中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张嘉鑫, 武雅蓉, 杨超, 康琳, 王景林, 崔玉军. 2020
- Zhang Jiaxin, Wu Yarong, Yang Chao, Kang Lin, Wang Jinglin, Cui Yujun. 2020
- 创伤弧菌毒力相关因子的全基因组关联分析研究
- Genome-wide association study on virulence factors of Vibrio vulnificus
- 微生物学报, 60(7): 1488-1495
- Acta Microbiologica Sinica, 60(7): 1488-1495
-
文章历史
- 收稿日期:2019-12-31
- 修回日期:2020-02-05
- 网络出版日期:2020-03-12
2. 军事科学院军事医学研究院微生物流行病研究所, 病原微生物生物安全国家重点实验室, 北京 100071
2. State Key Laboratory of Pathogen and Biosecurity, Institute of Microbiology and Epidemiology, Academy of Military Medical Sciences, Beijing 100071, China
创伤弧菌(Vibrio vulnificus),与霍乱弧菌、副溶血弧菌并称为人类三大致病性弧菌[1]。创伤弧菌可引发原发性败血症,是目前发现的致死率最高的致病弧菌[2],美国95%以上因食用海鲜而致死的病例是该物种引起的[3]。该病原对男性患者(约85%)和老年患者(> 40岁)的影响尤为严重,特别是患有肝病、糖尿病和免疫紊乱等潜在疾病的人群[2]。创伤弧菌的分布与温度有关,随着全球变暖和海水温度升高,其感染的病例数和地理区域不断扩大,明确创伤弧菌的毒力因子、探索其致病机制具有重要意义[4]。
全基因组关联分析(genome-wide association study,GWAS)是近年来迅速兴起的一种遗传机制分析方法。该方法将基于全基因组范围鉴定获得的遗传多态性位点与可观测性状(即表型)进行群体水平统计分析,从而筛选出最有可能影响该性状的遗传变异位点[5]。与传统关联分析比较,GWAS具有如下优势:(1)不需要建立前提假设和筛选候选基因,可直接探查与某表型关联的遗传变异机制[6];(2)检测范围更全面,可在整个基因组水平检测跟表型相关的所有变异位点[7];(3)可同时对大量样本进行全基因组水平的关联分析,实现精细定位,节省时间和成本[8]。GWAS在人类复杂疾病遗传机制和动植物育种方面已取得丰硕成果,打开了基因组学研究的新大门。但限于克隆繁殖特性所致的种群结构,细菌全基因组关联研究(bacterial genome wide association study,BGWAS)开展较晚,目前主要应用于细菌宿主适应性、毒力和抗生素耐药等表型与基因型的相关性研究中[9]。
本研究首次将BGWAS引入创伤弧菌的研究领域,对分离自临床和环境样本中共计260株创伤弧菌全基因组序列与其分离来源进行分析,在全基因组的范围内筛选可能与创伤弧菌毒力相关的变异位点或功能基因,为进一步探索创伤弧菌的致病机理,以及该病原的毒力检测和防治工作提供科学依据。
1 材料和方法 1.1 菌株来源本研究共收集260株创伤弧菌的全基因组序列,包括139个新测序的全基因组序列和121个公共基因组序列。新测序的菌株来源于:台湾成功大学(26个)、温州医科大学附属第一医院(5个)、中国科学院动物研究所(1个)、北京出入境检验检疫局(32个)、广东检验检疫技术中心卫检室(7个)、中国海洋微生物菌种保藏管理中心(2个)、浙江疾病预防控制中心(33株)和上海交通大学(33株),所有菌株均保存于–80 ℃;公共序列下载于NCBI数据库(https://www.ncbi.nlm.nih.gov/genome/genomes/189)。菌株背景信息见https://doi.org/10.6084/m9.figshare.11758704.v1。
1.2 接种与培养将收集的菌种在哥伦比亚血琼脂(Columbia blood agar,CBA)平板划线后37 ℃生化培养箱中培养,挑取单菌落接种于3 mL心浸液肉汤(Heart infusion,HI)液体培养基中,过夜培养至指数生长期。
1.3 全基因组测序取1.8 mL过夜培养菌液,使用细菌基因组DNA提取试剂盒(QIAGEN UltraClean® Microbial DNA Isolation kit),参照试剂盒说明书提取全基因组DNA。经检测质量合格后由测序公司(诺禾致源,北京)进行细菌全基因组重测序,测序平台为Novaseq 6000,测序文库大小为500 bp,测序reads读长为双端150 bp,平均测序深度257X (80–290X)。
1.4 质控及组装使用FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/)软件中FASTQ Quality Filter模块过滤低质量数据,并采用shovill (https://github.com/seemann/shovill)软件对过滤后的reads进行组装。对组装结果进行统计,得到本次测序菌株的平均基因组大小为5.03 Mb,平均GC含量为46.71% (https://doi.org/10.6084/m9.figshare.11759601.v1)。组装得到的基因组草图序列已上传至NCBI,Bioproject编号为PRJNA588308,详细信息见https://doi.org/10.6084/m9.figshare.11758704.v1。
1.5 变异检测和注释单核苷酸多态性(single nucleotide polymorphism,SNP)位点的鉴定方法参照已发表的相关工作[10]:以创伤弧菌CECT4999株基因组完成图为参考序列,使用MUMmer软件包[11]将260个创伤弧菌基因组序列分别比对到参考株基因组,获得核心基因组(全部菌株中均能找到对应碱基状态)中的SNP状态信息。为保证SNP鉴定结果的可靠性,位于重复区和测序质量值低的SNP未纳入后续分析。
采用Prokka软件[12]对各个菌株基因组序列进行基因预测,将基因序列分别与COG库(clusters of orthologous groups)和KEGG库(kyoto encyclopedia of genes and genomes)进行蛋白质同源序列比对,完成功能注释。使用Roary软件[13]构建泛基因组序列和基因获得缺失矩阵,进而判定基因获得缺失。
1.6 系统发育分析使用TreeBest软件(http://treesoft.sourceforge.net/treebest.shtml)对SNP数据集构建邻接(Neighbour-Joining,NJ)树,bootstrap值设为1000,并使用在线工具iTOL[14]进行可视化展示。
1.7 已知毒力因子分析统计VFDB数据库(virulence factors database)[15]和已发表文献中毒力因子的存在缺失情况。基于开源R软件的分析工具包进行统计学检验,采用列联表x2检验来判断创伤弧菌已知毒力因子在环境组和临床组中的频率分布差异,P < 0.05认为有显著性差异。
1.8 全基因组关联分析采用pyseer软件[16],将创伤弧菌的分离来源信息(临床和环境)分别与全基因组范围SNPs、基因存在缺失以及Kmers (将全基因组序列切分成几十到上百bp的短片段)进行关联分析。首先,使用Mash软件[17]计算样本两两之间的遗传距离,用于校正种群结构。然后,分别从SNPs、基因以及Kmers三个水平,采用pyseer的固定效应模型,过滤掉最小等位基因频率(MAF,Minor Allele Frequency) < 5%的变异,利用多维尺度(multidimensional scaling,MDS)来评估每一个SNP、基因或Kmer对表型的影响,并计算相应P值。
2 结果和讨论 2.1 创伤弧菌种群结构及地理分布本研究共分析了260株创伤弧菌的全基因组序列,这些菌株分离自1977年–2018年间,覆盖4个大洲共10个国家。变异检测发现229462个SNP变异,其中103117个位于基因间区,124999个是同义SNPs (碱基突变不引起氨基酸编码改变),40165个是非同义SNPs (碱基突变引起氨基酸编码改变),1255个是无义SNPs(碱基突变引起氨基酸编码改变成终止密码子)。基于所有SNPs构建的系统发育树(图 1)表明:创伤弧菌可划分为5个系统发育谱系(L1、L2、L3、L4、L5),与过去报道一致[18];系统发育树内部分支较长且bootstraps值低,表明该物种具有较高的同源重组率。菌株集中分布于L1、L2谱系(占93.1%),地理分布呈现一定的地域聚集性:L1谱系主要包含中国分离株(占86.8%)且临床株占比高(62%);L2谱系主要包含美国分离株(占42.9%)且环境株占比高(62%)。相比L1谱系,L2谱系菌株的地理分布更为广泛,分离国家包括西班牙、法国、丹麦、日本、中国、德国、韩国、以色列和巴西等。目前L3、L4和L5谱系分离株总数很少,但临床株占比高(72.2%),因此值得在未来监测中加以关注。
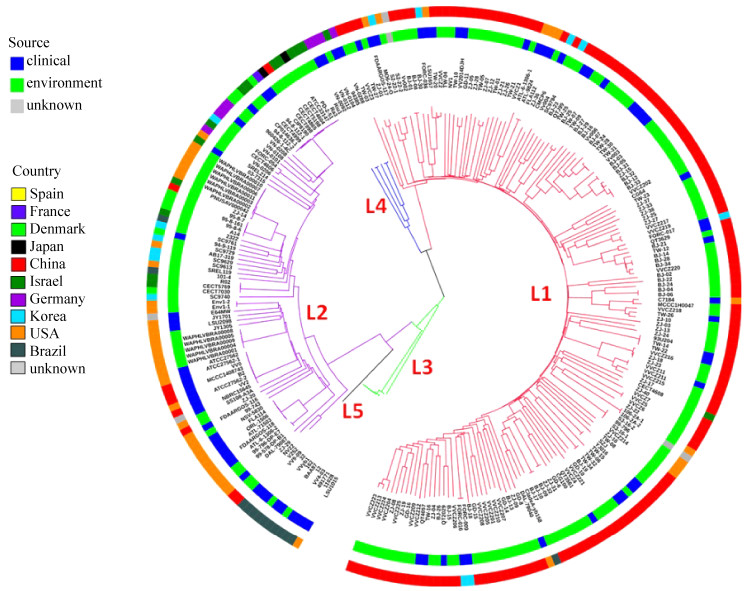
|
| 图 1 种群结构图 Figure 1 Population structure of V. vulnificus. A NJ tree of 260 V. vulnificus strains constructed based on 229462 SNPs. The ring colors from inner to outer indicate source of sampling and isolated location, respectively. Branch colors indicate different populations, pink for L1, purple for L2, green for L3, blue for L4, black for L5. |
2.2 已知毒力因子与创伤弧菌分离来源的关联分析
目前应用最为广泛的毒力因子数据库是刘波等在2005年建立的VFDB数据库,我们在该数据库的核心库(set A,仅包含经实验验证过的毒力基因)中,共找到226个存在等位基因的毒力因子。另外,我们通过文献调研找到56个其他的毒力因子。使用x2检验来计算282个已知毒力因子在创伤弧菌环境分离株和临床分离株之间的频率分布差异(https://doi.org/10.6084/m9.figshare.11760372.v1)。结果表明,仅有2个已知毒力因子pldA、vgrG在环境组和临床组中的分布有显著性差异(P < 0.05),因此,目前已知的大多数毒力因子无法解释创伤弧菌环境分离株与临床分离株之间的差异。
2.3 使用BGWAS方法鉴定毒力相关因子为全面挖掘创伤弧菌毒力相关元件,我们应用BGWAS方法,分别从SNPs、基因获得缺失和Kmers三个不同水平,在全基因组范围对260株创伤弧菌的分离来源表型与基因型进行关联分析,检测临床菌株中的特异性遗传元件。为降低假阳性率,我们结合先验知识,采用严格阈值对分析结果进行判定,即仅将统计检验P值低于已知毒力因子vgrG的遗传变异纳入最终结果[–log(P-value) > 10.4,图 2]。在该阈值下,BGWAS共鉴定出11个遗传变异位点,分别位于11个编码基因上,其中9个基因是本研究中首次发现与创伤弧菌毒力相关的遗传因子。全部11个基因中,有10个基因在SNPs和Kmers水平被同时检测到,提示统计检验结果是高度可靠的,值得注意的是,在基因获得缺失层面,未发现与分离来源相关的变异位点。这提示创伤弧菌的毒力主要与核心基因组相关,其附属基因组(包括基因组岛、质粒等)对其影响较弱。

|
| 图 2 菌株来源(环境/临床)与菌株SNPs/Gene/Kmers位点BGWAS分析的Manhattan图 Figure 2 Manhattan plots of BGWAS results for V. vulnificus. The X-axis of panel A and B separately indicate the position of chromosomeⅠand the chromosomeⅡof the reference genome (V. vulnificus CECT4999), and Y-axis is the –log10(P-value) score of BGWAS. Dash lines indicate the threshold used ot identify the significant results. Grey dots, the –log10(P-value) of the variation sites at the corresponding position of the chromosomes; Black diamond, gene that carried significant Kmers; Black dots, genes that carried both significant Kmers and SNPs detected in current study. |
2.4 显著相关基因的功能
我们对鉴定出的11个统计显著基因进行了注释和解读(表 1)。pldA和vgrG基因已被报道与创伤弧菌致病性相关:pldA基因编码磷脂酶A1,通过研究创伤弧菌细胞溶血素阴性突变株CVD707,发现磷脂酶A1是创伤弧菌的主要溶血和致死因子[19];vgrG基因是六型分泌系统(Type VI secretion system,T6SS)蛋白编码基因,在细菌感染宿主过程中发挥重要作用[20–21],该基因通过肌动蛋白交联域(actin cross-linking domains)发挥作用,以影响创伤弧菌毒力,在大肠杆菌和霍乱弧菌中也有相同的功能[22]。本研究新发现的9个基因可能以直接或间接方式影响创伤弧菌的致病能力。其中2个基因编码代谢蛋白:aroQ基因编码3-脱氢奎尼酸脱水酶,参与苯丙氨酸的合成,苯丙氨酸的寡聚作用可影响创伤弧菌毒力因子溶血素的细胞裂解活性和凋亡活性[23];glpA基因编码3-磷酸甘油醛脱氢酶亚基A,3-磷酸甘油醛脱氢酶(GAPDH)可修复DNA、调节组蛋白、促进细胞凋亡[24]、参与细菌黏附宿主组织、激发宿主保护性免疫反应[25],进而影响创伤弧菌毒力。5个基因编码结构蛋白:gspE基因是Ⅱ型分泌系统(Type Ⅱ Secretion System,T2SS)蛋白编码基因,T2SS是广泛分布于革兰氏阴性细菌中的蛋白质分泌系统,创伤弧菌T2SS可将多种蛋白转运到菌体细胞外,直接与宿主细胞发生相互作用而发挥其毒性[26];tolQ基因编码TolQ蛋白,与TolR、TolA和TolB可构成Tol系统,通过影响革兰氏阴性菌的生化和代谢机制,增强细菌毒性,在爱德华氏菌、小肠沙门氏菌、大肠杆菌、杜氏嗜血杆菌和霍乱弧菌中也有相同功能[27–28];flgJ、flgH、fliF基因编码鞭毛蛋白,是创伤弧菌鞭毛的组成成分,与细菌的侵袭、黏附能力和致病性有关[29]。2个基因编码信息传递蛋白:VVCECT4999_03045基因编码胰岛素酶M16,胰岛素酶可通过调控宿主内分泌信号为细菌提供有利生存条件,促进创伤弧菌的体内存活能力[30];tpbB基因编码二鸟苷酸环化酶,二鸟苷酸环化酶含有GGDEF结构域的蛋白质,可参与调控因子c-di-GMP的合成和降解,干扰创伤弧菌生物膜形成,影响粘附因子、运动性和毒力基因的表达[31],进而影响创伤弧菌毒力。
| Genes | Function | Supported variations (number) |
| pldA | Phospholipase A1 | K-mers (3), SNP (2) |
| vgrG | Type Ⅵ secretion system protein VgrG | K-mers (53), SNP (3) |
| aroQ | 3-dehydroquinate dehydratase | K-mers (2) |
| glpA | Glycerol-3-phosphate dehydrogenase subunit A | K-mers (2), SNP (3) |
| gspE | Type Ⅱ secretion system protein GspE | K-mers (2), SNP (3) |
| flgJ | Flagellar assembly peptidoglycan hydrolase FlgJ | K-mers (76), SNP (3) |
| flgH | Flagellar L-ring protein | K-mers (11), SNP (1) |
| fliF | Flagellar basal body M-ring protein FliF | K-mers (10), SNP (1) |
| tpbB | Diguanylate cyclase | K-mers (48), SNP (2) |
| tolQ | Protein TolQ | K-mers (8), SNP (2) |
| VVCECT4999_03045 | Insulinase M16 | K-mers (100), SNP (7) |
3 结论
本研究结合高通量测序技术和群体遗传学方法,对260株创伤弧菌分离株进行了系统发育重建和毒力相关因子挖掘研究。研究结果表明创伤弧菌由5个系统发育谱系组成,谱系与菌株地域分布存在关联:其中中国菌株主要聚集在L1分支,美国菌株主要聚集在L2分支。BGWAS分析发现11个遗传因子与临床或环境分离来源具有强相关性,表明这些基因可能与创伤弧菌临床毒力或致病性相关。其中9个基因的毒力相关性在本研究首次得到证明,为下一步深入探索创伤弧菌致病机制、设计新的疫苗或者药物提供了研究靶标。
| [1] |
Wang MY, Hu CJ. Pathogenicity and virulence factors of Vibrio vulnificus:research advances. Chinese Journal of Microecology, 2017, 29(12): 1470-1473.
(in Chinese) 王明义, 胡成进. 创伤弧菌致病性及其毒力因子研究进展. 中国微生态学杂志, 2017, 29(12): 1470-1473. |
| [2] | Baker-Austin C, Oliver JD. Vibrio vulnificus:new insights into a deadly opportunistic pathogen. Environmental Microbiology, 2018, 20(2): 423-430. DOI:10.1111/1462-2920.13955 |
| [3] | Hoffmann S, Maculloch B, Batz M. Economic burden of major foodborne illnesses acquired in the United States. US: US Department of Agriculture, Economic Research Service, 2015. |
| [4] | Yun NR, Kim DM. Vibrio vulnificus infection:a persistent threat to public health. The Korean Journal of Internal Medicine, 2018, 33(6): 1070-1078. DOI:10.3904/kjim.2018.159 |
| [5] |
He SB, Qu YP, Yu XQ, Cao J, Ye CR. Research status and prospect of genome-wide association study. Shanghai Journal of Animal Husbandry and Veterinary Medicine, 2017(3): 12-15.
(in Chinese) 何随彬, 瞿瑜萍, 俞向前, 曹杰, 叶承荣. 全基因组关联分析研究现状及展望. 上海畜牧兽医通讯, 2017(3): 12-15. |
| [6] | Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies:lessons from human GWAS. Nature Reviews Genetics, 2017, 18(1): 41-50. DOI:10.1038/nrg.2016.132 |
| [7] | Wang SB, Feng JY, Ren WL, Huang B, Zhou L, Wen YJ, Zhang J, Dunwell JM, Xu SZ, Zhang YM. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Scientific Reports, 2016, 6(1): 19444. DOI:10.1038/srep19444 |
| [8] | Xiao YJ, Liu HJ, Wu LJ, Warburton M, Yan JB. Genome-wide association studies in maize:praise and stargaze. Molecular Plant, 2017, 10(3): 359-374. DOI:10.1016/j.molp.2016.12.008 |
| [9] |
Yang C, Yang RF, Cui YJ. Bacterial genome-wide association study:methodologies and applications. Hereditas (Beijing), 2018, 40(1): 57-65.
(in Chinese) 杨超, 杨瑞馥, 崔玉军. 细菌全基因组关联研究的方法与应用. 遗传, 2018, 40(1): 57-65. |
| [10] | Cui YJ, Yu C, Yan YF, Li DF, Li YJ, Jombart T, Weinert LA, Wang ZY, Guo ZB, Xu LZ, Zhang YJ, Zheng HC, Qin N, Xiao X, Wu MS, Wang XY, Zhou DS, Qi ZZ, Du ZM, Wu HL, Yang XW, Cao HZ, Wang H, Wang J, Yao SS, Rakin A, Li YR, Falush D, Balloux F, Achtman M, Song YJ, Wang J, Yang RF. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(2): 577-582. DOI:10.1073/pnas.1205750110 |
| [11] | Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Current Protocols in Bioinformatics, 2003(1): 10. |
| [12] | Seemann T. Prokka:rapid prokaryotic genome annotation. Bioinformatics, 2014, 30(14): 2068-2069. DOI:10.1093/bioinformatics/btu153 |
| [13] | Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. Roary:rapid large-scale prokaryote pan genome analysis. Bioinformatics, 2015, 31(22): 3691-3693. DOI:10.1093/bioinformatics/btv421 |
| [14] | Letunic I, Bork P. Interactive tree of life (iTOL) v3:an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research, 2016, 44(W1): W242-W245. DOI:10.1093/nar/gkw290 |
| [15] | Liu B, Zheng DD, Jin Q, Chen LH, Yang J. VFDB 2019:a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Research, 2018, 47(D1): D687-D692. |
| [16] | Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. Pyseer:a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics, 2018, 34(24): 4310-4312. DOI:10.1093/bioinformatics/bty539 |
| [17] | Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. Mash:fast genome and metagenome distance estimation using MinHash. Genome Biology, 2016, 17(1): 132. DOI:10.1186/s13059-016-0997-x |
| [18] | Roig FJ, González-Candelas F, Sanjuán E, Fouz B, Feil EJ, Llorens C, Baker-Austin C, Oliver JD, Danin-Poleg Y, Gibas CJ, Kashi Y, Gulig PA, Morrison SS, Amaro C. Phylogeny of Vibrio vulnificus from the analysis of the core-genome:implications for intra-species taxonomy. Frontiers in Microbiology, 2018, 8: 2613. DOI:10.3389/fmicb.2017.02613 |
| [19] | Koo BS, Lee JH, Kim SC, Yoon HY, Kim KA, Kwon KB, Kim HR, Park JW, Park BH. Phospholipase A as a potent virulence factor of Vibrio vulnificus. International Journal of Molecular Medicine, 2007, 20(6): 913-918. |
| [20] |
Liu X, Wang SH, Meng QM, Han XG, Xu X, Yang DH, Ding C, Peng DX, Yu SQ. Effects of type Ⅵ secretion system 2 core component VgrG on the pathogenicity of avian Escherichia coli. Microbiology China, 2016, 43(9): 2106-2113.
(in Chinese) 刘新, 王少辉, 孟庆美, 韩先干, 许漩, 杨登辉, 丁铲, 彭大新, 于圣青. Ⅵ型分泌系统2核心组分VgrG对禽致病性大肠杆菌致病性的影响. 微生物学通报, 2016, 43(9): 2106-2113. |
| [21] | Church SR, Lux T, Baker-Austin C, Buddington SP, Michell SL. Vibrio vulnificus type 6 secretion system 1 contains anti-bacterial properties. PLoS One, 2016, 11(10): e0165500. DOI:10.1371/journal.pone.0165500 |
| [22] | Kudryashova E, Heisler DB, Kudryashov DS. Pathogenic mechanisms of actin cross-linking toxins: peeling away the layers//Mannherz HG. The Actin Cytoskeleton and Bacterial Infection. Cham: Springer, 2016: 87-112. |
| [23] | Kashimoto T, Ueno S, Ehara H, Fukudome S, Komai M, Susa N. Oligomerization is essential for apoptotic activity of Vibrio vulnificus hemolysin. Journal of Veterinary Medical Science, 2009, 71(10): 1403-1406. DOI:10.1292/jvms.001403 |
| [24] | Sukhanov S, Hou XW, Snarski P, Yoshida T, Delafontaine P. Smooth muscle specific glyceraldehyde-3'-phosphate dehydrogenase (GAPDH) reduces DNA damage, decreases cell apoptosis, suppresses atherosclerosis and promotes the stable plaque phenotype. The FASEB Journal, 2019, 33(S1): 522. |
| [25] | Rosinha GMS, Myioshi A, Azevedo V, Splitter GA, Oliveira SC. Molecular and immunological characterisation of recombinant Brucella abortus glyceraldehyde-3-phosphate-dehydrogenase, a T-and B-cell reactive protein that induces partial protection when co-administered with an interleukin-12-expressing plasmid in a DNA vaccine formulation. Journal of Medical Microbiology, 2002, 51(8): 661-671. DOI:10.1099/0022-1317-51-8-661 |
| [26] |
Ou P. Progress in Bacterium Type Ⅱ Secretion System. Journal of Hezhou University, 2007, 23(3): 137-140.
(in Chinese) 欧平. 细菌Ⅱ型分泌系统的研究进展. 贺州学院学报, 2007, 23(3): 137-140. DOI:10.3969/j.issn.1673-8861.2007.03.034 |
| [27] | Abdelhamed H, Lu JJ, Lawrence ML, Karsi A. Involvement of tolQ and tolR genes in Edwardsiella ictaluri virulence. Microbial Pathogenesis, 2016, 100: 90-94. DOI:10.1016/j.micpath.2016.09.011 |
| [28] | Song J, Hou HM, Wu HY, Li KX, Wang Y, Zhou QQ, Zhang GL. Transcriptomic analysis of Vibrio parahaemolyticus reveals different virulence gene expression in response to benzyl isothiocyanate. Molecules, 2019, 24(4): 761. DOI:10.3390/molecules24040761 |
| [29] | Phippen BL, Oliver JD, Payne SM. Role of Anaerobiosis in capsule production and biofilm formation in Vibrio vulnificus. Infection and Immunity, 2015, 83(2): 551-559. DOI:10.1128/IAI.02559-14 |
| [30] | Kim IH, Kim IJ, Wen YC, Park NY, Park JY, Lee KW, Koh A, Lee JH, Koo SH, Kim KS. Vibrio vulnificus secretes an insulin-degrading enzyme that promotes bacterial proliferation in vivo. Journal of Biological Chemistry, 2015, 290(30): 18708-18720. DOI:10.1074/jbc.M115.656306 |
| [31] | Nakhamchik A, Wilde C, Rowe-Magnus DA. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Applied and Environmental Microbiology, 2008, 74(13): 4199-4209. DOI:10.1128/AEM.00176-08 |
 2020, Vol. 60
2020, Vol. 60