示,海洋生境中的微生物可以通过苯乙酰CoA途径进行木质素分解代谢,以代替陆地上经典的β-酮己二酸芳香族分解代谢途径[13]。与β-酮己二酸代谢不同,苯乙酰CoA途径无需经过开环,可直接通过CoA与芳香族化合物连接,经环-环氧化或芳香族CoA硫酯的还原后实现环活化,能降解包括芳香族氨基酸及木质素类苯丙烷化合物在内的多种芳香族化合物[88-89]。此外,相比于依赖于双加氧酶的β-酮己二酸,苯乙酰CoA途径所产生的CoA硫酯能够更好地被细胞利用,尤其适用于处于海洋寡营养生境中的微生物,且CoA硫酯可以进入好氧和厌氧两种途径,使得兼性厌氧微生物在氧气波动或低氧条件下具有较高灵活性[90],故苯乙酰CoA途径可能更适用于海洋生境中木质素代谢。
| Ecosystem | Lignin-degrading microorganisms | Characteristic enzymes | Characteristic pathway |
| Land | Sphingomonas paucimobilis[56], Rhodococcus jostii[49] Burkholderia multivorans[84], Pseudomonas putida[48] Bacillus amyloliquefaciens[57], Comamonas serinivorans[53], Streptomyces viridosporus[46], Nocardia autotrophica[50] | Oxidative enzymes, etc. | Oxidative cleavage of protocatechuic acid, β-ketoadipate pathway, etc. |
| Marine | Bacillus ligniniphilus[85], Pseudomonas deceptionensis[86], Pseudomonas putida[17], Thalassospira sp.[87], Oceanimonas doudoroffii[16] | Oxidative enzymes, phenylacetyl- CoA ligase (PCL), etc. | Phenylacetic acid pathway, etc. |
综上所述,在海洋环境中,木质素的转化对于海洋碳库起着重要调节作用。除沉降到深海,长期埋藏在海底外,木质素还可经微生物转化成各种芳香族化合物。而这些芳香族化合物可能是潜在的RDOC,有助于海洋增汇。
3 木质素代谢研究前景已知海洋环境中存在多种木质素代谢微生物,但相较于陆地生态系统,对于海洋环境中木质素代谢微生物的认知仍然有限[11-12]。纯培养和高通量测序是目前研究海洋木质素代谢菌所采用的主要手段。纯培养是微生物研究的基石,微生物的生理或生态功能假说均需通过纯培养进行验证,但自然界中约有99%的微生物难以在现有实验条件和技术下进行培养。此外,分离培养工作量大,操作繁琐,通量低,大大限制了我们对海洋木质素代谢菌的认知。故发展快速、简便的适用于木质素代谢菌筛选和培养的新方法迫在眉睫。本课题组已基于木质素代谢菌的生理参数构建模型,建立了一种木质素利用菌快速筛选的方法,目前正在评审中。与此同时,宏基因组测序技术克服了依赖培养的微生物研究模式,直接以环境中微生物群体基因组为研究对象,借助于高通量测序和生物信息学技术获得新基因、新物种,探索自然生境中的微生物群落结构及功能[91]。但受限于现有测序和数据分析技术,使得在研究木质素代谢微生物群落方面具有以下几个难点。首先,对木质素降解基因认知不足,依赖于现有的公共基因组数据库对木质素代谢菌基因组功能注释成为一大难题。因此,构建针对木质素代谢的功能基因数据库,填补木质素代谢菌功能基因的空白是当前急需解决的难题。本课题组前期已构建一个专门用于宏基因组数据中氮循环功能基因挖掘的氮循环功能基因数据库NCycDB,以解决搜索效率低、基因家族同源性低以及参与氮循环基因的测序序列覆盖率低等问题[92]。该工作为今后建立木质素降解基因数据库奠定了良好基础。其次,参与木质素代谢基因的相关丰度可能较低,通过宏基因组技术极易忽视与之相关的低丰度基因,从而增加了数据分析的难度。借助现有的Geochip技术[93]开发针对木质素代谢的微阵列芯片,可将所有已知木质素代谢功能基因探针集中在芯片上,大大提高了目的基因的检测灵敏度,简化了数据分析的困难,两者相结合将能够更全面、准确地挖掘出参与木质素代谢的微生物功能基因。此外,结合宏转录组测序、宏蛋白质组以及质谱技术等可分别从转录水平、蛋白水平以及代谢物水平解析参与木质素代谢的成员、代谢途径及功能基因等。借助随机矩阵理论也有助于进一步探究菌株间、基因间的相互联系。同时,通过解析不同环境因子下参与木质素代谢的微生物群落结构和组成,将有助于发现环境因子对于微生物代谢调控的影响及微生物对于环境变化的应对策略。
探索海洋木质素代谢微生物,认知木质素在自然界中所参与的碳循环过程,为深入理解以木质素为代表的陆源POC在海洋碳汇中所发挥的作用以及海洋碳循环机制提供参考。
| [1] |
Jiao NZ. Carbon fixation and sequestration in the ocean, with special reference to the microbial carbon pump. Scientia Sinica Terrae, 2012, 42(10): 1473-1486.
(in Chinese) 焦念志. 海洋固碳与储碳——并论微型生物在其中的重要作用. 中国科学:地球科学, 2012, 42(10): 1473-1486. |
| [2] | Schlünz B, Schneider RR. Transport of terrestrial organic carbon to the oceans by rivers:re-estimating flux-and burial rates. International Journal of Earth Sciences, 2000, 88(4): 599-606. |
| [3] | 焦念志.海洋固碳与储碳.同济大学.第二届深海研究与地球系统科学学术研讨会论文集, 2012: 172. |
| [4] | Bianchi TS. The role of terrestrially derived organic carbon in the coastal ocean:A changing paradigm and the priming effect. Proceedings of the National Academy of Sciences, 2011, 108(49): 19473-19481. |
| [5] |
Lan HQ, Li XG, Zhang T, Sun SW. Biodegradation of lignin and its effect on the use of lignin as a biomarker of terrestrial organic carbon input. Transactions of Oceanology and Limnology, 2012, 10(1): 123-129.
(in Chinese) 兰海青, 李先国, 张婷, 孙书文. 木质素的生物降解及其对陆源有机碳指示作用的影响. 海洋湖沼通报, 2012, 10(1): 123-129. |
| [6] | McDonald N, Achterberg EP, Carlson CA, Gledhill M, Liu ST, Matheson-Barker JR, Nelson NB, Parsons RJ. The role of heterotrophic bacteria and archaea in the transformation of lignin in the open ocean. Frontiers in Marine Science, 2019, 6: 743. |
| [7] | 兰海青.木质素的生物降解及对其指示参数的影响.中国海洋大学硕士学位论文, 2012. |
| [8] |
Li HT, Yao K, He Q, Jia DY. Biodegradation and applications of lignin. Leather Science and Engineering, 2010(6): 27-31.
(in Chinese) 李海涛, 姚开, 何强, 贾冬英. 木质素的生物降解及其应用. 皮革科学与工程, 2010(6): 27-31. |
| [9] | Yu TT, Wu WC, Liang WY, Lever MA, Hinrichs KU, Wang FP. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proceedings of the National Academy of Sciences, 2018, 115(23): 6022-6027. |
| [10] | Chandra R, Raj A, Purohit HJ, Kapley A. Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere, 2007, 67(4): 839-846. |
| [11] |
Xie CX, Sun JZ, Li CL, Zhu DC. Exploring the lignin degradation by bacteria. Microbiology, 2015, 42(6): 1122-1132.
(in Chinese) 谢长校, 孙建中, 李成林, 朱道辰. 细菌降解木质素的研究进展. 微生物学通报, 2015, 42(6): 1122-1132. |
| [12] | Ohta Y, Nishi S, Haga T, Tsubouchi T. Screening and phylogenetic analysis of deep-sea bacteria capable of metabolizing lignin-derived aromatic compounds. Open Journal of Marine Science, 2012, 2(4): 177-187. |
| [13] | Woo HL, Hazen TC. Enrichment of bacteria from eastern mediterranean sea involved in lignin degradation via the phenylacetyl-CoA pathway. Frontiers in Microbiology, 2018, 9: 922. |
| [14] | Buchan A, Lecleir GR, Gulvik CA, González JM. Master recyclers:features and functions of bacteria associated with phytoplankton blooms. Nature Reviews Microbiology, 2014, 12(10): 686-698. |
| [15] | Jiao NZ, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo TW, Chen F, Azam F. Microbial production of recalcitrant dissolved organic matter:long-term carbon storage in the global ocean. Nature Reviews Microbiology, 2010, 8(8): 593-599. |
| [16] | Numata K, Morisaki K. Screening of marine bacteria to synthesize polyhydroxyalkanoate from lignin:contribution of lignin derivatives to biosynthesis by Oceanimonas doudoroffii. ACS Sustainable Chemistry and Engineering, 2015, 4(3): 569-573. |
| [17] | Lin L, Cheng YB, Pu YQ, Sun S, Li X, Jing MJ, Pierson EA, Gross DC, Dale BE, Dai SY, Ragauskas AJ, Yuan JS. Systems biology-guided biodesign of consolidated lignin conversion. Green Chemistry, 2016, 18(20): 5536-5547. |
| [18] | Wang XP, Lin L, Dong JD, Ling J, Wang WP, Wang HL, Zhang ZC, Yu XW. Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Applied and Environmental Microbiology, 2018, 84(18): e01469-18. |
| [19] | Lin L, Wang XP, Cao LF, Xu MY. Lignin catabolic pathways reveal unique characteristics of dye-decolorizing peroxidases in Pseudomonas putida. Environmental Microbiology, 2019, 21(5): 1847-1863. |
| [20] | Jiao NZ, Tang K, Cai HY, Mao YJ. Increasing the microbial carbon sink in the sea by reducing chemical fertilization on the land. Nature Reviews Microbiology, 2011, 9(1): 75. |
| [21] | Martínez ÁT, Speranza M, Ruiz-Dueñas FJ, Ferreira P, Camarero S, Guillen F, Martinez MJ, Gutierrez A, del Rio JC. Biodegradation of lignocellulosics:microbial, chemical, and enzymatic aspects of the fungal attack of lignin. International Microbiology, 2005, 8(3): 195-204. |
| [22] | Salvachúa D, Katahira R, Cleveland N S, Khanna P, Resch MG, Black BA, Purvine SO, Zink EM, Prieto A, Martinez MJ, Martinez AT, Simmons BA, Gladden JM, Beckham GT. Lignin depolymerization by fungal secretomes and a microbial sink. Green Chemistry, 2016, 18(22): 6046-6062. |
| [23] | Evans CS. Laccase activity in lignin degradation by Coriolus versicolor in vivo and in vitro studies. FEMS Microbiology Letters, 1985, 27(3): 339-343. |
| [24] | Kersten P, Cullen D. Extracellular oxidative systems of the lignin-degrading Basidiomycete Phanerochaete chrysosporium. Fungal Genetics and Biology, 2007, 44(2): 77-87. |
| [25] | Ong E, Pollock WBR, Smith M. Cloning and sequence analysis of two laccase complementary DNAs from the ligninolytic basidiomycete Trametes versicolor. Gene, 1997, 196(1-2): 113-119. |
| [26] |
Liu J, Liu JA, Zhou GY, Li H, Yang J. Isolation, screening and identifi cation of high effective bamboo-lignin degradation fungus. Journal of Central South University of Forestry and Technology, 2014(8): 48-52.
(in Chinese) 刘剑, 刘君昂, 周国英, 李河, 杨菁. 竹材木质素高效降解菌的分离筛选及鉴定. 中南林业科技大学学报, 2014(8): 48-52. |
| [27] | Reddy GV, Babu PR, Komaraiah P, Roy KRRM, Kothari IL. Utilization of banana waste for the production of lignolytic and cellulolytic enzymes by solid substrate fermentation using two Pleurotus species (P. ostreatus and P. sajor-caju). Process Biochemistry, 2003, 38(10): 1457-1462. |
| [28] | Hildén K, Martinez AT, Hatakka A, Lundell T. The two manganese peroxidases Pr-MnP2 and Pr-MnP3 of Phlebia radiata, a lignin-degrading basidiomycete, are phylogenetically and structurally divergent. Fungal Genetics and Biology, 2005, 42(5): 403-419. |
| [29] | Niemenmaa O, Uusi-Rauva A, Hatakka A. Demethoxylation of[O14CH3]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation, 2008, 19(4): 555-565. |
| [30] | Rothschild N, Novotny C, Sasek V, Dosoretz CG. Ligninolytic enzymes of the fungus Irpex lacteus (Polyporus tulipiferae):isolation and characterization of lignin peroxidase. Enzyme and Microbial Technology, 2002, 31(5): 627-633. |
| [31] |
Hao XR, Niu XL, Li Q, Pan J, Zhu XD. Difference in ligocellulose degradation of endophytic Chaetomium globosum isolates and related genes analysis. Letters in Biotechnology, 2014, 25(1): 1-8.
(in Chinese) 郝晓冉, 牛学良, 李强, 潘皎, 朱旭东. 球毛壳菌降解天然木质纤维素能力差异及酶系基因分析. 生物技术通讯, 2014, 25(1): 1-8. |
| [32] |
Wang YL, Sun X. Research progress of lignin biodegradation. Journal of Microbiology, 1998, 18(1): 48-51.
(in Chinese) 王宜磊, 孙迅. 木素生物降解研究进展. 微生物学杂志, 1998, 18(1): 48-51. |
| [33] | Lozovaya VV, Lygin AV, Zernova OV, Li S, Widholm JM, Hartman GL. Lignin degradation by Fusarium solani f. sp. glycines. Plant Disease, 2006, 90(1): 77-82. |
| [34] | Bonugli-Santos RC, Durrant LR, Silva MD, Sette LD. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzyme and Microbial Technology, 2010, 46(1): 32-37. |
| [35] | Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Natural Product Reports, 2011, 28(12): 1883-1896. |
| [36] | Rashid GMM, Taylor CR, Liu YQX, Zhang XY, Rea D, Fulop V, Bugg TDH. Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chemical Biology, 2015, 10(10): 2286-2294. |
| [37] | Ahmad M, Taylor C R, Pink D, Burton K, Eastwood D, Bending GD, Bugg TDH. Development of novel assays for lignin degradation:comparative analysis of bacterial and fungal lignin degraders. Molecular Biosystems, 2010, 6(5): 815-821. |
| [38] | Zhu DC, Zhang PP, Xie CX, Zhang WM, Sun JZ, Qian WJ, Yang B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnology for Biofuels, 2017, 10(1): 44. |
| [39] | Bugg TD, Rahmanpour R. Enzymatic conversion of lignin into renewable chemicals. Current Opinion in Chemical Biology, 2015, 29: 10-17. |
| [40] | Brown ME, Chang MCY. Exploring bacterial lignin degradation. Current Opinion in Chemical Biology, 2014, 19: 1-7. |
| [41] | Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Current Opinion in Biotechnology, 2011, 22(3): 394-400. |
| [42] | Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Natural Product Reports, 28(12): 1883-1896. |
| [43] | de Gonzalo G, Colpa DI, Habib MHM, Fraaije MW. Bacterial enzymes involved in lignin degradation. Journal of Biotechnology, 2016, 236: 110-119. |
| [44] | Zhou YY, Lin L, Wang H, Zhang ZC, Zhou JZ, Jiao NZ. Development of a CRISPR/Cas9n-based tool for metabolic engineering of Pseudomonas putida for ferulic acid-to-polyhydroxyalkanoate bioconversion. Communications Biology, 2020, 3(1): 98. |
| [45] | Ramachandra M, Crawford DL, Hertel G. Characterization of an extracellular lignin peroxidase of the lignocellulolytic actinomycete Streptomyces viridosporus. Applied and Environmental Microbiology, 1989, 54(12): 3057-3063. |
| [46] |
Pan MF, Jiang M, Zhou ZW. Latest research advances in biodegradation of lignin. Materials Review, 2011, 25(2): 372-377.
(in Chinese) 潘明凤, 姜曼, 周祚万. 木质素生物降解的最新研究进展. 材料导报:纳米与新材料专辑, 2011, 25(2): 372-377. |
| [47] | Ahmad M, Taylor C R, Pink D, Burton K, Eastwood D, Bending GD, Bugg TDH. Development of novel assays for lignin degradation:comparative analysis of bacterial and fungal lignin degraders. Molecular Biosystems, 2010, 6(5): 815-821. |
| [48] | Seto M, Kimbara K, Shimura M, Hatta T, Fukuda M, Yano K. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Applied and Environmental Microbiology, 1995, 61(9): 3353-3358. |
| [49] | Zimmermann W. Degradation of lignin by bacteria. Journal of Biotechnology, 1990, 13(2-3): 119-130. |
| [50] | Zhou C, Liu Z, Huang ZL, Yu XL, Ning P. A new strategy for co-composting dairy manure with rice straw:Addition of different inocula at three stages of composting. Waste Management, 2015, 40: 38-43. |
| [51] | Vasudevan N, Mahadevan A. Degradation of lignin and lignin derivatives by Acinetobacter sp.. Journal of Applied Microbiology, 2008, 70(2): 169-176. |
| [52] |
Zhang PP, Sun JZ, Xie CX, Zhu DC. Lignin degradation by Comamonas serinivorans C35. Microbiology China, 2017, 44(5): 1131-1137.
(in Chinese) 张佩佩, 孙建中, 谢长校, 朱道辰. Comamonas serinivorans C35木质素降解性能. 微生物学通报, 2017, 44(5): 1131-1137. |
| [53] | Kern HW, Kirk TK. Influence of molecular size and ligninase pretreatment on degradation of lignins by Xanthomonas sp. strain 99. Applied and Environmental Microbiology, 1987, 53(9): 2242-2246. |
| [54] |
Sun XF, Zhang ZJ, Cui HJ. Study on isoloation of lignin-degradation microorganisms of pulping liquor and degradation characteristics. Environmental Engineering, 2002, 20(3): 78-80.
(in Chinese) 孙先锋, 张志杰, 崔红军. 造纸黑液木质素降解微生物的分离和降解特性研究. 环境工程, 2002, 20(3): 78-80. |
| [55] | Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Bioscience Biotechnology and Biochemistry, 2007, 71(1): 1-15. |
| [56] |
Wang NX, Guo XJ, Li SN, Li CH, Zhu BC. Screening and identification on bacteria N13 with spore and capability of degrading cellulose. Animal Husbandry and Feed Science, 2009, 30(9): 7-10.
(in Chinese) 王纳贤, 郭晓军, 李术娜, 李春辉, 朱宝成. 产芽孢木质素降解菌株N13的分离与鉴定. 畜牧与饲料科学, 2009, 30(9): 7-10. |
| [57] | Hara H, Masai E, Miyauchi K, Katayama Y, Fukuda M. Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4, 5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6. Journal of Bacteriology, 2003, 185(1): 41-50. |
| [58] | Jimenez JI, Minambres B, Garcia JL, Diaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environmental Microbiology, 2002, 4(12): 824-841. |
| [59] | Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Bioence Biotechnology and Biochemistry, 2007, 71(1): 1-15. |
| [60] | Katayama Y, Nishikawa S, Nakamura M, Yano K, Yamasak M, Morohoshi N, Haraguchi T. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi, 1987, 33(1): 77-79. |
| [61] | Overhage J, Priefert H, Steinbuchel A. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Applied and Environmental Microbiology, 1999, 65(11): 4837-4847. |
| [62] | Trautwein K, Wilkes H, Rabus R. Proteogenomic evidence for β-oxidation of plant-derived 3-phenylpropanoids in "Aromatoleum aromaticum" EbN1. Proteomics, 2012, 12(9): 1402-1413. |
| [63] | Kasai D, Kamimura N, Tani K, Umeda S, Abe T, Fukuda M, Masai E. Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiology Letters, 2012, 332(1): 68-75. |
| [64] | Masai E, Harada K, Peng X. Cloning and characterization of the ferulic acid catabolic genes of Sphingomonas paucimobilis SYK-6. Applied and Environmental Microbiology, 2002, 68(9): 4416-4424. |
| [65] | Plaggenborg R, Overhage J, Loos A, Archer JAC, Lessard P, Sinskey AJ, Steinbuchel A, Priefert H. Potential of Rhodococcus strains for biotechnological vanillin production from ferulic acid and eugenol. Applied Microbiology and Biotechnology, 2006, 72(4): 745-755. |
| [66] | Yang WW, Tang HZ, Ni J, Wu QL, Hua DL, Tao F, Xu P. Characterization of two Streptomyces enzymes that convert ferulic acid to vanillin. PLoS One, 2013, 8(6): e67339. |
| [67] | Gallage NJ, Hansen EH, Kannangara R, Olsen CE, Motawia MS, Jørgensen K, Holme I, Hebelstrup K, Grisoni M, Møller BL. Vanillin formation from ferulic acid in vanilla planifolia is catalyzed by a single enzyme. Nature Communications, 2014, 19(5): 4037. |
| [68] | Vetting MW, D'Argenio DA, Ornston LN, Ohlendorf DH. Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 angstrom resolution:implications for the mechanism of an intradiol dioxygenase. Biochemistry, 2000, 39(27): 7943-7955. |
| [69] | Arciero DM, Orville AM, Lipscomb JD. Protocatechuate 4, 5-dioxygenase from Pseudomonas testosteroni. Methods in Enzymology, 1990, 188: 89-95. |
| [70] | Kasai D, Fujinami T, Abe T, Mase K, Katayama Y, Fukuda M, Masai E. Uncovering the protocatechuate 2, 3-cleavage pathway genes. Journal of Bacteriology, 2009, 191(21): 6758-6768. |
| [71] | Harwood CS, Parales RE. The β-ketoadipate pathway and the biology of self-identity. Annual Review of Microbiology, 1996, 50(1): 553-590. |
| [72] | Masai E, Katayama Y, Kubota S, Kawai S, Yamasaki M, Morohoshi N. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Letters, 1993, 323(1-2): 135-140. |
| [73] | Masai E, Katayama Y, Nishikawa S, Yamasaki M, Morohoshi N, Haraguchi T. Detection and localization of a new enzyme catalyzing the β-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Letters, 1989, 249(2): 348-352. |
| [74] | Vicuna R, Gonzalez B, Mozuch MD, Kirk TK. Metabolism of lignin model compounds of the arylglycerol-beta-aryl ether type by Pseudomonas acidovorans D3. Applied and Environmental Microbiology, 1987, 53(11): 2605-2609. |
| [75] | Sonoki T, Obi T, Kubota S, Higashi M, Masai E, Katayama Y. Coexistence of two different O demethylation systems in lignin metabolism by Sphingomonas paucimobilis SYK-6:cloning and sequencing of the lignin biphenyl-specific O-demethylase (LigX) gene. Applied and Environmental Microbiology, 2000, 66(5): 2125-2132. |
| [76] | Peng X, Masai E, Katayama Y, Fukuda M. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Applied and Environmental Microbiology, 1999, 65(6): 2789-2793. |
| [77] | Kishi K, Habu N, Samejima M, Yoshimoto T. Purification and some properties of the enzyme catalyzing the Cγ-elimination of a diarylpropane-type lignin model from Pseudomonas paucimobilis TMY1009. Agricultural and Biological Chemistry, 1991, 55(5): 1319-1323. |
| [78] | Kamoda S, Habu N, Samejima M, Yoshimoto T. Purification and some properties of lignostilbene-α, β-dioxygenase responsible for the Cα-Cβ cleavage of a diarylpropane type lignin model compound from Pseudomonas sp. TMY1009. Agricultural and Biological Chemistry, 2014, 53(10): 2757-2761. |
| [79] | Xu ZX, Lei P, Zhai R, Wen ZQ, Jin MJ. Recent advances in lignin valorization with bacterial cultures:microorganisms, metabolic pathways, and bio-products. Biotechnology for Biofuels, 2019, 12(1): 32. |
| [80] |
Li GB, Wang C, Li CT, Liu F, Li L. Photocatalytic degradation of syringol lignin model by titanium dioxide. Chemical Reaction Engineering and Technology, 2009, 25(3): 228-232.
(in Chinese) 李光壁, 王昶, 李晨陶, 刘芳, 李伶. 木素类模型物紫丁香醇TiO2光催化降解. 化学反应工程与工艺, 2009, 25(3): 228-232. |
| [81] | Kasai D, Masai E, Miyauchi K, Katayama Y, Fukuda M. Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6. Journal of Bacteriology, 2004, 186(15): 4951-4959. |
| [82] | Barry KP, Taylor EA. Characterizing the promiscuity of LigAB, a lignin catabolite degrading extradiol dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry, 2013, 52(38): 6724-6736. |
| [83] | Kasai D, Masai E, Katayama Y, Fukuda M. Degradation of 3-O-methylgallate in Sphingomonas paucimobilis SYK-6 by pathways involving protocatechuate 4, 5-dioxygenase. FEMS Microbiology Letters, 2013, 274(2): 323-328. |
| [84] | Akita H, Kimura Z, Mohd Yusoff MZM, Nakashima N, Hoshino T. Isolation and characterization of Burkholderia sp. strain CCA53 exhibiting ligninolytic potential. SpringerPlus, 2016, 5(1): 596. |
| [85] | Zhu DC, Zhang PP, Xie CX, Zhang WM, Sun JZ, Qian WJ, Qian WJ. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnology for Biofuels, 2017, 10(1): 44. |
| [86] | Ravi K, García-Hidalgo J, Nbel M, Gorwa-Grauslund MF, Liden G. Biological conversion of aromatic monolignol compounds by a Pseudomonas isolate from sediments of the Baltic Sea. AMB Express, 2018, 8(1): 32. |
| [87] | Woo HL, O'Dell KB, Utturkar S, McBride KR, Huntemann M, Clum A, Pillay M, Palaniappan K, Varghese N, Mikhailova N, Stamatis D, Reddy TBK, Ngan CY, Daum C, Shapiro N, Markowitz V, Ivanova N, Kyrpides N, Woyke T, Brown SD, Hazen TC. Near-complete genome sequence of Thalassospira sp. strain KO164 isolated from a lignin-enriched marine sediment microcosm. Genome Announcements, 2016, 4(6): e01297-16. |
| [88] | Ismail W, Gescher J. Epoxy coenzyme a thioester pathways for degradation of aromatic compounds. Applied and Environmental Microbiology, 2012, 78(15): 5043-5051. |
| [89] | Mohamed ES, Fuchs G. Purification and characterization of phenylacetate-coenzyme A ligase from a denitrifying Pseudomonas sp. an enzyme involved in the anaerobic degradation of phenylacetate. Archives of Microbiology, 1993, 159(6): 554-562. |
| [90] | Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds-from one strategy to four. Nature Reviews Microbiology, 2011, 9(11): 803-816. |
| [91] |
Chang Q, Li S. Microbial community comparison based on metagenomic data. Science Technology and Engineering, 2013, 13(9): 2457-2464.
(in Chinese) 常秦, 黎珊. 基于宏基因组数据的微生物群落比较方法. 科学技术与工程, 2013, 13(9): 2457-2464. |
| [92] | Tu QC, Lin L, Cheng L, Deng Y, He ZL. NCycDB:a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics, 2019, 35(6): 1040-1048. |
| [93] | Tu QC, Yu H, He ZL, Deng Y, Wu LY, Nostrand JDV, Zhou AF, Voordeckers J, Lee YJ, Qin YJ, Hemme CL, Shi Z, Xue K, Yuan T, Wang AJ, Zhou JZ. GeoChip 4:a functional gene-array-based high-throughput environmental technology for microbial community analysis. Molecular Ecology Resources, 2014, 14(5): 914-928. |
 2020, Vol. 60
2020, Vol. 60