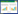中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 陈华枝, 熊翠玲, 祝智威, 王杰, 范小雪, 蒋海宾, 范元婵, 万洁琦, 卢家轩, 郑燕珍, 付中民, 徐国钧, 陈大福, 郭睿. 2020
- Chen Huazhi, Xiong Cuiling, Zhu Zhiwei, Wang Jie, Fan Xiaoxue, Jiang Haibin, Fan Yuanchan, Wan Jieqi, Lu Jiaxuan, Zheng Yanzhen, Fu Zhongmin, Xu Guojun, Chen Dafu, Guo Rui. 2020
- 基于small RNA组学分析揭示意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答机制
- Unraveling the mechanism underlying the immune responses of Apis mellifera ligustica to Nosema ceranae stress based on small RNA omics analyses
- 微生物学报, 60(7): 1458-1478
- Acta Microbiologica Sinica, 60(7): 1458-1478
-
文章历史
- 收稿日期:2019-10-12
- 修回日期:2019-12-24
- 网络出版日期:2020-05-19
意大利蜜蜂(Apis mellifera ligustica,简称意蜂)作为西方蜜蜂(Apis mellifera)的亚种之一,因具有采蜜量大、不易逃群和性情温顺等优良特性而广泛用于我国的养蜂生产,发挥着重要的经济价值和不可替代的生态价值[1]。东方蜜蜂微孢子虫(Nosema ceranae)是一种单细胞真菌病原,广泛感染世界各地的蜂群[2]。N. ceranae特异性侵染成年蜜蜂的中肠上皮细胞[3],可导致宿主寿命缩短[4–5]、能量胁迫[6–7]和免疫抑制[8–10]。接种感染N. ceranae 7 d的西方蜜蜂工蜂的累计死亡率显著上升[5];接种感染N. ceranae 14 d的西方蜜蜂工蜂死亡率接近100%[4]。Mayack等对未感染和感染N. ceranae的西方蜜蜂工蜂进行不同浓度蔗糖饲喂并测试其伸吻反应,发现N. ceranae感染的工蜂对梯度浓度蔗糖溶液的敏感程度和蔗糖溶液的消耗量均显著高于未感染组的工蜂[6]。近期,有研究报道N. ceranae对西方蜜蜂幼虫也具有侵染性[11–12]。
西方蜜蜂基因组信息[13]于2006年公布。Evans等基于该参考基因组,通过种间同源性比较分析发现西方蜜蜂存在Toll、Imd、JAK/STAT和JNK等4条免疫相关通路及6个抗菌肽编码基因,但相比于果蝇和按蚊,西方蜜蜂含有的免疫基因(涉及17个基因家族)数量下降了三分之二,表明西方蜜蜂的个体免疫能力有所下降,而群体防御在抵御病原入侵方面具有较高的重要性,因此西方蜜蜂具有被一些协同进化的专性病原感染的倾向性[14]。此后,人们利用分子生物学手段对西方蜜蜂响应N. ceranae的免疫应答进行了较多研究,发现N. ceranae能够通过抑制宿主的抗菌肽基因、细胞免疫相关基因的表达显著影响西方蜜蜂的免疫应答[8, 15];但也有研究表明N. ceranae会激活上述免疫基因的表达[16]。Li等研究发现N. ceranae侵染可导致西方蜜蜂naked cuticle基因上调表达,沉默naked cuticle基因会造成抗菌肽基因的上调、蜜蜂寿命延长且宿主肠道内病原孢子数量下降[17]。近年来,随着高通量测序技术的发展和普及,人们在mRNA组学层面对西方蜜蜂响应N. ceranae侵染的胁迫应答也进行了一些研究[10, 18],但总体仍较为有限。
微小RNA (microRNA, miRNA)是一类长度约23 nt的非编码RNA (non-coding RNA, ncRNA),其种子序列通过靶向完全或不完全匹配结合靶mRNA的3′ UTR区抑制或降解mRNA,从而调控基因的表达[19–21]。miRNA已被证明广泛参与动物、植物和微生物的众多生物学过程[22]。前人通过生物信息学分析对西方蜜蜂的miRNA进行了一些研究,涉及蜜蜂级型分化[23]、神经发育[24]和免疫应答[25]。Ashby等的研究结果表明,西方蜜蜂蜂王、雄峰和工蜂的miRNA表达谱在幼虫发育成蛹阶段具有显著差异,蛹期的miRNA可能参与调控西方蜜蜂的级型分化[23]。Huang等利用二代测序和生物信息学探究了N. ceranae感染1–6 d的西方蜜蜂工蜂miRNA的胁迫应答,发现宿主差异表达的miRNA (differentially expressed miRNA,DEmiRNA)可能通过参与调控物质代谢、能量代谢和酶类相关基因的表达,应对N. ceranae的侵染[26];该团队在此基础上进一步研究发现,miRNA可能介导N. ceranae与意蜂之间的跨界调控[27]。
目前,关于miRNA等ncRNA在蜜蜂响应和抵御N. ceranae侵染过程中的作用,相关研究仍然非常滞后。笔者团队前期已结合链特异性建库的RNA-seq技术对正常和N. ceranae胁迫的意蜂工蜂中肠进行测序,系统解析了意蜂工蜂响应N. ceranae胁迫的高表达基因(highly expressed gene,HEG)表达谱及HEG的潜在作用[28];揭示了宿主响应N. ceranae胁迫的基因差异表达谱和免疫应答[29];并解析了宿主响应胁迫的长链非编码RNA (long non-coding RNA,lncRNA)差异表达谱及竞争性内源RNA (competing endogenous RNA,ceRNA)调控网络[5]。对于蜜蜂响应N. ceranae胁迫的免疫应答机制,仍缺乏分子生物学水平的深入阐释。本研究利用small RNA-seq (sRNA-seq)技术对正常和N. ceranae胁迫7 d和10 d的意蜂工蜂中肠进行测序,通过深入分析宿主响应胁迫的DEmiRNA及其调控网络,以及宿主的细胞和体液免疫相关通路及富集mRNA,在miRNA组学层面揭示意蜂工蜂对N. ceranae的免疫应答机制,为阐明背后的分子机制提供必要的参考信息和数据基础。
1 材料和方法 1.1 供试昆虫和真菌供试意蜂工蜂取自福建农林大学动物科学学院(蜂学学院)教学蜂场,N. ceranae孢子[30]由动物科学学院(蜂学学院)蜜蜂保护实验室分离、纯化和保存。
1.2 意蜂工蜂的接种和人工饲养参照笔者团队前期已建立的方法[5]进行意蜂工蜂的接种和人工饲养,简述如下:(1)挑选外观健康、群势较强的6群意蜂,在每个蜂箱巢门口随机抓取6只工蜂,拉取中肠进行研磨,取少量研磨液进行显微制片和观察,对镜检不到孢子的样品进行PCR检测,N. ceranae特异性引物为Nc-F: CGGCGACGATGTGATATGAAAATATTAA, Nc-R: CCCGGTCATTCTCAAACAAAAAACCG, N. apis特异性引物Na-F: GGGGGCATGTCTTTG ACGTACTATGTA, Na-R: GGGGGGCGTTTAAAA TGTGAAACAACTATG[31],将PCR检测结果为阴性的意蜂蜂群作为本研究实验蜂群;(2)选取实验蜂群中封盖子较多的巢脾放入(34.0±0.5) ℃恒温培养箱,将刚出房的工蜂放入四周打孔的干净塑料盒(30头/盒),每个盒子上方插入一支装有50% (W/V)无菌糖水的饲喂器;(3) (34.0±0.5) ℃培养24 h,将上述工蜂饥饿处理2 h,然后给对照组的每只工蜂饲喂5 μL糖水(不含N. ceranae孢子),处理组的每只工蜂饲喂5 μL糖水(含1×106个N. ceranae孢子[5]),舍弃未食尽糖水的工蜂,将食尽糖水的工蜂用于后续实验,接种当天记为接种后0 d;(4)每日检查工蜂的存活情况,及时清理死亡的工蜂,每隔24 h更换糖水。本实验进行3次生物学重复。
1.3 测序样品制备及sRNA-seq测序蜜蜂中肠是N. ceranae寄生及其与宿主相互作用的主要部位,因而本研究选取意蜂工蜂中肠作为测序样品。N. ceranae在西方蜜蜂中肠上皮细胞中完成一轮生活史的周期为6 d[32],随后病原孢子数量可持续增长到20 d[33]。综合考虑,本研究选取正常和N. ceranae感染7 d和10 d的工蜂的中肠进行测序。分别拉取正常和N. ceranae感染7 d和10 d的意蜂工蜂中肠,每3只置于1个RNA-Free的EP管,立即放入液氮速冻,每次取样时间均控制在20 s以内。7 d对照组样品为Am7CK:Am7CK1、Am7CK2、Am7CK3;7 d处理组样品为Am7T:Am7T1、Am7T2、Am7T3;10 d对照组样品为Am10CK:Am10CK1、Am10CK2、Am10CK3;10 d处理组样品为Am10T:Am10T1、Am10T2、Am10T3。待取完全部样品,统一转移至–80 ℃超低温冰箱保存备用。利用Trizol法分别提取上述样品的总RNA,琼脂糖凝胶电泳切胶选择18–30 nt的片段。继而连接3′接头,连接产物以15%变性PAGE胶电泳分离,切胶选择36–44 nt目的条带;回收切胶产物,连接5′接头,然后对连接了两侧接头的小RNA样本进行反转录PCR。反转录产物以3.5%琼脂糖凝胶电泳分离,切胶回收140–160 bp区域条带,即为终文库。委托广州基迪奥生物技术有限公司对构建好的文库进行测序,测序平台Illumina HiSeqTM 2000。sRNA-seq的原始数据已上传NCBI SRA数据库,BioProject号:PRJNA408312。
1.4 测序数据质控参照郭睿等的方法[34]对测序数据进行过滤和质控,简述如下:(1)剔除原始读段(raw reads)中含5′接头序列、含polyA、低质量的reads和剪切掉3′接头序列后的短于18或长于30个核苷酸的序列,得到高质量的有效序列标签(clean tags);(2)利用Bowite软件[35]将获得的clean tags比对GenBank及Rfam (11.0)数据库,过滤比对上rRNA、scRNA、snoRNA和tRNA的clean tags,得到未注释的tags (unannotated tags);(3)比对N. ceranae参考基因组(assembly ASM98816v1) (https://www.ncbi.nlm.nih.gov/genome/931?genome_assembly_id=230435),去除比对上的数据(即为N. ceranae的数据);(4)将剩余数据继续比对西方蜜蜂参考基因组(assembly Amel_4.5) (http://www.ncbi.nlm.nih.gov/genome/48?genome_assembly_id=22683),剔除比对上基因组外显子、内含子和重复序列的clean tags,剩余比对上的数据(mapped tags)可用于后续分析。
1.5 miRNA的预测及差异分析利用miRDeep2软件[36]将上述剩余的mapped tags与miRBase数据库中收录的miRNA前体序列进行比对,获得已知miRNA序列。同时,将未比对上的tags比对基因组,得到可能的前体序列,根据tags在前体序列上的分布信息和前体结构能量信息,采用贝叶斯模型经打分实现novel miRNA的鉴定。利用TPM (tags per million)算法公式(TPM=T{ QUOTE 106/N,T表示单个miRNA的tags,N表示总miRNA的tags)对miRNA的表达量进行归一化。筛选DEmiRNA的标准:|log2(Fold change)|≥1,P{ QUOTE
≤0.05。上述筛选方法已广泛用于其他物种的miRNA组学研究[37–39]。利用OmicShare在线工具集合(www.omicshare.com)计算Pearson相关系数并作图,以及绘制DEmiRNA的火山图和热图,相关软件采用默认参数。
根据miRNA与西方蜜蜂参考基因组序列信息,利用TargetFinder软件[40]进行靶mRNA预测。将预测到的靶mRNA映射到GO和KEGG数据库,统计比对上GO条目或KEGG通路(pathway)的mRNA数。此外,利用Cytoscape软件[41]将DEmiRNA及其靶向结合的-免疫通路相关的差异表达mRNA (differentially expressed mRNA,DEmRNA)[29]的结合关系进行可视化。
1.7 Novel miRNA的Stem-loop RT-PCR验证参照Chen等[42]和郭睿等[34]的方法,随机挑选13个novel miRNA (novel-m0008-3p、novel-m0019-5p、novel-m0003-3p、novel-m0018-5p、novel-m0006-3p、novel-m0005-3p、novel-m0043-5p、novel-m0022-3p、novel-m0033-3p、novel-m0040-3p、novel-m0026-5p、novel-m0031-3p、novel-m0001-3p)进行表达验证,利用DNAMAN 8软件设计novel miRNA的Stem-loop引物、特异性上游和通用下游引物,委托生工生物工程(上海)股份有限公司合成,引物信息详见表 1。利用RNA抽提试剂盒(Promega公司,美国)提取Am7CK、Am7T、Am10CK和Am10T样品的总RNA,利用Stem-loop引物反转录得到相应的cDNA预混,作为PCR的模板。PCR反应体系(20 μL)包括cDNA 1 μL,上下游引物各1 μL,dH2O 7 μL,PCR mix 10 μL;PCR程序设置:94 ℃ 5 min;94 ℃ 30 s,50 ℃ 40 s,36个循环;72 ℃ 1 min。PCR产物经1.8%琼脂糖凝胶电泳检测,利用凝胶成像系统(上海培清,中国)观察和拍照。
| Primer name | Primer sequence (5′→3′) |
| novel-m0008-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCATCCTCC |
| novel-m0019-5p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATCTCGGA |
| novel-m0003-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACAACGG |
| novel-m0018-5p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCTACTG |
| novel-m0006-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCTCCCA |
| novel-m0005-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTAGGATG |
| novel-m0043-5p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGACGAAT |
| novel-m0022-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATCCTCTC |
| novel-m0033-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGATACC |
| novel-m0040-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCTAGCAC |
| novel-m0026-5p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCATCAG |
| novel-m0031-3p-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGAGAAAT |
| miR-8271-y-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCTATCT |
| miR-1-z-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATACATAC |
| miR-374-y-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATTACAA |
| miR-128-y-loop | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAGAGAC |
| miR-8271-y-F | GCGCGCGTATACTGAAT |
| miR-1-z-F | GCGCGCGTGGAATGTAAAGAA |
| miR-374-y-F | GCGCGCGCTTATCAGATTGTA |
| miR-128-y-F | GCGCGCGTCACAGTGAACCG |
| novel-m0008-3p-F | AGGGGATGGGAG |
| novel-m0019-5p-F | TGCAGGCGTGCAGA |
| novel-m0003-3p-F | TTTGAACAATTGTAC |
| novel-m0018-5p-F | GGCCCATTAGCT |
| novel-m0006-3p-F | CACAACCACTCTGA |
| novel-m0005-3p-F | ACACTCCAGCTGGGTTGACTATGATT |
| novel-m0043-5p-F | ACACTCCAGCTGGGTTAATTAAATACAT |
| novel-m0022-3p-F | ACACTCCAGCTGGGCGTGTCGAGGACA |
| novel-m0033-3p-F | ACACTCCAGCTGGGATATACCATTCGTC |
| novel-m0040-3p-F | ACACTCCAGCTGGGTCGGTTTTAAAGTCT |
| novel-m0026-5p-F | ACACTCCAGCTGGGTATCCACGATCATAA |
| novel-m0031-3p-F | ACACTCCAGCTGGGTGCGCGATCGATA |
| Universal R | CTCAACTGGTGTCGTGGA |
| U6-F | GTTAGGCTTTGACGATTTCG |
| U6-R | GGCATTTCTCCACCAGGTA |
1.8 DEmiRNA的qPCR验证
利用DNAMAN 8软件设计DEmiRNA的Stem-loop引物、特异性上游引物、通用下游引物,以及意蜂U6基因(DEmiRNA的内参)的特异性引物,委托生工生物工程(上海)股份有限公司合成。引物信息详见表 1。利用RNA抽提试剂盒(Promega公司,美国)分别提取Am7CK、Am7T、Am10CK和Am10T样品的总RNA,利用Stem-loop引物和随机引物反转录得到相应的cDNA分别作为DEmiRNA和U6基因的qPCR模板。qPCR反应体系(20 μL):cDNA 1 μL,上下游引物各1 μL,RNA-Free H2O 7 μL,SYBR Green Dye (上海翊圣,中国) 10 μL;ABI QuanStudio 3荧光定量PCR仪(ABI公司,美国)的qPCR程序设置:94 ℃ 5 min;94 ℃ 30 s,56 ℃ 40 s,40个循环。RT-qPCR每个反应进行3次平行重复和生物学重复。采用2–∆∆Ct法计算DEmiRNA的相对表达量,利用GraphPad Prism 7软件进行Student’s t-test检验和绘图。
2 结果和分析 2.1 测序数据质控与评估上述12组意蜂工蜂中肠样品共测得165895574条raw reads,经严格过滤和质控共得到132028990条clean tags,每组样品的clean tags占raw reads的比例都达到77.04%及以上(表 2)。此外,Am7CK、Am10CK、Am7T和Am10T的组内平均Pearson相关性分别达到87.92%、99.93%、91.14%和99.67% (图 1)。以上结果表明测序数据质量和样品重复性良好。
| Samples | Raw reads | Clean tags |
| Am7CK1 | 14021728 | 11355712 (83.38%) |
| Am7CK2 | 13479635 | 10077417 (77.04%) |
| Am7CK3 | 13033478 | 10828352 (85.55%) |
| Am7T1 | 13731512 | 10764025 (80.66%) |
| Am7T2 | 13195285 | 10728197 (83.57%) |
| Am7T3 | 14332490 | 11456414 (82.16%) |
| Am10CK1 | 13083648 | 10127041 (80.51%) |
| Am10CK2 | 13544596 | 10710261 (81.52%) |
| Am10CK3 | 13716280 | 11019319 (82.75%) |
| Am10T1 | 14346575 | 12065860 (87.31%) |
| Am10T2 | 14861853 | 11591319 (80.99%) |
| Am10T3 | 14548494 | 11305073 (80.68%) |
|
| 图 1 各组不同生物学重复间的Pearson相关性 Figure 1 Pearson correlation coefficients among various biological replicas within every groups. |
2.2 意蜂工蜂中肠miRNA的鉴定与分析
利用miRDeep2软件共预测出984个miRNA,包含928个已知miRNA和56个新miRNA;它们长度介于18–28 nt,大部分的miRNA的长度为18 nt (263,27%)和22 nt (264,27%) (图 2-A)。此外,不同长度的miRNA的首位碱基偏向性存在明显差异,19–25 nt的miRNA首位碱基以U为主,18 nt和26 nt的miRNA首位碱基以A为主,28 nt的miRNA首位碱基以G为主(图 2-B)。从上述56个新miRNA中随机挑选了13个novel miRNA进行Stem-loop RT-PCR,琼脂糖凝胶电泳结果显示其中12个(92.31%)均能扩增出符合预期的特异性条带(图 3)。

|
| 图 2 意蜂工蜂中肠miRNA的结构特征 Figure 2 Structural properties of miRNA in the midgut of A. m. ligustica worker. A: Length distribution of miRNA; B: First base bias of miRNA. |

|
| 图 3 意蜂工蜂中肠新miRNA的Stem-loop RT-PCR鉴定 Figure 3 Stem-loop RT-PCR identification of novel miRNAs in A. m. ligustica worker's midgut. |
2.3 意蜂工蜂中肠DEmiRNA的预测和分析
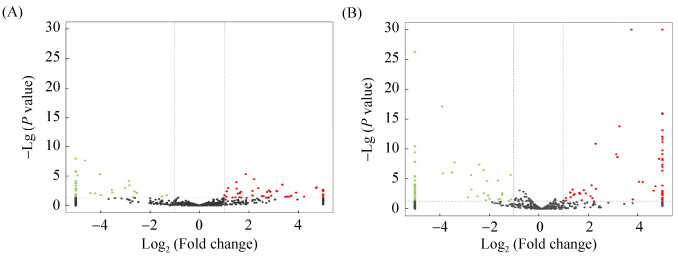
结果显示,Am7CK vs. Am7T比较组中共有84个DEmiRNA,包括48个上调miRNA和36个下调miRNA (图 4-A);Am10CK vs. Am10T比较组中共有107个DEmiRNA,包括56个上调miRNA和51个下调miRNA (图 4-B)。Am7CK vs. Am7T中上调幅度最大的为miR-9075-y[log2(Fold change)=12.29],其次是miR-2188-x[log2(Fold change)=12.21]和ame-miR-6052[log2(Fold change)=11.73],下调幅度最大的为miR-8191-x[log2(Fold change)=–14.22],其次是miR-298-x[log2(Fold change)=–13.64]和miR-434-y[log2(Fold change)=–13.48] (表 3)。Am10CK vs. Am10T比较组中上调幅度最大的为miR-8503-x[log2(Fold change)=15.97],其次是miR-2965-y[log2(Fold change)=15.21]和miR-4700-x [log2(Fold change)=14.75],下调幅度最大的为miR-462-x[log2(Fold change)=–16.42],其次是miR-142-y [log2(Fold change)=–14.75]和miR-144-y [log2(Fold change)=–14.51] (表 4)。
|
| 图 4 各比较组中DEmiRNA的火山图 Figure 4 Volcano plots of DEmiRNAs in every comparison groups. A: Volcano plot of DEmiRNAs in Am7CK vs. Am7T; B: Volcano plot of DEmiRNAs in Am10CK vs. Am10T. Green dots indicate downregulated miRNAs and red dots indicate upregulated miRNAs. |
| miRNA name | TPM in Am7CK group | TPM in Am7T group | Log2(Fold change) | P value |
| Up-regulated miRNAs | ||||
| miR-9075-y | 0.001 | 5.020 | 12.293 | 0.003 |
| miR-2188-x | 0.001 | 4.740 | 12.211 | 0.021 |
| ame-miR-6052 | 0.001 | 3.390 | 11.727 | 0.002 |
| miR-1983-y | 0.001 | 3.040 | 11.570 | 0.004 |
| miR-8212-y | 0.001 | 2.833 | 11.468 | 0.004 |
| miR-224-x | 0.001 | 2.327 | 11.184 | 0.013 |
| miR-92-x | 0.001 | 1.647 | 10.685 | 0.023 |
| miR-501-y | 0.001 | 1.273 | 10.314 | 0.047 |
| miR-374-y | 0.203 | 5.397 | 4.730 | 0.001 |
| miR-767-x | 0.203 | 5.347 | 4.717 | 0.001 |
| Down-regulated miRNAs | ||||
| miR-8191-x | 19.120 | 0.001 | –14.223 | 0.000 |
| miR-298-x | 12.743 | 0.001 | –13.637 | 0.000 |
| miR-434-y | 11.440 | 0.001 | –13.482 | 0.000 |
| miR-541-x | 10.293 | 0.001 | –13.329 | 0.000 |
| miR-5100-y | 9.750 | 0.001 | –13.251 | 0.000 |
| miR-291-y | 7.853 | 0.001 | –12.939 | 0.000 |
| miR-292-y | 6.880 | 0.001 | –12.748 | 0.001 |
| miR-4700-x | 5.990 | 0.001 | –12.548 | 0.022 |
| miR-293-y | 4.937 | 0.001 | –12.269 | 0.003 |
| miR-381-y | 4.377 | 0.001 | –12.096 | 0.001 |
2.4 意蜂工蜂中肠DEmiRNA靶mRNA的预测和分析
Am7CK vs. Am7T比较组包含的84个DEmiRNA共预测出由4003个基因转录形成的9827个靶mRNA,它们富集在50条功能条目(图 5-A),涉及19条与细胞组分相关条目,例如细胞(1189个mRNA)和细胞组件(1189个mRNA);11条与分子功能相关条目,例如结合(2727个mRNA)和催化活性(1794个mRNA);20条生物学进程相关条目,例如细胞进程(2617个mRNA)和代谢进程(2276个mRNA)。

|
| 图 5 各比较组中DEmiRNA靶mRNA的GO分类 Figure 5 GO classifications of DEmiRNA target mRNAs in every comparison groups. A: GO classifications of DEmiRNA target mRNAs in Am7CK vs. Am7T; B: GO classifications of DEmiRNA target mRNAs in Am10CK vs. Am10T. |
Am10CK vs. Am10T比较组包含的107个DEmiRNA共预测出由4301个基因转录形成的10720个靶mRNA,它们富集在47条GO条目(图 5-B),涉及细胞(1342个mRNA)和细胞组件(1342个mRNA)等17条细胞组分相关条目;结合(2996个mRNA)和催化活性(1934个mRNA)等11条与分子功能相关条目;细胞进程(2865个mRNA)和代谢进程(2427个mRNA)等19条与生物学进程相关条目。其中,Am7CK vs. Am7T中分别有675、7和2个靶mRNA注释在应激反应、免疫系统进程和细胞杀伤;Am10CK vs. Am10T中分别有753和7个靶mRNA注释在应激反应和免疫系统进程。
| miRNA name | TPM in Am10CK group | TPM in Am10T group | Log2(Fold change) | P value |
| Up-regulated miRNAs | ||||
| miR-8503-x | 0.001 | 64.363 | 15.974 | 0.000 |
| miR-2965-y | 0.001 | 37.877 | 15.209 | 0.000 |
| miR-4700-x | 0.001 | 27.610 | 14.753 | 0.000 |
| miR-8797-x | 0.001 | 25.680 | 14.648 | 0.000 |
| miR-7572-y | 0.001 | 22.733 | 14.473 | 0.000 |
| miR-3232-x | 0.001 | 18.357 | 14.164 | 0.000 |
| miR-4577-y | 0.001 | 18.353 | 14.164 | 0.000 |
| miR-301-x | 0.001 | 17.167 | 14.067 | 0.000 |
| miR-5001-x | 0.001 | 14.707 | 13.844 | 0.000 |
| miR-2172-x | 0.001 | 7.387 | 12.851 | 0.001 |
| Down-regulated miRNAs | ||||
| miR-462-x | 87.560 | 0.001 | –16.418 | 0.000 |
| miR-142-y | 27.487 | 0.001 | –14.746 | 0.000 |
| miR-144-y | 23.357 | 0.001 | –14.512 | 0.000 |
| miR-144-x | 17.910 | 0.001 | –14.128 | 0.000 |
| miR-223-y | 17.157 | 0.001 | –14.066 | 0.000 |
| miR-8159-x | 12.397 | 0.001 | –13.598 | 0.000 |
| miR-7132-y | 9.860 | 0.001 | –13.267 | 0.000 |
| miR-5112-x | 8.670 | 0.001 | –13.082 | 0.000 |
| let-7-z | 8.143 | 0.001 | –12.991 | 0.000 |
| miR-2184-x | 7.807 | 0.001 | –12.930 | 0.000 |
2.5 意蜂工蜂中肠DEmiRNA的靶mRNA的通路分析
Am7CK vs. Am7T比较组中DEmiRNA的靶mRNA共富集在138条通路,富集数最多的前5位分别是Wnt信号通路(173个mRNA)、内吞作用(139个mRNA)、Hippo信号通路(118个mRNA)、嘌呤代谢(97个mRNA)和泛素介导的蛋白水解(90个mRNA) (表 5)。Am10CK vs. Am10T比较组中DEmiRNA的靶mRNA共富集在135条通路(图 6-B),富集数最多的前5位分别是Wnt信号通路(200个mRNA)、内吞作用(120个mRNA)、嘌呤代谢(112个mRNA)、Hippo信号通路(110个mRNA)和泛素介导的蛋白水解(100个mRNA) (表 6)。
| Pathway name | Pathway ID | Number of target mRNA | P value |
| Wnt signaling pathway | ko04310 | 173 | 0.000 |
| Endocytosis | ko04144 | 139 | 0.000 |
| Hippo signaling pathway | ko04391 | 118 | 0.000 |
| Purine metabolism | ko00230 | 97 | 0.612 |
| Ubiquitin mediated proteolysis | ko04120 | 90 | 0.009 |
| Neuroactive ligand-receptor interaction | ko04080 | 89 | 0.000 |
| Phototransduction | ko04745 | 83 | 0.000 |
| mRNA surveillance pathway | ko03015 | 78 | 0.000 |
| Protein processing in endoplasmic reticulum | ko04141 | 75 | 0.516 |
| FoxO signaling pathway | ko04068 | 68 | 0.000 |

|
| 图 6 意蜂工蜂中肠DEmiRNA靶向免疫通路相关DEmRNA的调控网络 Figure 6 Regulatory network of midgut DEmiRNAs and their target DEmRNAs associated with immune pathway in A. m. ligustica worker. A: Regulatory network of DEmiRNA-DEmRNA in Am7CK vs. Am7T comparison group; B: Regulatory network of DEmiRNA-DEmRNA in Am10CK vs. Am10T comparison group. |
| Pathway name | Pathway ID | Number of target mRNA | P value |
| Wnt signaling pathway | ko04310 | 200 | 0.000 |
| Endocytosis | ko04144 | 120 | 0.074 |
| Purine metabolism | ko00230 | 112 | 0.194 |
| Hippo signaling pathway | ko04391 | 110 | 0.000 |
| Ubiquitin mediated proteolysis | ko04120 | 100 | 0.001 |
| Phototransduction | ko04745 | 85 | 0.000 |
| Neuroactive ligand-receptor interaction | ko04080 | 83 | 0.000 |
| Spliceosome | ko03040 | 83 | 0.689 |
| Protein processing in endoplasmic reticulum | ko04141 | 75 | 0.777 |
| RNA transport | ko03013 | 75 | 0.989 |
进一步分析发现,Am7CK vs. Am7T和Am10CK vs. Am10T中DEmiRNA靶mRNA均富集在果糖和甘露糖代谢(30个mRNA和29个mRNA)、其他多糖降解(28个mRNA和25个mRNA)、淀粉和蔗糖代谢(29个mRNA和32个mRNA)、半乳糖代谢(20个mRNA和18个mRNA)、三羧酸循环(24个mRNA和24个mRNA)和氧化磷酸化(36个mRNA和44个mRNA)等物质和能量代谢通路;Am7CK vs. Am7T和Am10CK vs. Am10T比较组的DEmiRNA靶mRNA均富集于内吞作用和泛素介导的蛋白水解等5条细胞免疫相关通路(表 7);MAPK信号通路和FoxO信号通路等5条体液免疫相关通路(表 8)。
| Pathway | Pathway ID | Num of target mRNAs in Am7CK vs. Am7T | Num of target mRNAs in Am10CK vs. Am10T |
| Endocytosis | ko04144 | 139 | 120 |
| Ubiquitin mediated proteolysis | ko04120 | 90 | 100 |
| Lysosome | ko04142 | 42 | 54 |
| Phagosome | ko04145 | 38 | 39 |
| Insect hormone biosynthesis | ko00981 | 8 | 4 |
| Pathways | Pathway ID | Num of target mRNAs in Am7CK vs. Am7T | Num of target mRNAs in Am10CK vs. Am10T |
| FoxO signaling pathway | ko04068 | 68 | 71 |
| MAPK signaling pathway - fly | ko04013 | 19 | 20 |
| Jak-STAT signaling pathway | ko04630 | 16 | 20 |
| Drug metabolism - cytochrome P450 | ko00982 | 6 | 4 |
| Metabolism of xenobiotics by cytochrome P450 | ko00980 | 6 | 4 |
结合前期分析得到的DEmRNA结果[29],构建DEmiRNA与DEmRNA之间的调控网络,分析结果显示Am7CK vs. Am7T中有26个DEmiRNA靶向结合与内吞作用、泛素介导的蛋白水解、吞噬体、FoxO信号通路和MAPK信号通路等免疫相关的10个DEmRNA (XM_006570195.2、XM_006564220.2和XM_006570873.2等) (图 6-A);Am10CK vs. Am10T中有15个DEmiRNA靶向结合与溶酶体、泛素介导的蛋白水解、内吞作用、MAPK信号通路和FoxO信号通路等免疫相关10个DEmRNA (XM_006567428.2、XM_006570714.2和XM_006561133.2等) (图 6-B)。
2.6 意蜂工蜂中肠DEmiRNA和mRNA的RT-qPCR验证随机挑选图 6中的4个DEmiRNA进行RT-qPCR验证,结果显示它们的表达趋势与测序数据一致(图 7),证实了测序数据和上述miRNA差异表达趋势的可靠性。

|
| 图 7 意蜂工蜂中肠DEmiRNA的RT-qPCR验证 Figure 7 RT-qPCR verification of DEmiRNA in the midgut of A. m. ligustica worker. *: p < 0.05; **: p < 0.01. |
3 讨论
miRNA作为基因表达的关键调控因子,在昆虫细胞发育和免疫等生理过程中扮演重要角色[43]。目前,西方蜜蜂对N. ceranae的胁迫应答的组学研究主要集中在mRNA层面[4, 18, 44],但在ncRNA层面的研究极为有限[5, 26]。Dussaubat等研究发现,N. ceranae接种后14 d的西方蜜蜂死亡率接近100%;作者结合石蜡切片和RNA-seq技术对感染肠道进行分析,结果显示N. ceranae侵染导致宿主肠道结构受到破坏,调控细胞更新的相关基因表达,导致受损肠道无法修复[4]。本研究对正常和N. ceranae胁迫7 d和10 d的意蜂工蜂中肠进行测序,通过生物信息学方法鉴定到984个长度介于18–28 nt的miRNA,其中多数miRNA的首位碱基偏向U,与中华蜜蜂(Apis cerana cerana,简称中蜂)[25]和菜蛾盘绒茧蜂(Cotesia vestalis)[45]的miRNA结构特征相似。前期研究中,笔者团队从意蜂幼虫肠道的sRNA-seq数据中预测出560个miRNA,同样发现这些miRNA的长度介于17–27 nt,首位碱基多偏向U[46]。上述结果说明不同昆虫物种的miRNA,以及意蜂不同虫态的miRNA,具有类似的结构特征。此外,本研究鉴定到56个novel miRNA,为miRBase数据库中西方蜜蜂的miRNA信息提供了有益补充。miRNA表达具有时序表达特异性[47–48]。本研究Am7CK vs. Am7T和Am10CK vs. Am10T比较组中分别筛选到84和107个DEmiRNA,18个DEmiRNA为二者共有,二者特有的DEmiRNA分别为66和89个,表明意蜂工蜂中肠在N. ceranae胁迫的不同阶段通过差异表达不同的miRNA对病原入侵产生应答。
蜜蜂微孢子虫病是一种慢性病,N. ceranae对蜜蜂宿主的侵染是一个较为长期的过程,可能持续1个月甚至更长的时间,而在侵染的全过程,宿主和病原都在时刻进行相互作用;而且,意蜂工蜂对N. ceranae的胁迫应答是一个复杂而持续的过程,伴随着非常复杂的分子事件发生。因此,深入解析宿主的胁迫应答机制需要从多个角度尽可能获得更多的信息。Huang等[26]曾对正常和N. ceranae感染1–6 d的西方蜜蜂工蜂中肠进行深度测序,首次筛选到17个DEmiRNA,可靶向结合413个mRNA;作者进一步分析发现这些靶mRNA注释到了铁结合、信号转导、细胞核、DNA结合、跨膜转运活性等5条功能条目,以及嘌呤代谢、硫胺素新陈代谢、氨基苯甲酸酯降解、抗生素生物合成、氧化磷酸化、多糖降解、核黄素的新陈代谢、氨基糖和核苷酸糖代谢、嘧啶代谢等9条物质和能量代谢通路,未发现有靶mRNA涉及细胞和体液免疫相关通路。通过与本研究结果的比较分析,发现N. ceranae感染后1–6 d和感染后10 d的宿主中肠中共有的DEmiRNA仅有1个(ame-miR-1),表明大部分DEmiRNA具有时序表达特异性;此外,Am7CK vs. Am7T和Am10CK vs. Am10T的DEmiRNA靶mRNA分别注释到了50和47个功能条目,以及138和135条通路,包括5条细胞免疫通路和5条体液免疫通路,但未发现有靶mRNA注释到铁离子结合功能条目、氨基苯甲酸酯降解和抗生素生物合成通路。鉴于miRNA具有时序表达特异性[47–48],上述比较结果表明蜜蜂宿主的不同miRNA在N. ceranae感染的不同阶段产生不同的应答,从而发挥不同的作用;此外,比较结果还进一步丰富了西方蜜蜂响应N. ceranae的胁迫应答的miRNA相关信息。但需要强调的是,Huang等[26]的研究中选择的是西方蜜蜂,但没有具体说明为西方蜜蜂的哪一个具体蜂种;而本研究选择的是意蜂,两项研究选择的蜂种或许存在差异,可能对分析结果产生一定影响。
miRNA通过miRNA种子序列靶向结合mRNA的3′-UTR发挥抑制或降解作用[19–21]。本研究对两个比较组中DEmiRNA靶向结合的mRNA进行预测,结果显示Am7CK vs. Am7T和Am10CK vs. Am10T的DEmiRNA分别靶向9827和10720个mRNA。进一步对DEmiRNA及其调控的免疫通路相关DEmRNA进行调控网络构建(图 6),分析发现Am7CK vs. Am7T中有16个DEmiRNA (ame-miR-193、ame-miR-2788和ame-miR-6052等)靶向结合10个DEmRNA,Am10CK vs. Am10T有25个DEmiRNA (miR-1-z、ame-miR-3727和ame-miR-980等)靶向结合10个DEmRNA。前人研究发现,miR-1不仅可抑制癌细胞增殖[49],还可促进细胞凋亡[50]。昆虫中,miR-1已被证明参与调控黑腹果蝇(Drosophila melanogaster)[51]和埃及伊蚊(Aedes aegypti)[52]的免疫进程。此外,Huang等研究发现ame-miR-1在N. ceranae侵染6 d后的西方蜜蜂工蜂中肠中显著下调[26]。本研究中,miR-1-z (与miR-1高度同源)在Am10CK vs. Am10T中显著下调[log2(Fold change)=–11.573, P=0.02],靶向4个富集在溶酶体通路的DEmRNA (XM_006567428.2、XM_006567431.2、XM_016914010.1和XM_016914011.1),表明N. ceranae在侵染意蜂工蜂的过程可能通过抑制miR-1-z的表达上调溶酶体通路基因的表达水平,对宿主细胞的增殖、凋亡和免疫进程进行调控。在另一项研究中,笔者团队发现在N. ceranae侵染7 d的中蜂工蜂中肠中,miR-1-x的表达也受到了一定抑制[53]。以上结果说明miR-1可能同时参与调控意蜂工蜂和中蜂工蜂对N. ceranae的胁迫应答中的细胞更新和免疫进程。
N. ceranae不含线粒体,增殖所需能量全部窃取自宿主细胞[6–7]。被N. ceranae寄生的西方蜜蜂因受到能量胁迫而增加蔗糖溶液的摄入量[6]。本研究发现,上述两个比较组中DEmiRNA的靶mRNA均有部分注释到果糖和甘露糖代谢(30个mRNA和29个mRNA)、其他多糖降解(28个mRNA和25个mRNA)、半乳糖代谢(20个mRNA和18个mRNA)、淀粉和蔗糖代谢(29个mRNA)等糖类代谢相关通路;此外,两个比较组中DEmiRNA的靶mRNA均有部分注释到氧化磷酸化(36个mRNA和44个mRNA)和三羧酸循环(24个mRNA和24个mRNA)等能量代谢相关通路。Huang等也发现N. ceranae感染1–6 d的西方蜜蜂工蜂中肠的DEmiRNA参与调节宿主氧化磷酸化通路相关基因的表达[26]。氧化磷酸化是昆虫细胞线粒体合成ATP的主要方式[54]。笔者团队前期对N. ceranae胁迫7 d和10 d的意蜂工蜂中肠DEmRNA和DElncRNA进行了系统分析,发现有1个DEmRNA (XM_001123191.4)富集在氧化磷酸化通路[29],有3个DElncRNA (XR_001702895.1、XR_411776.2和XR_001704989.1)涉及该通路的调控[5]。本研究中,对于Am7CK vs. Am7T和Am10CK vs. Am10T比较组中的DEmiRNA,分别有36和44个靶mRNA注释到氧化磷酸化通路。上述结果表明,氧化磷酸化通路在意蜂与N. ceranae相互作用中扮演特殊角色,N. ceranae可能通过调控该通路对宿主造成能量胁迫,以满足自身增殖需要,而意蜂工蜂可能通过调节该通路对病原进行胁迫应答;本研究发现的相关DEmiRNA及前期发现的DElncRNA或许能作为新型的分子靶点,用于蜜蜂微孢子虫病的治疗,这是下一步的研究重点。蜜蜂球囊菌(Ascosphaera apis,简称球囊菌)是另一种常见的蜜蜂真菌病原,其引发的蜜蜂白垩病是长期困扰养蜂生产的一大顽疾[55]。前期研究中,笔者团队通过比较转录组学分析探究中蜂幼虫和意蜂幼虫对球囊菌的抗性差异,发现受到强烈正向选择的意蜂单拷贝基因富集在氧化磷酸化等4条代谢通路,推测球囊菌主要通过影响意蜂幼虫能量代谢和物质代谢降低宿主的抵御作用[56]。综上,推测氧化磷酸化通路在意蜂不同虫态响应真菌病原胁迫应答的过程中发挥特殊作用,该通路也是不同真菌病原的重要调控靶点。
本研究发现,Am7CK vs. Am7T和Am10CK vs. Am10T中DEmiRNA的靶mRNA均有部分注释到应激反应和细胞杀伤等免疫相关的功能条目,此外Am7CK vs. Am7T中DEmiRNA的靶mRNA还富集在免疫系统进程,表明意蜂工蜂的DEmiRNA参与了免疫防御的调控。昆虫识别病原体是免疫应答的第一步,由二类模式识别受体(pattern recognition receptors,PRRs)激活,一类是参与细胞免疫识别的相关基因,如Nimrod、SRCB和TEP等;另一类是参与体液免疫识别的相关基因,如PGRP、βGBP和GNBP等[57]。本研究中,分别有5个DEmiRNA (Am7CK vs. Am7T中的miR-294-y和miR-295-y;Am10CK vs. Am10T中的ame-miR-981、miR-142-y和miR-316-x)和1个DEmiRNA (Am10CK vs. Am10T中的miR-186-x)靶向结合SRCB和PGRP的转录本,说明上述DEmiRNA参与调控宿主对N. ceranae的免疫识别。昆虫的细胞免疫主要方式包括血淋巴介导的吞噬、集结和包裹等方式消除病原;体液免疫的主要方式为识别病原、胞内信号通路激活以及诱导抗菌肽等效应因子基因的诱导表达[58]。本研究的2个比较组中DEmiRNA的靶mRNA皆有部分注释到内吞作用(139个mRNA和120个mRNA)、溶酶体(42个mRNA和54个mRNA)、吞噬体(42个mRNA和54个mRNA)、泛素介导的蛋白水解(90个mRNA和100个mRNA)和昆虫激素的生物合成(8个mRNA和4个mRNA)等5条细胞免疫通路;另有部分靶mRNA注释到5条体液免疫相关通路,包括FoxO信号通路(68个mRNA和71个mRNA)、MAPK信号通路(19个mRNA和20个mRNA)、Jak-STAT信号通路(16个mRNA和20个mRNA)、药物代谢-细胞色素P450 (6个mRNA和4个mRNA)和细胞色素P450对外源生物的代谢作用(6个mRNA和4个mRNA)。然而,未发现靶向6个抗菌肽基因(abaecin、apidaecin、apisimin、hymenoptaecin、defensin-1和defensin-2)的miRNA表达量发生变化。因此,笔者推测DEmiRNA在N. ceranae胁迫的免疫应答中主要参与调控病原识别、细胞免疫通路和部分体液免疫通路,但不参与调控抗菌肽基因的表达及抗菌肽的合成。
近年来,miRNA介导的跨界调控已成为国内外的研究热点[59–60]。植物来源的miRNA可以跨界调控动物体内mRNA的表达,从而影响动物的各种生理过程[59–60]。2012年,张辰宇团队在摄入大米的小鼠血液和器官中检测到来源于大米的MIR168a,并证明MIR168a能够导致小鼠肝脏功能损伤,摄入MIR168a的抑制物可恢复肝脏功能,首次证实了miRNA具有跨界调控功能[59]。2018年,Huang等基于生物信息学分析报道了N. ceranae的miRNA与西方蜜蜂的mRNA之间存在潜在的跨界调控关系[27],仍需要进一步的实验验证。目前,笔者团队正在探究意蜂工蜂中肠DEmiRNA对于N. ceranae的mRNA的靶向结合关系,以期更深入地理解和揭示意蜂对N. ceranae的胁迫应答机制和二者的互作机制。
综上所述,本研究利用高通量测序技术和生物信息学方法从意蜂工蜂中肠鉴定到928个已知miRNA和56个新miRNA,在Am7CK vs. Am7T和Am10CK vs. Am10T中分别鉴定到84和107个DEmiRNA;通过靶mRNA和调控网络分析揭示意蜂工蜂中肠的DEmiRNA可能通过负调控靶mRNA对部分糖代谢、能量代谢、病原体识别、细胞免疫和体液免疫通路进行调控,以应对N. ceranae的入侵;miR-1-z可能在宿主响应N. ceranae胁迫中参与细胞增殖、凋亡和免疫进程;氧化磷酸化通路可能在意蜂工蜂与N. ceranae的相互作用中扮演特殊角色。
| [1] | Bromenshenk JJ, Henderson CB, Seccomb RA, Welch PM, Debnam SE, Firth DR. Bees as biosensors:chemosensory ability, honey bee monitoring systems, and emergent sensor technologies derived from the pollinator syndrome. Biosensors, 2015, 5(4): 678-711. |
| [2] | Shumkova R, Georgieva A, Radoslavov G, Sirakova D, Dzhebir G, Neov B, Bouga M, Hristov P. The first report of the prevalence of Nosema ceranae in Bulgaria. PeerJ, 2018, 6(5): e4252. |
| [3] | Higes M, García-Palencia P, Urbieta A, Nanetti A, Martín-Hernández R. Nosema apis and Nosema ceranae tissue tropism in worker honey bees (Apis mellifera). Veterinary Pathology, 2020, 57(1): 132-138. |
| [4] | Dussaubat C, Brunet JL, Higes M, Colbourne JK, Lopez J, Choi JH, Martín-Hernández R, Botías C, Cousin M, McDonnell C, Bonnet M, Belzunces LP, Moritz RFA, Le Conte Y, Alaux C. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS One, 2012, 7(5): e37017. |
| [5] | Chen DF, Chen HZ, Du Y, Zhou DD, Geng SH, Wang HP, Wan JQ, Xiong CL, Zheng YZ, Guo R. Genome-wide identification of long non-coding RNAs and their regulatory networks involved in Apis mellifera ligustica response to Nosema ceranae infection. Insects, 2019, 10(8): 245. |
| [6] | Mayack C, Naug D. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. Journal of Invertebrate Pathology, 2009, 100(3): 185-188. |
| [7] | Martín-Hernández R, Botías C, Barrios L, Martínez-Salvador A, Meana A, Mayack C, Higes M. Comparison of the energetic stress associated with experimental Nosema ceranae and Nosema apis infection of honeybees (Apis mellifera). Parasitology Research, 2011, 109(3): 605-612. |
| [8] | Antúnez K, Martín-Hernández R, Prieto L, Meana A, Zunino P, Higes M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environmental Microbiology, 2009, 11(9): 2284-2290. |
| [9] | Paris L, El Alaoui H, Delbac F, Diogon M. Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Current Opinion in Insect Science, 2018, 26: 149-154. |
| [10] | Aufauvre J, Misme-Aucouturier B, Viguès B, Texier C, Delbac F, Blot N. Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS One, 2014, 9(3): e91686. |
| [11] | Eiri DM, Suwannapong G, Endler M, Nieh JC. Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS One, 2015, 10(5): e0126330. |
| [12] | BenVau LR, Nieh JC. Larval honey bees infected with Nosema ceranae have increased vitellogenin titers as young adults. Scientific Reports, 2017, 7(1): 14144. |
| [13] | The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature, 2006, 443(7114): 931-949. |
| [14] | Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Molecular Biology, 2006, 15(5): 645-656. |
| [15] | Chaimanee V, Chantawannakul P, Chen YP, Evans JD, Pettis JS. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. Journal of Insect Physiology, 2012, 58(8): 1090-1095. |
| [16] | Sinpoo C, Paxton RJ, Disayathanoowat T, Krongdang S, Chantawannakul P. Impact of Nosema ceranae and Nosema apis on individual worker bees of the two host species (Apis cerana and Apis mellifera) and regulation of host immune response. Journal of Insect Physiology, 2018, 105: 1-8. |
| [17] | Li WF, Evans JD, Huang Q, Rodríguez-García C, Liu J, Hamilton M, Grozinger CM, Webster TC, Su SK, Chen YP. Silencing the honey bee (Apis mellifera) naked cuticle gene (nkd) improves host immune function and reduces Nosema ceranae infections. Applied and Environmental Microbiology, 2016, 82(22): 6779-6787. DOI:10.1128/AEM.02105-16 |
| [18] | Badaoui B, Fougeroux A, Petit F, Anselmo A, Gorni C, Cucurachi M, Cersini A, Granato A, Cardeti G, Formato G, Mutinelli F, Giuffra E, Williams JL, Botti S. RNA-sequence analysis of gene expression from honeybees (Apis mellifera) infected with Nosema ceranae. PLoS One, 2017, 12(3): e0173438. DOI:10.1371/journal.pone.0173438 |
| [19] | Wang L, Jiang P, He Y, Hu HY, Guo Y, Liu XY, Qiu HH, Ma QL, Ouyang F. A novel mechanism of smads/miR-675/TGFβR1 axis modulating the proliferation and remodeling of mouse cardiac fibroblasts. Journal of Cellular Physiology, 2019, 234(11): 20275-20285. DOI:10.1002/jcp.28628 |
| [20] | Li K, Wu Y, Yang H, Hong PY, Fang XD, Hu YJ. H19/miR-30a/C8orf4 axis modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. Journal of Cellular Physiology, 2019, 234(11): 20925-20934. |
| [21] | Kim EJ, Kim JS, Lee S, Lee H, Yoon JS, Hong JH, Chun SH, Sun S, Won HS, Hong SA, Kang K, Jo JY, Choi M, Shin DH, Ahn YH, Ko YH. QKI, a miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. International Journal of Cancer, 2019, 145(6): 1585-1595. |
| [22] | Bartel DP. MicroRNAs:target recognition and regulatory functions. Cell, 2009, 136(2): 215-233. |
| [23] | Ashby R, Forêt S, Searle I, Maleszka R. MicroRNAs in honey bee caste determination. Scientific Reports, 2016, 6: 18794. |
| [24] | Hori S, Kaneko K, Saito TH, Takeuchi H, Kubo T. Expression of two microRNAs, ame-mir-276 and -1000, in the adult honeybee (Apis mellifera) brain. Apidologie, 2011, 42(1): 89-102. DOI:10.1051/apido/2010032 |
| [25] |
Du Y, Tong XY, Zhou DD, Chen DF, Xiong CL, Zheng YZ, Xu GJ, Wang HP, Chen HZ, Guo YL, Long Q, Guo R. MicroRNA responses in the larval gut of Apis cerana cerana to Ascosphaera apis stress. Acta Microbiologica Sinica, 2019, 59(9): 1747-1764.
(in Chinese) 杜宇, 童新宇, 周丁丁, 陈大福, 熊翠玲, 郑燕珍, 徐国钧, 王海朋, 陈华枝, 郭意龙, 隆琦, 郭睿. 中华蜜蜂幼虫肠道响应球囊菌胁迫的microRNA应答分析. 微生物学报, 2019, 59(9): 1747-1764. |
| [26] | Huang Q, Chen YP, Wang RW, Schwarz RS, Evans JD. Honey bee microRNAs respond to infection by the microsporidian parasite Nosema ceranae. Scientific Reports, 2015, 5: 17494. DOI:10.1038/srep17494 |
| [27] | Evans JD, Huang Q. Interactions among host-parasite microRNAs during Nosema ceranae proliferation in Apis mellifera. Frontiers in Microbiology, 2018, 9: 698. DOI:10.3389/fmicb.2018.00698 |
| [28] |
Guo R, Dao C, Xiong CL, Zheng YZ, Fu ZM, Geng SH, Chen DF. Analysis of the highly expressed genes in the midgut of Apis mellifera ligustica worker under the stress of Nosema ceranae. Journal of Environmental Entomology, 2018, 40(5): 1106-1112.
(in Chinese) 郭睿, 刀晨, 熊翠玲, 郑燕珍, 付中民, 耿四海, 陈大福. 意大利蜜蜂工蜂中肠响应Nosema ceranae胁迫的高表达基因分析. 环境昆虫学报, 2018, 40(5): 1106-1112. |
| [29] |
Fu ZM, Chen HZ, Liu SY, Zhu ZW, Fan XX, Fan YC, Wan JQ, Zhang L, Xiong CL, Xu GJ, Chen DF, Guo R. Immune responses of Apis mellifera ligustica to Nosema ceranae stress. Scientia Agricultura Sinica, 2019, 52(17): 3069-3082.
(in Chinese) 付中民, 陈华枝, 刘思亚, 祝智威, 范小雪, 范元婵, 万洁琦, 张璐, 熊翠玲, 徐国钧, 陈大福, 郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答. 中国农业科学, 2019, 52(17): 3069-3082. |
| [30] | Guo R, Chen DF, Xiong CL, Hou CS, Zheng YZ, Fu ZM, Liang Q, Diao QY, Zhang L, Wang HQ, Hou ZX, Kumar D. First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie, 2018, 49(5): 660-670. |
| [31] | VanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, Frazier M, Frazier J, Cox-Foster D, Chen YP, Underwood R, Tarpy DR, Pettis JS. Colony collapse disorder:a descriptive study. PLoS One, 2009, 4(8): e6481. |
| [32] | Huang Q, Chen YP, Wang RW, Cheng S, Evans JD. Host-parasite interactions and purifying selection in a microsporidian parasite of honey bees. PLoS One, 2016, 11(2): e0147549. |
| [33] | Huang WF, Solter LF. Comparative development and tissue tropism of Nosema apis and Nosema ceranae. Journal of Invertebrate Pathology, 2013, 113(1): 35-41. |
| [34] |
Guo R, Du Y, Xiong CL, Zheng YZ, Fu ZM, Xu GJ, Wang HP, Chen HZ, Geng SH, Zhou DD, Shi CY, Zhao HX, Chen DF. Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Scientia Agricultura Sinica, 2018, 51(21): 4197-4209.
(in Chinese) 郭睿, 杜宇, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 王海朋, 陈华枝, 耿四海, 周丁丁, 石彩云, 赵红霞, 陈大福. 意大利蜜蜂幼虫肠道发育过程中的差异表达microRNA及其调控网络. 中国农业科学, 2018, 51(21): 4197-4209. |
| [35] | Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 2009, 10(3): R25. |
| [36] | Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research, 2012, 40(1): 37-52. |
| [37] | Cheng HY, Wang Y, Tao X, Fan YF, Dai Y, Yang H, Ma XR. Genomic profiling of exogenous abscisic acid-responsive microRNAs in tomato (Solanum lycopersicum). BMC Genomics, 2016, 17: 423. DOI:10.1186/s12864-016-2591-8 |
| [38] | Liu H, Yang X, Zhang ZK, Zou WC, Wang HN. miR-146a-5p promotes replication of infectious bronchitis virus by targeting IRAK2 and TNFRSF18. Microbial Pathogenesis, 2018, 120: 32-36. |
| [39] | Li SY, Yang J, Wang LY, Du F, Zhao JL, Fang R. Expression profile of microRNAs in porcine alveolar macrophages after Toxoplasma gondii infection. Parasites & Vectors, 2019, 12(1): 65. |
| [40] | Allen E, Xie ZX, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 2005, 121(2): 207-221. |
| [41] | Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8:new features for data integration and network visualization. Bioinformatics, 2011, 27(3): 431-432. |
| [42] | Chen CF, Ridzon DA, Broomer AJ, Zhou ZH, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research, 2005, 33(20): e179. |
| [43] | Asgari S. MicroRNA functions in insects. Insect Biochemistry and Molecular Biology, 2013, 43(4): 388-397. |
| [44] | Holt HL, Aronstein KA, Grozinger CM. Chronic parasitization by Nosema microsporidia causes global expression changes in core nutritional, metabolic and behavioral pathways in honey bee workers (Apis mellifera). BMC Genomics, 2013, 14: 799. DOI:10.1186/1471-2164-14-799 |
| [45] | Wang ZZ, Ye XQ, Shi M, Li F, Wang ZH, Zhou YN, Gu QJ, Wu XT, Yin CL, Guo DH, Hu RM, Hu NN, Chen T, Zheng BY, Zou JN, Zhan LQ, Wei SJ, Wang YP, Huang JH, Fang XD, Strand MR, Chen XX. Parasitic insect-derived miRNAs modulate host development. Nature Communications, 2018, 9(1): 2205. |
| [46] |
Xiong CL, Du Y, Chen DF, Zheng YZ, Fu ZM, Wang HP, Geng SH, Chen HZ, Zhou DD, Wu SZ, Shi CY, Guo R. Bioinformatic prediction and analysis of miRNAs in the Apis mellifera ligustica larval gut. Chinese Journal of Applied Entomology, 2018, 55(6): 1023-1033.
(in Chinese) 熊翠玲, 杜宇, 陈大福, 郑燕珍, 付中民, 王海朋, 耿四海, 陈华枝, 周丁丁, 吴素珍, 石彩云, 郭睿. 意大利蜜蜂幼虫肠道的miRNAs的生物信息学预测及分析. 应用昆虫学报, 2018, 55(6): 1023-1033. |
| [47] | He L, Hannon GJ. MicroRNAs:small RNAs with a big role in gene regulation. Nature Reviews Genetics, 2004, 5(7): 522-531. |
| [48] | Ha MJ, Kim VN. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology, 2014, 15(8): 509-524. |
| [49] | Yan HB, Huang JC, Chen YR, Yao JN, Cen WN, Li JY, Jiang YF, Chen G, Li SH. Role of miR-1 expression in clear cell renal cell carcinoma (ccRCC):a bioinformatics study based on GEO, ArrayExpress microarrays and TCGA database. Pathology-Research and Practice, 2018, 214(2): 195-206. |
| [50] | Tang YH, Zheng JY, Sun Y, Wu ZG, Liu ZM, Huang GZ. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. International Heart Journal, 2009, 50(3): 377-387. |
| [51] | Fullaondo A, Lee SY. Identification of putative miRNA involved in Drosophila melanogaster immune response. Developmental & Comparative Immunology, 2012, 36(2): 267-273. |
| [52] | Shrinet J, Jain S, Jain J, Bhatnagar RK, Sunil S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Neglected Tropical Diseases, 2014, 8(1): e2616. DOI:10.1371/journal.pntd.0002616 |
| [53] | Chen DF, Du Y, Chen HZ, Fan YC, Fan XX, Zhu ZW, Wang J, Xiong CL, Zheng YZ, Hou CS, Diao QY, Guo R. Comparative identification of microRNAs in Apis cerana cerana workers' midguts in response to Nosema ceranae invasion. Insects, 2019, 10(9): 258. DOI:10.3390/insects10090258 |
| [54] | Li YY, Zhang R, Liu SL, Donath A, Peters RS, Ware J, Misof B, Niehuis O, Pfrender ME, Zhou X. The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evolutionary Biology, 2017, 17: 269. |
| [55] | Evison SE. Chalkbrood:epidemiological perspectives from the host-parasite relationship. Current Opinion in Insect Science, 2015, 10: 65-70. |
| [56] |
Xiong CL, Du Y, Wang HQ, Zheng YZ, Fu ZM, Wang HP, Zhang L, Chen DF, Guo R. Unraveling the mechanism regulating the Ascosphaera apis-resistance difference between Apis cerana cerana and Apis mellifera ligustica larvae based on comparative transcriptome analysis. Journal of China Agricultural University, 2019, 24(5): 106-114.
(in Chinese) 熊翠玲, 杜宇, 王鸿权, 郑燕珍, 付中民, 王海朋, 张璐, 陈大福, 郭睿. 基于比较转录组学分析揭示中华蜜蜂及意大利蜜蜂幼虫的球囊菌抗性差异机制. 中国农业大学学报, 2019, 24(5): 106-114. |
| [57] | Keehnen NLP, Rolff J, Theopold U, Wheat CW. Insect antimicrobial defences:a brief history, recent findings, biases, and a way forward in evolutionary studies. Advances in Insect Physiology, 2017, 52: 1-33. |
| [58] | Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annual Review of Immunology, 2007, 25: 697-743. |
| [59] | Zhang L, Hou DX, Chen X, Li DH, Zhu LY, Zhang YJ, Li J, Bian Z, Liang XY, Cai X, Yin Y, Wang C, Zhang TF, Zhu DH, Zhang DM, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen JQ, Wang J, Wang M, Zhang QP, Zhang JF, Zen K, Zhang CY. Exogenous plant MIR168a specifically targets mammalian LDLRAP1:evidence of cross-kingdom regulation by microRNA. Cell Research, 2012, 22(1): 107-126. |
| [60] | Zhu KG, Liu MH, Fu Z, Zhou Z, Kong Y, Liang HW, Lin ZG, Luo J, Zheng HQ, Wan P, Zhang J, Zen KF, Chen J, Hu FL, Zhang CY, Ren J, Chen X. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genetics, 2017, 13(8): e1006946. |
 2020, Vol. 60
2020, Vol. 60