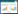中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张新旭, 李猛. 2020
- Xinxu Zhang, Meng Li. 2020
- 海洋沉积物中几类常见古菌类群的分布与代谢特征
- Distribution and metabolic features of some common archaeal groups in marine sediments
- 微生物学报, 60(9): 1907-1921
- Acta Microbiologica Sinica, 60(9): 1907-1921
-
文章历史
- 收稿日期:2020-03-21
- 修回日期:2020-04-28
- 网络出版日期:2020-07-21
古菌(Archaea),又称古细菌、古生菌或太古生物。古菌与细菌有很多相似之处,即它们都是单细胞生物,并且没有细胞核与膜结构的细胞器。因此,古菌长期以来被认为是细菌的一种。自1977年Woese和Fox根据16S rRNA分析提出生命三域理论以来,古菌便逐渐被认为是与细菌和真核生物并列存在的一个独立的生命域[1]。最初,科学家们分离培养得到的大部分古菌都拥有产甲烷、硫氧化等特殊的生理生化功能,而且它们几乎都来源于盐湖、热泉、海底热液等高盐、高温、高压或厌氧等极端环境[2-3];这些环境与地球的早期环境十分相似,因此古菌被认为可能是地球上最古老的生命形式之一[4-5]。
海洋占据了地球表面约70%的区域,其中蕴含着大量的古菌微生物资源。2019年,根据Hoshino和Inagaki的估算,在海洋沉积物的深部生物圈中含有大量的古菌细胞(约1.1×1029个),其含量约占沉积物中微生物总量的37.3%[6]。近年来,随着分子生物学和高通量测序技术的快速发展,科学家们利用荧光原位杂交[7]、荧光定量PCR[8]、宏基因组学[9]等技术方法对环境中的未培养微生物展开研究,人们对于古菌的认识也从最初的两个大的门类(广古菌门和泉古菌门)[1, 10]逐渐增加到目前的4个超级门(广古菌、TACK古菌、阿斯加德古菌和DPANN古菌),近30个门类(图 1)[11-15]。因此,人们逐渐认识到古菌作为海洋微生物的重要组分广泛分布于各种海洋环境,在碳、氮、硫等元素的生物地球化学循环和地球生命演化过程中扮演着极为重要的角色[1, 11, 16-18]。2019年,东秀珠和黄力等综述了我国科学家在古菌研究中取得的主要成果,进一步阐释了古菌研究的重要意义[18]。然而,目前尚未有报道详细介绍过海洋沉积物中的古菌群落的分布与代谢特征,它们的生理生化机制及其生态学意义也都没有被充分研究。因此,人们对于海洋古菌的科学认识存在较多不足,还有很多未知的“微生物暗物质”亟待进一步的深入挖掘和研究。
为了更加系统全面地了解海洋沉积物中的古菌群落,本文对广泛分布于近岸或深渊等沉积环境中的四类常见的古菌类群(深古菌门、乌斯古菌门、阿斯加德古菌超门和底栖古菌目)进行综述,内容涵盖其分类地位、系统发育、生态分布以及潜在生理代谢机制等,以期阐明古菌对驱动碳、氮、硫等地球元素循环的重要贡献,同时也为研究古菌在近岸与深渊沉积物环境中的生态效应与环境适应性机制提供新的思路。
1 深古菌门(Bathyarchaeota)深古菌最初被划分为泉古菌门,原名为Miscellaneous Crenarchaeotal Group(MCG)古菌[19-21]。2012年,李平一等对3个深古菌的fosmids序列进行分析,预测深古菌具有独特的进化位置[22]。2013年,基于深古菌单细胞基因组测序结果,Lloyd等对深古菌的部分单拷贝基因进行系统进化分析,推测深古菌是介于奇古菌门和曙古菌门之间的一个新的古菌门类[23]。2014年,孟君等通过多个串联基因的系统发育分析,发现深古菌在系统进化树上显著区别于以前确定的所有古菌门类,属于一类较老的古菌,因此提议将其确立为一个新门,即深古菌门[24]。2016年,何莹等收集了所有已知的深古菌基因组,利用串联基因组中的53个单拷贝保守基因进行系统发育分析,进一步确定深古菌是一个新的古菌门[25]。作为一个独立的古菌门类,深古菌含有众多内部分支亚群(subgroup)。根据16S rRNA基因的系统发育分析,深古菌可以细分为17个亚群[26]。随后,深古菌被进一步划分为21个亚群,各个亚群内部之间的16S rRNA基因序列的最小相似度仅为75%[27]。2018年,周之超等针对已有的深古菌16S rRNA基因(共获得24902条序列)进行了系统分析,发现深古菌至少可以分为25个不同的亚群,各个亚群内部的16S rRNA基因序列的相似度均为92%,表明深古菌具有极高的种群多样性[28]。
深古菌不仅具有较高的种群多样性,它们在自然界中的分布也非常广泛,且含量极为丰富。研究表明,深古菌在土壤、热泉、湖泊、河口、近岸湿地、海洋深部生物圈以及深渊沉积物等各类生态环境中均能被检测到[19-20, 22, 26-27, 29-49]。前期的研究发现,深古菌是红树林湿地等近岸环境中最主要的古菌类群,其相对丰度可占据总古菌的30%–70%,且与沉积物中的总有机碳含量和pH密切相关[28, 47, 50-51]。在海洋深部生物圈中,深古菌作为最主要且最活跃的微生物类群之一,通常占据了环境中总古菌数量的30%–60%[26, 28, 52]。而在深渊沉积环境中,目前仅在雅浦海沟6500 m水深的表层沉积物中有检测到深古菌[49]。
由于深古菌在不同环境中的种类和含量相差较大,人们对不同亚群的深古菌在自然界的分布规律及其环境适应性也开展了一系列的研究。前人研究发现,深古菌亚群5a和5b主要分布在静止、缺氧、富含硫化物和还原性有机物的底层水体中,亚群6广泛分布于不同层的水体和沉积物[53]。Lazar等分析了White Oak河口沉积物的深古菌多样性分布及其与环境因子之间的相互关系,发现深古菌亚群6主要分布于硫化氢消耗层,而亚群1、5和8则倾向出现于更深层的沉积物[44]。在我国珠江口,深古菌作为主要古菌种类,其比例也随着沉积物深度的增加而增加,其中以亚群15尤为明显[36]。Fillol等对深古菌21个亚群的复杂集群系统发育(complex intragroup phylogeny)进行分析,发现海洋和淡水沉积物中具有显著不同的深古菌亚群。同时,该研究进一步表明不同亚群的深古菌具有独特的环境适应性,能特异性地分布于不同的生态环境[27]。最近,周之超等对深古菌的生态分布进行了系统分析,发现深古菌各个亚群在不同自然环境中的生态分布差异较大,其中亚群1、8和15主要分布于海洋沉积物,亚群5b、5bb和11主要分布在陆地和淡水沉积物,而亚群6则广泛分布于各类沉积物[28]。
尽管深古菌在环境中具有极高的种群多样性、分布广泛且含量丰富,但目前尚未获得深古菌的纯培养菌株,对其生态功能和生理代谢机制还知之甚少。利用单细胞测序或宏基因组分析手段,科学家们发现深古菌不仅具有利用有机碳的异养代谢机制(如利用蛋白质、芳香烃化合物、几丁质、纤维素和木质素等复杂大分子有机化合物)[23-25, 54-57],还具备利用二氧化碳和氢气的自养代谢途径[25, 54-55]。2006年,Biddle等通过对沉积物中的稳定同位素和古菌脂类分析,发现深古菌可能利用沉积物中的有机物作为碳源[58]。随后,孟君等发现深古菌含有编码细菌叶绿素合成酶的基因bchG,为古菌参与光合作用的起源进化过程提供了有力依据[59]。Webster等和Seyler等分别推测河口沉积物和盐沼泽地的深古菌可能属于一类能利用低分子有机酸或高分子生物聚合物等有机物作为碳源的异养型微生物[60-61]。Gagen等进一步通过富集培养实验测试了深古菌对不同种类有机物的利用情况,发现添加0.02%的甘油可以显著抑制其生长[62]。Meador等通过对河口沉积物中的古菌脂类进行定量分析,推测深古菌也有可能是一类自养型微生物或可利用含13C亏损的有机物的异养型微生物[63]。Evans等在两个深古菌的基因组中首次发现了甲烷代谢途径和新的甲基辅酶M还原酶(MCR)编码基因,推测深古菌具有甲烷代谢的功能[55]。近年来,王风平课题组对深古菌展开了系统深入的研究:(1)发现深古菌多个亚群同时拥有利用无机碳自养合成乙酸和降解复杂有机化合物的异养代谢途径,提出深古菌是海洋沉积物中碳循环的核心驱动者[25];(2)通过系统进化分析显示深古菌可能起源于海底热液或陆地热泉等热环境[64];(3)通过富集培养和稳定同位素标记实验证明了深古菌可以降解木质素等大分子难降解有机质[57]。
在此基础上,周之超等基于已发表的所有深古菌基因组及相关信息,对深古菌的生理代谢进行系统分析,确定深古菌具有丰富多样的碳代谢机制,具备利用各类有机碳和无机碳作为底物进行生长的潜力,在全球碳循环中可能具有不可忽视的作用[28]。最近,潘杰等发现深古菌具备利用各种高价含氮化合物(羟胺、亚硝酸盐、三甲胺和氮气)生成铵盐并转换为尿素的能力,暗示深古菌在氮循环中也扮演着重要的角色。值得一提的是,潘杰等还发现亚群6 (Bathy-6)具有转录活性的钴胺素(cobalamin)和视紫红质(rhodopsins)合成基因,以及好氧代谢相关基因,这暗示了深古菌亚群6是一类微好氧、对光敏感且能合成钴胺素的古菌类群[65]。此外,研究还发现深古菌亚群6常分布于低pH和高TOC含量的沉积物中,进一步确定了深古菌亚群6具有独特的生理代谢机制和环境适应性[47, 66]。
2 乌斯古菌门(Woesearchaeota)乌斯古菌最初被称为Deep-sea Hydrothermal Vent Euryarchaeota Group-6 (DHVEG-6),主要是由于其广泛分布于深海热液生态系统[67-68]。2015年,Castelle等通过宏基因组分箱组装技术构建了乌斯古菌的完整基因组(完整度100%的闭环DNA);经过基因组水平的系统发育分析,他们发现乌斯古菌明显区别于已知的任何一个古菌门类,因此将其命名为一个独立的新古菌门——乌斯古菌(Woesearchaeota),以此纪念古菌的研究先驱、著名微生物学家Carl Woese教授[69]。2018年,刘小波等基于已报道的乌斯古菌的16S rRNA基因序列,采用生物信息学手段将乌斯古菌划分为26个亚群,并进一步发现生境中的氧气含量是决定乌斯古菌分布与多样性的关键因素[70]。除了深海热液区,乌斯古菌还广泛分布于深渊海沟[71-77]、淡水河湖沉积物[69, 78-79]、废水污泥[80]、冻土[81]和生物反应器[82]等厌氧环境中。例如,在地球表面最深处的马里亚纳海沟(水深 > 10000 m),沉积物中的乌斯古菌是相对丰度第二高的古菌(1%–22%),仅次于奇古菌(10%–60%)[71, 73-74];乌斯古菌也分别存在于雅浦海沟沉积物(6578 m)[49, 77]、日本海沟沉积物(7963 m)[74]、克马德克海沟沉积物(6011–9177 m)[72]以及伊豆-小笠原海沟沉积物(9776 m)[74],而且其相对丰度随着沉积物深度的增加而升高[71-72]。此外,邢鹏等在2016年发现磷浓度的升高可能有利于太湖底泥中乌斯古菌的生长[78]。
由于Castelle等构建出的乌斯古菌基因组(0.8 Mb)明显小于其他物种,其中缺失了一些核酸合成和糖代谢相关基因,并存在一些分解肽聚糖的壁质转糖类似基因,他们推测乌斯古菌可能与革兰氏阴性菌存在共生或者寄生关系[69]。Ortiz-Alvarez等也发现乌斯古菌的丰度与环境中细菌的多样性呈正相关关系,这进一步佐证了乌斯古菌可能具有共生或寄生习性[83]。随后,刘小波等采用共现性网络分析发现乌斯古菌与产甲烷古菌存在潜在的共生关系,并发现乌斯古菌具有发酵有机物产生乙酸、氢气及甲基化合物的潜能,而这些发酵产物可以进一步为产甲烷古菌提供底物[70]。此外,李猛和王勇等也分别从雅浦海沟和马里亚纳海沟的沉积物宏基因组样品中获得了一些乌斯古菌的基因组[75, 77]。然而,目前人们对于乌斯古菌的研究主要集中在多样性和生态分布,对其生理代谢机制研究还非常有限,因此还需要更深入研究其生理生化途径,才能最终了解乌斯古菌在近岸与深渊环境中的生态功能及其环境适应性机制。
3 阿斯加德古菌超门(Asgard archaea)早在1999年,Vetriani等利用克隆文库的方法首先在大西洋深海沉积物中发现了一类古菌,命名为Marine Benthic Group B (MBG-B)[84]。随后,Inagaki等在深海沉积物中也找到了一类与MBG-B相似的高丰度古菌,并将它命名为Deep-Sea Archaeal Group (DSAG)[20, 85]。2012年,Durbin等发现MBG-B/DSAG古菌广泛存在于深海沉积物中,且其相对丰度与沉积物中的有机质含量相关[86]。随后,Jorgensen等通过系统发育分析和定量PCR研究发现,MBG-B/DSAG古菌在某些区域的深海沉积物中属于优势群落(可占据总微生物量的50%),并进一步推测MBG-B/DSAG古菌可能参与了沉积物的有机碳循环和铁锰循环等[87-88]。2015年,Spang等通过对北冰洋中脊附近Loki’ Castle热液区的深海沉积物宏基因组测序分析,获得了3个MBG-B/DSAG古菌的基因组,并将其命名为洛基古菌(Lokiarchaeota)[67]。随后,Seitz等和刘杨等也分别在美国的河口和中国的红树林湿地沉积物中获得索尔古菌(Thorarchaeota)的基因组[89-90]。2017年初,包括洛基古菌、索尔古菌、奥丁古菌(Odinarchaeota)和海姆达尔古菌(Heimdallarchaeota)在内的一个新的古菌超级门被命名为阿斯加德古菌(Asgard)[14],它与广古菌、TACK和DPANN古菌并列(图 1)[11]。另外,由于海姆达尔古菌内部的16S rRNA序列的相似度只有65%,Eme等进一步将Heimdallarchaeota分为Heimdallarchaeota AAG (Ancient Archaeal Group)及Heimdallarchaeota MHVG (Marine Hydrothermal Vent Group)两个类别,并且将原来属于MHVG的另一部分序列预测为Asgard古菌的第6个新的分支[91]。2018年,Karst等发现了更多的Asgard古菌16S rRNA序列,其中包括8个可能归属于Asgard古菌的新分支(DAS1到DAS8)[92]。最近,蔡铭伟等通过对我国近海区域阿斯加德古菌的大规模调查分析,发现了阿斯加德古菌超门的第6个门,并将其命名为葛德古菌(Gerdarchaeota)[93]。这些研究结果表明,阿斯加德古菌的多样性极为丰富,其确切的系统分类还有待进一步的深入研究。
长期以来,人们推测原始真核生物逐渐进化形成现存真核生物的路径是:前真核细胞通过内吞作用捕获了一类古阿尔法变形菌,然后被捕获的细菌逐渐演变为真核细胞的线粒体。近些年,阿斯加德古菌和真核生物的进化关系受到了科学界的广泛关注[91, 94-97]。经过对阿斯加德古菌基因组的系统分析,Ettema等发现阿斯加德超古菌门中的所有成员的基因组中都拥有一部分真核生物特有的细胞结构特征蛋白基因(Eukaryotic signature proteins),如与膜转运、细胞区隔和细胞骨架构建相关的蛋白等[14, 67, 91]。基于核糖体蛋白和其他保守蛋白的系统发育分析,他们还发现阿斯加德古菌(尤其是海姆达尔古菌)是目前距离真核生物起源最近的原核生物,这暗示了原始的真核生物可能起源于阿斯加德古菌[14, 67]。如果这个推论被证实,那么地球生命最初的起源应该只包含细菌和古菌这两类最基础的门类,这对于Woese等早在1977年建立起来的细菌-古菌-真核生物三域理论[1]将是一个重大的挑战,这有可能改变整个地球生命演化的理论体系。然而,关于这方面的研究还具有一定的争议[98-100]:Burns等基于基因预测的营养模式分析认为阿斯加德古菌不能进行吞噬过程,因此其对真核生命起源的作用尚未明确[97];Sousa等对洛基古菌进行比较基因组学分析认为其属于氢营养型古菌,并进一步推测洛基古菌可能与真核生物起源的线粒体“氢假说”有关[101]。对此,周之超等发表一篇综述概览了长时间来学界关于二域或三域的争论,着重论述了自阿斯加德古菌发现以来对于这一问题带来的新认识和突破[102]。2020年,Imachi等首次获得了一类洛基古菌的富集物(占微生物总量的87.5%),将其命名为Candidatus Prometheoarchaeum syntrophicum;这类古菌的基因组中同样含有真核生物特征蛋白基因,并且具有类似“触角”的细胞结构。因此,他们提出了真核细胞演化的缠绕-包裹-奴役(Entangle-Engulf-Enslave)的E3模型[103]。
阿斯加德古菌广泛分布于厌氧环境中,包括河口、热泉、盐湖、红树林湿地、湖泊沉积物,以及海底热液、冷泉、深部生物圈和深渊海沟沉积物等环境[14, 48]。例如,Karst等和Seitz等分别在湖泊与河口沉积物中检测到阿斯加德古菌[89, 92],而刘杨等在红树林湿地沉积物中也获得了3个相对完整的阿斯加德古菌的基因组[90]。此外,在海底以下330 m的深部生物圈沉积物以及深渊海沟沉积物中也能检测到阿斯加德古菌[48, 104]。在目前已命名的6个阿斯加德古菌门类中,洛基古菌的分布最广且通常在深海沉积物中具有非常高的丰度[86-87],索尔古菌和海姆达尔古菌是沉积物中的少数类群[11],奥丁古菌主要来源于陆地热泉或深海热液环境[11],而葛德古菌主要分布于河流、湖泊和海洋的沉积物[93]。
目前,阿斯加德古菌还没有成功分离到纯培养菌株,其代谢途径主要是基于宏基因组分箱组装获得的基因组推测得出。例如,洛基古菌可能是一种自养型微生物,在厌氧环境下可能可以利用H2作为能源供给[101]。索尔古菌可能利用丙酮酸盐及蛋白质水解产物等作为能源供给,并且可能参与到厌氧发酵过程的产乙酸阶段[89];同时,索尔古菌也可能是一类混合营养型微生物,具有固氮和发酵产乙醇等代谢通路,并且可以通过合成四氢甲烷蝶呤和四氢叶酸两种中间产物来固定CO2[90]。而葛德古菌不仅拥有好氧呼吸和厌氧发酵的代谢通路,还具备利用有机质和无机碳的混合营养代谢潜能;此外,葛德古菌与其他大部分已报道的阿斯加德古菌均有降解多肽、氨基酸和脂肪酸等有机质的转录活性[93]。然而,由于缺乏阿斯加德古菌的纯培养菌株,对其参与碳、氮、硫等元素的生物地球化学循环中的代谢机制和作用也有待进一步验证。
4 底栖古菌目(Thermoprofundales)底栖古菌曾被称为MBG-D (Marine Benthic Group D)或DHVEG-1 (Deep-sea Hydrothermal Vent Euryarchaeota Group 1)。早在1999年,Verniani等通过建立16S rRNA基因克隆文库的手段对几个深海沉积物样品中的古菌多样性进行调查,发现沉积物中有一类叫做MBG-D的古菌[84];同年,Takai等也在深海热液口附近发现了一类叫做DHVEG-1的古菌[105]。2017年,Adam等曾将MBG-D/DHVEG-1古菌命名为Izemarchaea,并认为它可能属于广古菌门下的一个纲,但该推断缺乏实验数据和理论支持[15]。2018年,周之超等结合现有MBG-D/DHVEG-1古菌基因组的多个串联基因及16S rRNA基因的系统发育分析,确定MBG-D/DHVEG-1古菌隶属于广古菌门热源体纲(Euryarchaeota:Thermoplastmata)下的一个新目;根据其生存环境和生理代谢潜能特性,最终将其命名为底栖古菌(Thermoprofundales,图 1)[106]。通过对目前已报道的底栖古菌16S rRNA基因序列进行系统分类,发现底栖古菌至少可以细分为16个不同的亚群(Subgroup),且各个亚群内部的16S rRNA基因序列相似度仅为91.5%,这表明底栖古菌具有极高的多样性[106]。
底栖古菌在自然界中的分布非常广泛,在近岸、河口、红树林湿地、深海、热液和冷泉等环境中都有报道;其中,底栖古菌主要分布于海洋沉积物、海底热液和红树林湿地沉积物,而从这三类环境中所检测到的底栖古菌的16S rRNA基因序列占据了目前已报道的底栖古菌序列的70%左右[106]。此外,底栖古菌是海洋沉积物中最主要的古菌类群之一,其相对丰度通常可占据环境中总古菌数量的10%–100%[23, 106]。2006年,Inagaki等发现底栖古菌和厌氧甲烷氧化古菌(ANME)在深海冷泉区可能具有共生关系[107],然而底栖古菌的16S rRNA基因的丰度似乎与硫酸盐的减少和产甲烷的生物地球化学过程无关[23, 108]。在一个淡水湖泊的沉积物中,底栖古菌的相对丰度随着沉积物的深度加深而增加,并逐渐取代产甲烷菌[109]。此外,底栖古菌在高盐环境中也是常见的高丰度菌群,并且其种群结构随着碳含量的变化仅会出现一些微小波动[110]。最近,周之超等发现底栖古菌的亚群3和亚群4可能是高盐环境的指示类群,亚群8c属于中盐环境的指示类群,而亚群9b和9c则是淡水环境的指示类群[106]。此外,还发现亚群6和亚群7常分布于甲烷渗漏的冷泉区,而亚群9c则更多出现于非冷泉环境。微生物群落的共现性分析还表明,底栖古菌与洛基古菌(Lokiarchaeota)[67]和Hadesarchaea[111]有显著的关联性,这些关联表明这些古菌群之间可能存在相互作用[106]。
近年来,科学家们通过单细胞基因组和宏基因组测序分析方法对底栖古菌的基因组和代谢潜能展开了初步的研究[23, 112]。2013年,Lloyd等采用单细胞基因组测序发现底栖古菌可能会分泌胞外蛋白酶来降解沉积物中的蛋白质[23]。另一项宏基因组调查显示,底栖古菌和深古菌在白栎河河口沉积物中大量共存,且都具有乙酸生成和蛋白质降解的代谢潜能[54, 112]。最近,周之超等从香港米埔自然保护区的红树林和潮间带泥滩的地下沉积物中解析了4个几乎完整的底栖古菌基因组,发现它们可能运输和同化多肽,并通过发酵过程产生乙酸和乙醇[106]。同时,转录组学分析表明,底栖古菌基因组中与乙酸、氨基酸利用以及肽酶相关的基因高度表达,尤其是M09B型细胞外肽酶家族的细胞外肽酶(胶原酶)基因。除了含有编码异养碳代谢的核心基因,底栖古菌还含有合成四氢甲烷蝶呤和四氢叶酸两种中间产物的代谢途径,通过它们可以进行二氧化碳固定以及不完全的二羧酸盐/4-羟基丁酸盐循环[106]。然而,目前对于底栖古菌在近岸和深渊生态系统中的时空变化规律、种群结构、碳代谢多样性及其在碳、氮等生物地球化学循环中的贡献等仍缺乏深入认识。
5 总结和展望古菌是海洋沉积物中碳、氮、硫等地球元素循环过程的重要参与者。本文介绍的四大类古菌类群均为近岸和深渊等海洋沉积环境中常见的或丰度较高的古菌,也是近些年国内外古菌生态学研究领域的热点或潜在新亮点(图 2)。这些研究结果初步显示,不同种类的古菌可能具备差异的生态位和功能,协同推动自然环境中的碳氮等元素转化过程,对元素的生物地球化学循环有重要的贡献。然而,我们还需要更多的证据和实验数据来确定这个科学推论。对此,我们提出以下3个未来的研究方向以供参考。

|
| 图 2 广泛分布于近岸与深渊沉积物环境中的四大类古菌的代谢特征 Figure 2 Metabolic features of the four archaeal groups which are commonly distributed in coastal or hadal sediments. |
(1) 未培养古菌的富集培养一直是微生物学界关注的焦点和难点之一。以组学数据结果为指导,尝试应用新的富集培养方法和分离纯化技术来获得这些古菌的纯培养体或富集物,将为深入了解海洋沉积物古菌的生物学特性提供最直接的证据。
(2) 研制或改进一批适用于原位检测海洋沉积物古菌的生物量、代谢速率或细胞活性的仪器或方法,并建立这些古菌在海洋沉积物中的生态学模型,为揭示这些古菌的生理生化特征、进一步估算其对碳、氮、硫等地球元素循环的贡献提供技术和理论支持。
(3) 古菌是目前距离真核生物最近的原核生物,其与真核生命起源的关系是目前备受关注的重大科学问题。此问题的解答需要深入研究这些古菌中的真核特征蛋白与真核生物的相似蛋白功能的异同,这将为完善生命与地球环境相互作用与协同演化的理论提供重要的科学依据。
综上所述,深入开展这几类古菌的研究,不仅可以丰富人们对海洋古菌的科学认识、有助于解答“水圈生境中微生物驱动地球元素循环的机制”这一核心科学问题,也可为改善和保护海洋生态系统功能、应对全球气候变化、保障海洋生态环境安全、揭示真核生命起源等提供新的科学思路。
| [1] | Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain:the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74(11): 5088-5090. DOI:10.1073/pnas.74.11.5088 |
| [2] | Britschgi TB, Giovannoni SJ. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Applied and Environmental Microbiology, 1991, 57(6): 1707-1713. DOI:10.1128/AEM.57.6.1707-1713.1991 |
| [3] | DeLong EF. Archaea in coastal marine environments. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(12): 5685-5689. DOI:10.1073/pnas.89.12.5685 |
| [4] | Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature, 2006, 440(7083): 516-519. DOI:10.1038/nature04584 |
| [5] | Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution:insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evolutionary Biology, 2004, 4(1): 44. DOI:10.1186/1471-2148-4-44 |
| [6] | Hoshino T, Inagaki F. Abundance and distribution of Archaea in the subseafloor sedimentary biosphere. The ISME Journal, 2019, 13(1): 227-231. DOI:10.1038/s41396-018-0253-3 |
| [7] | Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and Archaea in the deep ocean. Applied and Environmental Microbiology, 2004, 70(7): 4411-4414. DOI:10.1128/AEM.70.7.4411-4414.2004 |
| [8] | Takai K, Oida H, Suzuki Y, Hirayama H, Nakagawa S, Nunoura T, Inagaki F, Nealson KH, Horikoshi K. Spatial distribution of marine crenarchaeota group Ⅰ in the vicinity of deep-sea hydrothermal systems. Applied and Environmental Microbiology, 2004, 70(4): 2404-2413. DOI:10.1128/AEM.70.4.2404-2413.2004 |
| [9] | DeLong EF, Wu KY, Prézelin BB, Jovine RVM. High abundance of Archaea in Antarctic marine picoplankton. Nature, 1994, 371(6499): 695-697. DOI:10.1038/371695a0 |
| [10] | Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms:proposal for the domains Archaea, Bacteria, and Eucarya. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(12): 4576-4579. DOI:10.1073/pnas.87.12.4576 |
| [11] | Spang A, Caceres EF, Ettema TJG. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science, 2017, 357(6351): eaaf3883. DOI:10.1126/science.aaf3883 |
| [12] | Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. A new view of the tree of life. Nature Microbiology, 2016, 1(5): 16048. DOI:10.1038/nmicrobiol.2016.48 |
| [13] | Jay ZJ, Beam JP, Dlakic M, Rusch DB, Kozubal MA, Inskeep WP. Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nature Microbiology, 2018, 3(6): 732-740. DOI:10.1038/s41564-018-0163-1 |
| [14] | Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckstrom D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature, 2017, 541(7637): 353-358. DOI:10.1038/nature21031 |
| [15] | Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. The growing tree of Archaea:new perspectives on their diversity, evolution and ecology. The ISME Journal, 2017, 11(11): 2407-2425. DOI:10.1038/ismej.2017.122 |
| [16] | Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Annual Review of Microbiology, 2013, 67(1): 437-457. DOI:10.1146/annurev-micro-092412-155614 |
| [17] | Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF. Community-wide analysis of microbial genome sequence signatures. Genome Biology, 2009, 10(8): R85. DOI:10.1186/gb-2009-10-8-r85 |
| [18] |
Dong XZ, Li M, Xiang H, Xu J, Wang FP, Shen YL, Zhang ZF, Han J, Li J, Li M, Huang L. Unraveling the secrets of the third form of life-A review and future perspective on archaeal research in China. SCIENTIA SINICA Vitae, 2019, 49(11): 1520-1542.
(in Chinese) 东秀珠, 李猛, 向华, 徐俊, 王风平, 申玉龙, 张臻峰, 韩静, 李洁, 李明, 黄力. 探秘生命的第三种形式——我国古菌研究之回顾与展望. 中国科学:生命科学, 2019, 49(11): 1520-1542. |
| [19] | Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. Archaeal diversity in waters from deep South African gold mines. Applied and Environmental Microbiology, 2001, 67(12): 5750-5760. DOI:10.1128/AEM.67.21.5750-5760.2001 |
| [20] | Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, Nealson KH, Horikoshi K. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Applied and Environmental Microbiology, 2003, 69(12): 7224-7235. DOI:10.1128/AEM.69.12.7224-7235.2003 |
| [21] | Reed DW, Fujita Y, Delwiche ME, Blackwelder DB, Sheridan PP, Uchida T, Colwell FS. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Applied and Environmental Microbiology, 2002, 68(8): 3759-3770. DOI:10.1128/AEM.68.8.3759-3770.2002 |
| [22] | Li PY, Xie BB, Zhang XY, Qin QL, Dang HY, Wang XM, Chen XL, Yu J, Zhang YZ. Genetic structure of three fosmid-fragments encoding 16S rRNA genes of the Miscellaneous Crenarchaeotic Group (MCG):implications for physiology and evolution of marine sedimentary archaea. Environmental Microbiology, 2012, 14(2): 467-479. DOI:10.1111/j.1462-2920.2011.02637.x |
| [23] | Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB. Predominant archaea in marine sediments degrade detrital proteins. Nature, 2013, 496(7444): 215-218. DOI:10.1038/nature12033 |
| [24] | Meng J, Xu J, Qin D, He Y, Xiao X, Wang FP. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. The ISME Journal, 2014, 8(3): 650-659. DOI:10.1038/ismej.2013.174 |
| [25] | He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nature Microbiology, 2016, 1(6): 16035. DOI:10.1038/nmicrobiol.2016.35 |
| [26] | Kubo K, Lloyd KG, Biddle JF, Amann R, Teske A, Knittel K. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. The ISME Journal, 2012, 6(10): 1949-1965. DOI:10.1038/ismej.2012.37 |
| [27] | Fillol M, Auguet JC, Casamayor EO, Borrego CM. Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage. The ISME Journal, 2016, 10(3): 665-677. DOI:10.1038/ismej.2015.143 |
| [28] | Zhou ZC, Pan J, Wang FP, Gu JD, Li M. Bathyarchaeota:globally distributed metabolic generalists in anoxic environments. FEMS Microbiology Reviews, 2018, 42(5): 639-655. DOI:10.1093/femsre/fuy023 |
| [29] | Parkes RJ, Webster G, Cragg BA, Weightman AJ, Newberry CJ, Ferdelman TG, Kallmeyer J, Jørgensen BB, Aiello IW, Fry JC. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature, 2005, 436(7049): 390-394. DOI:10.1038/nature03796 |
| [30] | Vetriani C, Reysenbach AL, Doré J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiology Letters, 1998, 161(1): 83-88. DOI:10.1111/j.1574-6968.1998.tb12932.x |
| [31] | Kasai Y, Takahata Y, Hoaki T, Watanabe K. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environmental Microbiology, 2005, 7(6): 806-818. DOI:10.1111/j.1462-2920.2005.00754.x |
| [32] | Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, Nealson KH, Horikoshi K, D'Hondt S, Jorgensen BB. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(8): 2815-2820. DOI:10.1073/pnas.0511033103 |
| [33] | Castro H, Ogram A, Reddy KR. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida Everglades. Applied and Environmental Microbiology, 2004, 70(11): 6559-6568. DOI:10.1128/AEM.70.11.6559-6568.2004 |
| [34] | Li QQ, Wang FP, Chen ZW, Yin XJ, Xiao X. Stratified active archaeal communities in the sediments of Jiulong River estuary, China. Frontiers in Microbiology, 2012, 3: 311. |
| [35] | Chen JQ, Wang FP, Jiang LJ, Yin XJ, Xiao X. Stratified communities of active archaea in shallow sediments of the Pearl River Estuary, Southern China. Current Microbiology, 2013, 67(1): 41-50. DOI:10.1007/s00284-013-0320-y |
| [36] | Liu JW, Yang HM, Zhao MX, Zhang XH. Spatial distribution patterns of benthic microbial communities along the Pearl Estuary, China. Systematic and Applied Microbiology, 2014, 37(8): 578-589. DOI:10.1016/j.syapm.2014.10.005 |
| [37] | Jiang LJ, Zheng YP, Chen JQ, Xiao X, Wang FP. Stratification of archaeal communities in shallow sediments of the Pearl River Estuary, Southern China. Antonie Van Leeuwenhoek, 2011, 99(4): 739-751. DOI:10.1007/s10482-011-9548-3 |
| [38] | Jurgens G, Glöckner FO, Amann R, Saano A, Montonen L, Likolammi M, Munster U. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiology Ecology, 2000, 34(1): 45-56. |
| [39] |
Jiang LJ, Peng XT, Zhou HY, Wang FP. Vertical distribution and diversity of Archaea in coastal sediments of Qi'ao Island (Zhujiang River Estuary), southern China, estimated by 16S rDNA analysis. Acta Oceanologica Sinica, 2008, 30(4): 114-122.
(in Chinese) 姜丽晶, 彭晓彤, 周怀阳, 王风平. 非培养手段分析珠江口淇澳岛海岸带沉积物中的古菌多样性. 海洋学报, 2008, 30(4): 114-122. |
| [40] |
Jiao L, Su X, Wang YY, Zhang Y, Chen F, Jiang HC. Archaeal diversity in sediments of core BY1 from Shenhu Area, northern South China Sea. Acta Oceanologica Sinica, 2012, 34(4): 181-187.
(in Chinese) 焦露, 苏新, 王媛媛, 张勇, 陈芳, 蒋宏忱. 南海神狐海域BY1钻孔岩心沉积物古菌多样性分析. 海洋学报, 2012, 34(4): 181-187. |
| [41] |
Zhang XY, Xu J, Xiao J, Xu J. Archaeal lineages detected in mangrove sediment in the Zhangjiang estuary of Fujian province. Journal of Shandong University (Natural Science), 2009, 44(3): 1-6, 10.
(in Chinese) 张熙颖, 许静, 肖静, 徐俊. 福建漳江口红树林沉积物中的古菌类群. 山东大学学报(理学版), 2009, 44(3): 1-6, 10. |
| [42] | Yan B, Hong K, Yu ZN. Archaeal communities in mangrove soil characterized by 16S rRNA gene clones. Journal of Microbiology, 2006, 44(5): 566-571. |
| [43] | Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 2013, 41(1): e1. DOI:10.1093/nar/gks808 |
| [44] | Lazar CS, Biddle JF, Meador TB, Blair N, Hinrichs KU, Teske AP. Environmental controls on intragroup diversity of the uncultured benthic archaea of the miscellaneous Crenarchaeotal group lineage naturally enriched in anoxic sediments of the White Oak River estuary (North Carolina, USA). Environmental Microbiology, 2015, 17(7): 2228-2238. DOI:10.1111/1462-2920.12659 |
| [45] | Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(17): 9188-9193. DOI:10.1073/pnas.93.17.9188 |
| [46] | Yu TT, Liang QY, Niu MY, Wang FP. High occurrence of Bathyarchaeota (MCG) in the deep-sea sediments of South China Sea quantified using newly designed PCR primers. Environmental Microbiology Reports, 2017, 9(4): 374-382. DOI:10.1111/1758-2229.12539 |
| [47] | Pan J, Chen YL, Wang YM, Zhou ZC, Li M. Vertical distribution of Bathyarchaeotal communities in mangrove wetlands suggests distinct niche preference of Bathyarchaeota subgroup 6. Microbial Ecology, 2019, 77(2): 417-428. DOI:10.1007/s00248-018-1309-7 |
| [48] | Tarn J, Peoples LM, Hardy K, Cameron J, Bartlett DH. Identification of free-living and particle-associated microbial communities present in hadal regions of the Mariana Trench. Frontiers in Microbiology, 2016, 7: 665. |
| [49] | Fu LL, Li D, Mi TZ, Zhao J, Liu CG, Sun CJ, Zhen Y. Characteristics of the archaeal and bacterial communities in core sediments from Southern Yap Trench via in situ sampling by the manned submersible Jiaolong. Science of the Total Environment, 2020, 703: 134884. DOI:10.1016/j.scitotenv.2019.134884 |
| [50] |
Chen YL, Pan J, Zhou ZC, Wang FP, Li M. Progress in studies on Bathyarchaeota in coastal ecosystems. Microbiology China, 2017, 44(7): 1690-1698.
(in Chinese) 陈玉连, 潘杰, 周之超, 王风平, 李猛. 滨海深古菌的研究进展. 微生物学通报, 2017, 44(7): 1690-1698. |
| [51] | Zhou ZC, Meng H, Liu Y, Gu JD, Li M. Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S rRNA gene sequencing. Frontiers in Microbiology, 2017, 8: 2148. DOI:10.3389/fmicb.2017.02148 |
| [52] | Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiology Ecology, 2008, 66(2): 181-196. DOI:10.1111/j.1574-6941.2008.00566.x |
| [53] | Fillol M, Sànchez-Melsió A, Gich F, Borrego CM. Diversity of Miscellaneous Crenarchaeotic Group archaea in freshwater karstic lakes and their segregation between planktonic and sediment habitats. FEMS Microbiology Ecology, 2015, 91(4): fiv020. |
| [54] | Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs KU, Teske AP. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environmental Microbiology, 2016, 18(4): 1200-1211. DOI:10.1111/1462-2920.13142 |
| [55] | Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science, 2015, 350(6259): 434-438. DOI:10.1126/science.aac7745 |
| [56] | Zhang WP, Ding W, Yang B, Tian RM, Gu S, Luo HW, Qian PY. Genomic and transcriptomic evidence for carbohydrate consumption among microorganisms in a cold seep brine pool. Frontiers in Microbiology, 2016, 7: 1825. |
| [57] | Yu TT, Wu WC, Liang WY, Lever MA, Hinrichs KU, Wang FP. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(23): 6022-6027. DOI:10.1073/pnas.1718854115 |
| [58] | Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sorensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs KU. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(10): 3846-3851. DOI:10.1073/pnas.0600035103 |
| [59] | Meng J, Wang FP, Wang F, Zheng YP, Peng XT, Zhou HY, Xiao X. An uncultivated crenarchaeota contains functional bacteriochlorophyll a synthase. The ISME Journal, 2009, 3(1): 106-116. DOI:10.1038/ismej.2008.85 |
| [60] | Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiology Ecology, 2010, 72(2): 179-197. DOI:10.1111/j.1574-6941.2010.00848.x |
| [61] | Seyler LM, McGuinness LM, Kerkhof LJ. Crenarchaeal heterotrophy in salt marsh sediments. The ISME Journal, 2014, 8(7): 1534-1543. DOI:10.1038/ismej.2014.15 |
| [62] | Gagen EJ, Huber H, Meador T, Hinrichs KU, Thomm M. Novel cultivation-based approach to understanding the Miscellaneous Crenarchaeotic Group (MCG) Archaea from sedimentary ecosystems. Applied and Environmental Microbiology, 2013, 79(20): 6400-6406. DOI:10.1128/AEM.02153-13 |
| [63] | Meador TB, Bowles M, Lazar CS, Zhu C, Teske A, Hinrichs KU. The archaeal lipidome in estuarine sediment dominated by members of the Miscellaneous Crenarchaeotal Group. Environmental Microbiology, 2015, 17(7): 2441-2458. DOI:10.1111/1462-2920.12716 |
| [64] | Feng XY, Wang YZ, Zubin R, Wang FP. Core metabolic features and hot origin of Bathyarchaeota. Engineering, 2019, 5(3): 498-504. DOI:10.1016/j.eng.2019.01.011 |
| [65] | Pan J, Zhou ZC, Béjà O, Cai MW, Yang YC, Liu Y, Gu JD, Li M. Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome, 2020, 8: 43. DOI:10.1186/s40168-020-00820-1 |
| [66] | Zou DY, Pan J, Liu ZB, Zhang CL, Liu HB, Li M. The distribution of Bathyarchaeota in surface sediments of the Pearl River Estuary along salinity gradient. Frontiers in Microbiology, 2020, 11: 285. DOI:10.3389/fmicb.2020.00285 |
| [67] | Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature, 2015, 521(7551): 173-179. DOI:10.1038/nature14447 |
| [68] | Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, Takai K. Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing. Microbes and Environments, 2012, 27(4): 382-390. DOI:10.1264/jsme2.ME12032 |
| [69] | Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, Taylor RC, Williams KH, Banfield JF. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Current Biology, 2015, 25(6): 690-701. DOI:10.1016/j.cub.2015.01.014 |
| [70] | Liu XB, Li M, Castelle CJ, Probst AJ, Zhou ZC, Pan J, Liu Y, Banfield JF, Gu JD. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome, 2018, 6(1): 102. DOI:10.1186/s40168-018-0488-2 |
| [71] | Nunoura T, Nishizawa M, Hirai M, Shimamura S, Harnvoravongchai P, Koide O, Morono Y, Fukui T, Inagaki F, Miyazaki J, Takaki Y, Takai K. Microbial diversity in sediments from the bottom of the Challenger Deep, the Mariana Trench. Microbes and Environments, 2018, 33(2): 186-194. DOI:10.1264/jsme2.ME17194 |
| [72] | Peoples LM, Grammatopoulou E, Pombrol M, Xu XX, Osuntokun O, Blanton J, Allen EE, Nunnally CC, Drazen JC, Mayor DJ, Bartlett DH. Microbial community diversity within sediments from two geographically separated hadal trenches. Frontiers in Microbiology, 2019, 10: 347. DOI:10.3389/fmicb.2019.00347 |
| [73] | Cui GJ, Li J, Gao ZM, Wang Y. Spatial variations of microbial communities in abyssal and hadal sediments across the Challenger Deep. PeerJ, 2019, 7(2): e6961. |
| [74] | Hiraoka S, Hirai M, Matsui Y, Makabe A, Minegishi H, Tsuda M, Juliarni, Rastelli E, Danovaro R, Corinaldesi C, Kitahashi T, Tasumi E, Nishizawa M, Takai K, Nomaki H, Nunoura T. Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments. The ISME Journal, 2020, 14(3): 740-756. DOI:10.1038/s41396-019-0564-z |
| [75] | Gao ZM, Huang JM, Cui GJ, Li WL, Li J, Wei ZF, Chen J, Xin YZ, Cai DS, Zhang AQ, Wang Y. In situ meta-omic insights into the community compositions and ecological roles of hadal microbes in the Mariana Trench. Environmental Microbiology, 2019, 21(11): 4092-4108. DOI:10.1111/1462-2920.14759 |
| [76] | Jing HM, Zhu WD, Liu HB, Zheng LP, Zhang Y. Particle-attached and free-living archaeal communities in the benthic boundary layer of the Mariana Trench. Frontiers in Microbiology, 2018, 9: 2821. DOI:10.3389/fmicb.2018.02821 |
| [77] | Zhang XX, Xu W, Liu Y, Cai MW, Luo ZH, Li M. Metagenomics reveals microbial diversity and metabolic potentials of seawater and surface sediment from a hadal biosphere at the Yap Trench. Frontiers in Microbiology, 2018, 9: 2402. DOI:10.3389/fmicb.2018.02402 |
| [78] | Fan XF, Xing P. The vertical distribution of sediment archaeal community in the "Black Bloom" disturbing Zhushan Bay of Lake Taihu. Archaea, 2016, 2016: 8232135. |
| [79] | Casamayor EO, Triado-Margarit X, Castaneda C. Microbial biodiversity in saline shallow lakes of the Monegros Desert, Spain. FEMS Microbiology Ecology, 2013, 85(3): 503-518. DOI:10.1111/1574-6941.12139 |
| [80] | Kuroda K, Hatamoto M, Nakahara N, Abe K, Takahashi M, Araki N, Yamaguchi T. Community composition of known and uncultured archaeal lineages in anaerobic or anoxic wastewater treatment sludge. Microbial Ecology, 2015, 69(3): 586-596. DOI:10.1007/s00248-014-0525-z |
| [81] | Shcherbakova V, Yoshimura Y, Ryzhmanova Y, Taguchi Y, Segawa T, Oshurkova V, Rivkina E. Archaeal communities of Arctic methane-containing permafrost. FEMS Microbiology Ecology, 2016, 92(10): fiw135. DOI:10.1093/femsec/fiw135 |
| [82] | Bandara WMKRTW, Kindaichi T, Satoh H, Sasakawa M, Nakahara Y, Takahashi M, Okabe S. Anaerobic treatment of municipal wastewater at ambient temperature:analysis of archaeal community structure and recovery of dissolved methane. Water Research, 2012, 46(17): 5756-5764. DOI:10.1016/j.watres.2012.07.061 |
| [83] | Ortiz-Alvarez R, Casamayor EO. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environmental Microbiology Reports, 2016, 8(2): 210-217. DOI:10.1111/1758-2229.12370 |
| [84] | Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Applied and Environmental Microbiology, 1999, 65(10): 4375-4384. DOI:10.1128/AEM.65.10.4375-4384.1999 |
| [85] | Inagaki F, Takai K, Komatsu T, Kanamatsu T, Fujioka K, Horikoshi K. Archaeology of Archaea:geomicrobiological record of Pleistocene thermal events concealed in a deep-sea subseafloor environment. Extremophiles, 2001, 5(6): 385-392. DOI:10.1007/s007920100211 |
| [86] | Durbin AM, Teske A. Archaea in organic-lean and organic-rich marine subsurface sediments:an environmental gradient reflected in distinct phylogenetic lineages. Frontiers in Microbiology, 2012, 3: 168. |
| [87] | Jørgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C. Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Frontiers in Microbiology, 2013, 4: 299. |
| [88] | Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, Øvreås L, Steen IH, Thorseth IH, Pedersen RB, Schleper C. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(42): E2846-E2855. DOI:10.1073/pnas.1207574109 |
| [89] | Seitz KW, Lazar CS, Hinrichs KU, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. The ISME Journal, 2016, 10(7): 1696-1705. DOI:10.1038/ismej.2015.233 |
| [90] | Liu Y, Zhou ZC, Pan J, Baker BJ, Gu JD, Li M. Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. The ISME Journal, 2018, 12(4): 1021-1031. DOI:10.1038/s41396-018-0060-x |
| [91] | Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. Archaea and the origin of eukaryotes. Nature Reviews Microbiology, 2017, 15(12): 711-723. DOI:10.1038/nrmicro.2017.133 |
| [92] | Karst SM, Dueholm MS, McIlroy SJ, Kirkegaard RH, Nielsen PH, Albertsen M. Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nature Biotechnology, 2018, 36(2): 190-195. DOI:10.1038/nbt.4045 |
| [93] | Cai MW, Liu Y, Yin XR, Zhou ZC, Friedrich MW, Richter-Heitmann T, Nimzyk R, Kulkarni A, Wang XW, Li WJ, Pan J, Yang YC, Gu JD, Li M. Diverse Asgard archaea including the novel phylum Gerdarchaeota participate in organic matter degradation. Science China Life Sciences, 2020, 63(6): 886-897. |
| [94] | Nasir A, Kim KM, Caetano-Anollés G. Lokiarchaeota:eukaryote-like missing links from microbial dark matter?. Trends in Microbiology, 2015, 23(8): 448-450. DOI:10.1016/j.tim.2015.06.001 |
| [95] | Saw JH, Spang A, Zaremba-Niedzwiedzka K, Juzokaite L, Dodsworth JA, Murugapiran SK, Colman DR, Takacs-Vesbach C, Hedlund BP, Guy L, Ettema TJG. Exploring microbial dark matter to resolve the deep archaeal ancestry of eukaryotes. Philosophical Transactions of The Royal Society B: Biological Sciences, 2015, 370(1678): 20140328. DOI:10.1098/rstb.2014.0328 |
| [96] | Archibald JM. Endosymbiosis and eukaryotic cell evolution. Current Biology, 2015, 25(19): R911-R921. DOI:10.1016/j.cub.2015.07.055 |
| [97] | Burns JA, Pittis AA, Kim E. Gene-based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nature Ecology & Evolution, 2018, 2(4): 697-704. |
| [98] | Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genetics, 2017, 13(6): e1006810. DOI:10.1371/journal.pgen.1006810 |
| [99] | Da Cunha V, Gaia M, Nasir A, Forterre P. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genetics, 2018, 14(3): e1007215. DOI:10.1371/journal.pgen.1007215 |
| [100] | Spang A, Eme L, Saw JH, Caceres EF, Zaremba-Niedzwiedzka K, Lombard J, Guy L, Ettema TJG. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genetics, 2018, 14(3): e1007080. DOI:10.1371/journal.pgen.1007080 |
| [101] | Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nature Microbiology, 2016, 1(5): 16034. DOI:10.1038/nmicrobiol.2016.34 |
| [102] | Zhou ZC, Liu Y, Li M, Gu JD. Two or three domains:a new view of tree of life in the genomics era. Applied Microbiology and Biotechnology, 2018, 102(7): 3049-3058. DOI:10.1007/s00253-018-8831-x |
| [103] | Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song CH, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature, 2020, 577(7791): 519-525. DOI:10.1038/s41586-019-1916-6 |
| [104] | Nunoura T, Inagaki F, Delwiche ME, Colwell FS, Takai K. Subseafloor microbial communities in methane hydrate-bearing sediment at two distinct locations (ODP Leg204) in the cascadia margin. Microbes and Environments, 2008, 23(4): 317-325. DOI:10.1264/jsme2.ME08514 |
| [105] | Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics, 1999, 152(4): 1285-1297. |
| [106] | Zhou ZC, Liu Y, Lloyd KG, Pan J, Yang YC, Gu JD, Li M. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). The ISME Journal, 2019, 13(4): 885-901. DOI:10.1038/s41396-018-0321-8 |
| [107] | Inagaki F, Kuypers MMM, Tsunogai U, Ishibashi JI, Nakamura KI, Treude T, Ohkubo S, Nakaseama M, Gena K, Chiba H, Hirayama H, Nunoura T, Takai K, Jørgensen BB, Horikoshi K, Boetius A. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(38): 14164-14169. DOI:10.1073/pnas.0606083103 |
| [108] | Lipp JS, Morono Y, Inagaki F, Hinrichs KU. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature, 2008, 454(7207): 991-994. DOI:10.1038/nature07174 |
| [109] | Borrel G, Lehours AC, Crouzet O, Jézéquel D, Rockne K, Kulczak A, Duffaud E, Joblin K, Fonty G. Stratification of Archaea in the deep sediments of a freshwater meromictic lake:vertical shift from methanogenic to uncultured archaeal lineages. PLoS One, 2012, 7(8): e43346. DOI:10.1371/journal.pone.0043346 |
| [110] | Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Applied and Environmental Microbiology, 2010, 76(3): 757-768. |
| [111] | Baker BJ, Saw JH, Lind AE, Lazar CS, Hinrichs KU, Teske AP, Ettema TJG. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nature Microbiology, 2016, 1: 16002. DOI:10.1038/nmicrobiol.2016.2 |
| [112] | Lazar CS, Baker BJ, Seitz KW, Teske AP. Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. The ISME Journal, 2017, 11(5): 1118-1129. DOI:10.1038/ismej.2016.189 |
 2020, Vol. 60
2020, Vol. 60