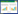中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张婷, 杨梦华. 2020
- Zhang Ting, Yang Menghua. 2020
- 副溶血弧菌的毒力基因表达调控的分子机制
- Molecular mechanisms of virulence genes expression in Vibrio parahaemolyticus
- 微生物学报, 60(7): 1345-1357
- Acta Microbiologica Sinica, 60(7): 1345-1357
-
文章历史
- 收稿日期:2019-10-15
- 修回日期:2020-01-01
- 网络出版日期:2020-06-11
副溶血弧菌是一种革兰氏阴性嗜盐细菌,广泛存在于海洋、河口和河流环境中,通过受污染的水源或食物进入人体可导致水样性腹泻[1];也可通过开放性伤口感染,引起败血症,在免疫低下或具有潜在肝脏疾病的患者中会危及生命[2-3]。除了对人类宿主的影响以外,副溶血弧菌还能感染无脊椎动物,如甲壳类动物[4]、珊瑚[5]和软体动物[6],以及非人类脊椎动物,如海獭[7]、鳍鱼[8]等。根据是否含有相关致病功能的蛋白质的基因,副溶血弧菌可分为致病性菌株和非致病性菌株[9]。致病性副溶血弧菌都可产生耐热直接溶血素(the thermostable direct hemolysin,TDH)或耐热相关溶血素(the Tdh-related hemolysin,TRH)或两者兼有。目前研究发现,高达90%的临床分离菌株都具有tdh和/或trh基因,而这两种基因在环境分离株中却很少存在[10]。由副溶血弧菌引起的感染通常与多种血清型相关,自1996年以来,O3︰K6、O4︰K68、O1︰K25、O1︰26和O1︰KUT血清型菌株是引起疾病暴发的主要群体[11-13]。
已知TDH阳性的副溶血弧菌对其宿主具有溶血性、肠毒性、心脏毒性和细胞毒性的危害[14-15]。这些毒性作用由细菌多种毒力因子引起,包括细胞溶菌素、蛋白酶、脂肪酶、铁载体、胞外多糖和效应蛋白等,它们主要通过细菌的III型分泌系统(Type III secretion systems,T3SS)和VI型分泌系统(Type VI secretion systems,T6SS),被输送到宿主细胞,直接影响宿主细胞蛋白的活性[16-19]。
T3SS是副溶血弧菌主要的毒力因子,副溶血弧菌有2个T3SS系统,分别位于3.3 Mb的大圆形染色体和1.9 Mb的小圆形染色体上,分别命名为T3SS1和T3SS2[20]。T3SS1在副溶血弧菌菌株中普遍存在,其主要功能是引起细胞毒性,也会对生物被膜(biofilm)的形成、运动性等细菌表型产生影响,有助于副溶血弧菌在环境中的生存[20-22]。T3SS2基因簇位于副溶血弧菌80 kb致病岛内,仅存在于从败血症和其他疾病患者中分离出的致病性副溶血弧菌及其衍生株中,与其他细菌的T3SS的相似度较低。对副溶血弧菌的致病性具有重要作用的TDH和TRH基因就是位于T3SS2的基因簇内[16, 23]。T3SS2具有肠毒性和对肠道细胞系具有细胞毒性。与T3SS相似,致病性副溶血弧菌的大小基因组上分别编码2个T6SS,位于大基因组上的是T6SS1(vp1386-vp1420),而位于小基因组上的是T6SS2(vpa1030-vpa1043)[24]。T6SS2在副溶血弧菌黏附宿主细胞的过程中起作用,而T6SS1的作用尚未得到证实[19, 25]。
尽管致病性副溶血弧菌具有多种毒力因子,然而,在体外水生环境中,这些毒力因子几乎是不被激活的,只有当副溶血弧菌进入宿主,或在特定的环境中生存时,这些毒力因子才被大量表达并引起宿主致病[26]。因此,副溶血弧菌毒力基因的激活受到环境信号因子的严格调控,环境信号因子通过与细菌本身的信号调控通路相结合,共同调节与致病相关的毒力基因的表达。目前的研究表明,副溶血弧菌T3SS1的毒力基因的表达主要受到ExsA/C/D/E系统和H-NS的复杂调控,使得T3SS1相关毒力因子在不同的环境条件下表达。而VtrA/B/C在有胆盐存在的条件下可以激活T3SS2基因簇的表达。另外,调控因子ToxR与群体感应系统同时参与多种毒力因子的调控,包括T3SS、T6SS、tdh等。然而,对于副溶血弧菌各种毒力基因表达的调控机制,还存在许多问题,尤其是关于哪些环境因子对副溶血弧菌毒力基因的表达起到诱导调控作用,目前还不是很清楚。对这些环境信号因子的分析研究必将有助于我们更好地理解副溶血弧菌的致病机制。
本文主要对调控副溶血弧菌主要毒力因子表达的分子机制进行综述,为今后进一步研究副溶血性弧菌基因表达调控的分子机制,以及更好地理解宿主与病原体的相互作用对副溶血弧菌的致病机制的影响提供理论参考。
1 ExsACDE级联调控通路对T3SS1基因簇的表达调控副溶血弧菌的T3SS1由42个基因(vp1656-vp1697)组成,其中30个基因与耶尔森氏菌(Yersinia)的基因具有序列同源性,而其余12个可编码效应蛋白[27]。在T3SS1基因簇的末端,有vp1699、vp1698、vp1701和vp1702,它们分别与假单胞菌(Pseudomonas)中的exsA、exsD、exsC、exsE有序列同源性。因此,副溶血弧菌vp1699、vp1698、vp1701和vp1702也被命名为exsA、exsD、exsC、exsE。
ExsA位于T3SS1基因簇的末端区域,是激活T3SS1基因的转录主要调控因子。ExsA属于AraC/XylR转录激活因子家族的成员[28],两个ExsA单体通过氨基末端结构域相互作用以形成功能性ExsA同型二聚体,通过ExsA同型二聚体内在的螺旋转角螺旋(HTH) DNA识别结构域与T3SS1相关基因的转录起始位点上游约50 bp的保守序列(TNAAANA)结合,从而激活依赖于ExsA的T3SS1相关基因的转录[29]。T3SS1基因簇中转录依赖于ExsA的基因有vp1667、vp1688、vp1662、vp1668、vp1670、vp1695、vp1696、vp1686、vp1687、vp1656、vp1659、vp1698、vp1699,这些基因包括T3SS1的结构基因、调控基因和效应蛋白基因[27]。当副溶血弧菌的ExsA失去功能时,T3SS1的功能也几乎丧失。因此,调控ExsA蛋白的功能是调控T3SS1功能的关键。人们发现在实验室的常规培养条件下(含3% NaCl的LB培养基,简称为MLB培养基),副溶血弧菌的T3SS1基因簇几乎不被表达,然而当副溶血弧菌与宿主细胞(如HeLa或Caco-2细胞)接触时[30],或在细胞培养基DMEM (Dulbecco’s modified Eagle’s medium,DMEM)中培养时,T3SS1基因簇中的表达量极大地增加。因此将在DMEM和MLB培养条件分别称为T3SS1基因转录的诱导条件和非诱导条件[27]。但也有研究发现,在普通的LB培养基中添加Mg2+和EGTA,且在30 ℃进行培养,T3SS1蛋白分泌也会增加[31]。
通过研究在诱导条件和非诱导条件下生长的副溶血弧菌ExsA的蛋白功能发现,在非诱导条件下,ExsA与细胞内的ExsD单体结合形成1︰1复合物。ExsD是抗活化剂,与ExsA结合后可以阻止ExsA与DNA结合,进而抑制ExsA依赖性基因的转录[27]。因此,ExsD是副溶血弧菌中T3SS1基因转录的负调节因子。另外,当副溶血弧菌在诱导条件下生长时,ExsC蛋白被表达,ExsC与ExsD结合的亲和力要比ExsD与ExsA结合的更高,因此,通过ExsC与ExsD形成复合物,使ExsA从ExsA/ExsD复合物中释放出来,从而激活ExsA依赖性基因的转录[27]。在非诱导条件下生长时,副溶血弧菌还表达另外一个重要的调控蛋白ExsE,ExsE可以与ExsC形成2︰1的复合物ExsC/ExsE[32-33],这使得ExsC不能与ExsD的蛋白结合,从而促进ExsD进一步抑制ExsA的功能。而且,在非诱导条件下,由于ExsC和ExsE的表达量较低,而ExsD的表达量较高,更容易形成抑制性ExsA/ExsD和ExsC/ExsE复合物,从而使ExsA的活性受到最大的限制[34]。因此,在非诱导条件下生长的副溶血弧菌,其T3SS1基因簇的表达量是极低的。当副溶血弧菌在诱导条件下生长时,随着T3SS1功能的增强,ExsE通过T3SS1被分泌到胞外,由此导致细胞内ExsE浓度的降低,这有利于ExsC/ExsD复合物的形成,从而进一步从ExsA/ExsD复合物中释放ExsA以激活T3SS1基因的表达[35]。有研究发现,缺失exsE基因的副溶血弧菌突变株尽管在非诱导条件下,T3SS1的基因也能被表达,这说明ExsE对T3SS1基因簇的表达起抑制作用。ExsE除了具有调控ExsA活性的功能以外,它也有助于副溶血弧菌对HeLa细胞的黏附作用[35]。ExsACDE调控通路对副溶血性弧菌T3SS1基因的表达调控的作用示意图如图 1所示。
研究发现exsA基因本身的表达也受到VP0529和H-NS等多种调控因子的多重调控。VP0529是一个DNA结合蛋白,可形成稳定的具有翼状螺旋-转角-螺旋(wHTH)结构域的同源二聚体。与弧菌中的HlyU蛋白有序列同源性[36]。HlyU是许多弧菌中常见的转录调节因子,可激活包括霍乱弧菌(Vibrio cholerae)的溶血素基因hlyA、创伤弧菌(Vibrio vulnificus)的rtxA1操纵子以及鳗弧菌(Vibrio anguillarum)的plp-vah1和rtxACHBDE基因簇的基因[37-39]。H-NS是一种核酸相关的DNA结合蛋白,广泛存在于革兰氏阴性细菌中,可与靶基因中富含A+T的DNA序列的启动子区结合,从而沉默靶基因的转录[40]。研究发现副溶血弧菌的VP0529调控因子可以解除H-NS沉默的作用,使得表达受H-NS抑制的基因被激活[40]。VP0529可以从exsA启动子中置换H-NS,从而解除H-NS对exsA基因表达的沉默作用[41]。在不表达H-NS的情况下,尽管没有VP0529,exsA启动子效率也显著增加,这表明在副溶血弧菌中VP0529不是严格激活exsA启动子的调控因子,而主要起到置换负调节子的作用[41](图 2)。

|
| 图 2 ToxR与H-NS/HlyU对exsA基因的调控作用c Figure 2 ToxR, HlyU and H-NS regulate the expression of exsA. |
综上所述,在副溶血弧菌中ExsACDE信号通路与其他转录调控因子协同作用,通过感知不同的环境信号对T3SS1基因簇的转录表达进行调控。然而对于这一信号通路调控的分子机制还有许多问题有待进一步的探讨。比如当副溶血弧菌与HeLa细胞接触时,exsA基因表达增加,从而诱导T3SS1基因表达[42],那么究竟是什么样的分子机制使得副溶血弧菌感知其与宿主细胞的接触从而激活ExsACDE系统?还有,当副溶血弧菌在含有Mg2+和EGTA的非诱导条件培养基中(LB)并在30 ℃的条件下生长,exsA基因表达也可达到最大表达量;而当培养基中加入高Ca2+时,exsA基因几乎不表达[31]。那么Mg2+与Ca2+是通过什么样的分子机制调控exsA基因的转录?除了这两种金属离子以外,还有哪些环境信号因子对T3SS1基因簇的表达起调控作用呢?它们与ExsACDE信号通路的关系又是如何?目前,这些问题都还有待更深入的研究,而对这些问题的解答无疑为阐明副溶血弧菌T3SS1基因转录调控机制具有重要的生物学意义。
2 ToxR对副溶血弧菌毒力因子的表达调控作用ToxR是弧菌中普遍存在的一个跨膜调节蛋白,是细菌在不利环境条件下存活所必需的,在调节细菌对胁迫的耐受和毒力表达方面起着重要作用[43]。在霍乱弧菌中,ToxR是毒力基因转录表达的关键调控因子,在胆汁酸存在条件下,ToxR可直接激活霍乱毒素ctx的转录[44-45],除此之外,ToxR也参与调控外膜蛋白(Omps),如ompU和ompT,以及生物膜形成[46-47]。副溶血弧菌ToxR与霍乱弧菌ToxR高度相似[48]。目前对于ToxR参与调节副溶血弧菌TDH和T3SS1相关基因表达的研究比较详细[49-50]。
在副溶血弧菌中ToxR和另一个重要的调节因子CalR协同调控T3SS1部分基因的表达。CalR是由VP0350编码LysR调控因子家族的转录调节因子,是霍乱弧菌中LeuO同源蛋白,在副溶血弧菌中将其命名为CalR[51]。ToxR与calR的启动子区域结合后,会激活calR基因的转录,CalR可与T3SS1基因簇中3个操纵子(vp1667-vp1655,vp1687-1686和exsBAD-vscBCD)的启动子区结合以抑制它们的转录,从而抑制副溶血弧菌依赖于T3SS1的细胞毒性[52]。
另外,ToxR也参与调节TDH的表达。TDH是副溶血弧菌的主要毒力因子,具有溶血活性、细胞毒性和肠毒性[53]。副溶血弧菌中TDH由tdh1(vpa1378)和tdh2(vpa1314)两个基因编码,两者均位于染色体2上的致病岛Vp-PAI中,由于tdh2的转录水平远高于tdh1的转录水平(> 25倍),因此tdh2是TDH活性的主要决定基因[54]。研究发现,ToxR能直接结合到tdh2的启动子上,对tdh2的转录具有正调控作用[55],tdh2的转录还受到CalR的调控,在tdh2的启动子区含有CalR的结合位点,当CalR与tdh2的启动子区的-178 bp至-318 bp结合后,tdh2的转录水平显著降低,表明CalR对tdh2转录是负调控的[56]。由于ToxR结合位点位于CalR结合位点的5′端,因此推测CalR对tdh2转录有具有负调控作用,一方面可能因为CalR对tdh2转录有直接抑制作用,另一方面CalR阻断了ToxR与tdh2启动子的结合,从而抑制tdh2的转录,然而这个猜测有待进一步验证[56]。
研究发现,ToxR还可以与群体感应的调节因子AphA和OpaR协同作用,抑制副溶血弧菌的T6SS1的表达。ToxR可以直接与T6SS1基因簇中vp1400-1406和vp1409-1407的启动子区结合,直接抑制其转录[57]。同时,ToxR间接地负调节T6SS1基因簇中vp1388-1390和vp1393-1406的转录[57],但其具体的分子机制仍然不是很清楚。
此外,也有报道指出ToxR是副溶血弧菌T3SS2相关基因表达所必需的[50]。推测ToxR很可能通过增强T3SS2基因簇关键调控因子VtrA的活性,从而影响其下游毒力相关基因的表达[50]。然而其分子机制还有待进一步探究。ToxR参与副溶血性弧菌毒力基因表达调控的示意图如图 3所示。

|
| 图 3 ToxR对副溶血性弧菌毒力因子的表达调控作用 Figure 3 Regulation of virulence factors expression by ToxR in V. parahaemolyticus. |
综上所述,与在霍乱弧菌中的调控功能相类似,跨膜调节蛋白ToxR参与副溶血弧菌多种毒力因子的表达调控,对于副溶血弧菌的致病性与其在不同环境中的生存至关重要。
3 VtrABC信号调控通路对T3SS2的调控作用研究证明VtrA、VtrC和VtrB是副溶血弧菌T3SS2基因簇关键基因表达的重要调控因子[58]。VtrA (VPA1332)位于T3SS2基因簇中间的位置,是由253个氨基酸组成的跨内膜蛋白,其N端1-133氨基酸位于细胞质中,具有OmpR蛋白家族的翼-螺旋-转角-螺旋(winged-helix-turn-helix,wHTH) DNA结合结构域,氨基酸残基134-156形成一个跨膜螺旋,C末端(157-253 aa)位于周质空间中[58-59]。vtrC(vpa1333)位于vtrA基因下游,与vtrA基因处于同一个操纵元,这两个基因的开放阅读框(open reading frame,ORF)具有17个核苷酸的重叠。VtrC是由161个氨基酸组成的内膜蛋白。VtrA和VtrC的周质结构域会形成1︰1异二聚体VtrA/VtrC[60]。当副溶血弧菌进入宿主肠道后,肠道中的胆汁酸分子会分别结合到VtrA和VtrC周质空间结构域的β-桶结构的疏水内部,结合了胆汁酸的VtrA蛋白功能被激活,从而诱导下游vtrB基因的转录与表达[60]。受VtrA诱导表达的VtrB也是一个跨内膜调控蛋白,其胞内的DNA结合结构域与VtrA的结构相似,也是属于OmpR蛋白家族,含有wHTH的DNA结合结构域[59]。被激活的VtrB进而诱导T3SS2基因簇相关基因的表达。
研究发现副溶血弧菌的VtrA与霍乱弧菌的TcpP蛋白功能十分相似,如果单独表达VtrA的胞内DNA结合结构域,并不能激活下游毒力基因的表达,VtrA必须以跨膜蛋白的形式发挥功能[59],而在含有胆汁酸的条件下,VtrA与VtrC各自的周质空间结构域可以形成二聚体,从而激活VtrA的胞内DNA结合结构域的功能,进而诱导下游基因的表达[59]。这种VtrA/VtrC寡聚化后的信号调控通路在弧菌中是普遍存在的,例如无霍乱毒素(CT)和毒素共调节菌毛(TCP)的致病性霍乱弧菌AM19226的VttRA/VttRB,以及霍乱弧菌致病毒株El Tor中的TcpP/TcpH和ToxR/ToxS调控因子[61]。我们的研究发现,VtrA蛋白的第43位半胱氨酸残基对VtrA形成同源二聚体及其激活下游毒力基因表达的功能至关重要。当把VtrA的第43位半胱氨酸突变为丝氨酸,VtrA便不能形成二聚体,也不能激活下游毒力基因的表达[62]。因此,T3SS2关键调控因子VtrB的转录受到多种调控因子与环境信号因子的相互作用,对其具体的分子机制的探究必将有助于我们更好地了解致病性副溶血弧菌的致病机制。
4 副溶血弧菌群体感应调控通路对毒力因子的表达调控作用群体感应(quorum sensing)是细菌通过产生自体诱导因子感知细胞密度的变化从而调节相关基因的表达,以此来适应不同的生存环境的一种基因表达调控机制[63-64]。副溶血弧菌的群体感应系统参与调控多种细胞功能,包括毒力因子的产生、生物被膜的形成、耐药性和运动性等[64]。目前发现副溶血弧菌产生三类自诱导物,分别称为哈氏自诱导物1 (Harveyi autoinducer,HAI-1)、2型自诱导物(AI-2)和霍乱自诱导物1 (Cholera autoinducer,CAI-1),其受体蛋白分别是LuxN、LuxP/LuxQ和CqsS[65]。而AphA和OpaR是副溶血弧菌中与群体感应调控相关的2个重要的调控因子[65-66]。
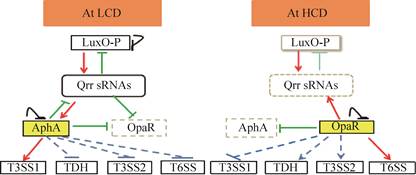
在低细胞密度(low cell density,LCD)下,自体诱导物浓度比较低,自体诱导物的受体蛋白LuxN和LuxU启动级联磷酸化,导致LuxO蛋白的磷酸化[66]。磷酸化的LuxO激活Qrr sRNAs (Qrr1-5)的转录,Qrr1-5会促进AphA的表达却抑制OpaR的表达[66](图 4)。受群体感应调控的AphA蛋白一方面可激活与毒力、运动性和生物被膜形成相关的基因表达,另一方面又会反馈抑制OpaR和qrr1-5的转录[67],从而对AphA蛋白本身的表达进行反馈调控,这也许是群体感应调控的一种作用机制。目前研究发现,AphA对T3SS1、Vp-PAI (pathogenicity island)和T6SS1相关基因的表达都具有调控作用[68]。在感染的初始阶段,副溶血弧菌的细胞密度比较低,受群体感应系统诱导表达的AphA与exsBAD-vscBCD的启动子区结合,从而激活T3SS1相关基因的转录,增强病原体的细胞毒性。AphA对存在于Vp-PAI中的vtrA、vopB2和tdh2的转录具有负调控的作用,而且还可以间接地抑制T6SS1基因簇中vp1388-1390,vp1400-1406和vp1407-1409基因的表达[66],但其详细的分子机制仍不清楚。
|
| 图 4 副溶血性弧菌群体感应调控通路对毒力因子的表达调控作用[68] Figure 4 Quorum sensing system regulates the expression of the major virulence loci in V. parahaemolyticus[68]. The red arrow line represents positive regulation. The green vertical lines represents negative regulation. The blue dotted arrow represents indirect positive regulation. The blue dotted line represents indirect negative regulation. |
在高细胞密度(HCD)下,高浓度的自体诱导物导致LuxN和LuxU级联磷酸化通路被抑制,使得LuxO蛋白去磷酸化,从而使得Qrr1-5的产生减少,而OpaR的活性增强[65]。OpaR是LuxR蛋白家族的调控因子,在高细胞密度条件下对副溶血弧菌的多种细胞功能具有重要的调控作用。OpaR的调控作用与AphA的作用相反,OpaR一方面可以激活qrr1-5的转录,另一方面可以抑制毒力基因以及生物被膜相关基因的转录[65, 69]。在副溶血弧菌感染的后期,细菌细胞密度升高,受群体感应系统调控的OpaR被大量表达,OpaR可直接与exsBAD-vscBCD启动子区结合,从而抑制T3SS1相关基因的转录;另一方面,OpaR也可以与T3SS2基因簇的vpa1332-1333(vtrAC)基因的启动子区结合,激活T3SS2相关基因的转录,增强病原体的肠毒性,引起副溶血弧菌的临床症状[66]。对与OpaR结合的启动子序列分析发现,T6SS1和T6SS2基因簇的多个启动子区都含有OpaR的结合序列,因此推测OpaR也可能对T6SS1和T6SS2相关基因的表达具有调控作用[70](图 4)。
尽管AphA与OpaR分别在副溶血弧菌不同的细胞密度下发挥调控作用,然而它们之间并非是完全独立的调控通路,相反,AphA与OpaR的调控作用相互之间有着非常密切的联系。比如,编码AphA与OpaR基因的转录都受到sRNA Qrr1-5的调控;受AphA调控的下游基因同时也受OpaR的调控作用。因此,与霍乱弧菌相似,副溶血弧菌的群体感应信号调控通路在不同的细胞密度条件下都可发挥作用,其调控作用多样且复杂。
5 小结和展望副溶血弧菌是人类食用了污染的未煮熟或生的海产品而引起的肠胃炎的主要病原菌,其毒力因子的表达受到多种信号因子与调控通路的共同激活。副溶血弧菌的ExsACDE信号通路对T3SS1基因簇的转录调控有重要作用。而CalR作为T3SS1的直接抑制因子,可直接结合到T3SS1基因簇中的VP1656-1667 (T3SS1结构蛋白)、ExsBAD-VscBCD (T3SS1调控蛋白)以及VP1686-1687 (T3SS1效应蛋白)的启动子区,从而抑制T3SS1的活性。目前研究主要还是针对ExsACDE信号调控通路对T3SS1基因转录的研究较多,而对于非依赖ExsA的信号通路调控的研究还比较少。ToxR作为副溶血弧菌的毒力调节因子,可以直接或间接调控T3SS1基因的功能,然而,其具体的调控机制有待进一步探究。ToxR也可以与T3SS2基因簇中的VPA1332-1333的启动子区以及TDH2的启动子直接结合激活它们的转录[48]。此外,群体感应调控系统在副溶血弧菌的毒力基因表达激活过程中也发挥着重要的作用。副溶血弧菌群体感应系统的2个重要的调控因子AphA和OpaR对下游多个信号通路具有调控作用,如exsACDE、toxR、vtrA等调控蛋白基因的表达都直接或间接受群体感应系统的调控。另外,我们最近的研究发现,副溶血弧菌毒力因子除了在转录水平上受到多种信号调控通路的影响以外,其蛋白质还受到二硫键氧还蛋白(Disulfide bondoxidoredutase,Dsb)翻译后的修饰作用,从而影响细菌的毒力与致病性[71]。
综上所述,副溶血弧菌毒力因子的激活受到ExsACDE级联调控通路、VtrABC调控通路、ToxR调控通路以及群体感应等多种信号通路的控制。这些信号调控通路构成了一个复杂的调控网络,在不同的信号因子的刺激下,发挥不同的功能。目前的研究主要都是集中在毒力因子单个蛋白功能的研究,而对毒力调控网络的研究较少,对于与这些调控网络相关的信号因子的探究更少。因此,今后应该进一步拓展副溶血性弧菌毒力基因的表达调控网络分子机制的研究,尤其需要加强对激活这些调控通路的信号因子的研究,这必将促进我们对副溶血性弧菌的致病机制的了解,为今后预防和治疗副溶血弧菌所引起的疾病提供理论参考。
| [1] | Su YC, Liu C. Vibrio parahaemolyticus:A concern of seafood safety. Food Microbiology, 2007, 24(6): 549-558. DOI:10.1016/j.fm.2007.01.005 |
| [2] | Park MS, Park KH, Bahk GJ. Interrelationships between multiple climatic factors and incidence of foodborne diseases. International Journal of Environmental Research and Public Health, 2018, 15(11): 2482. DOI:10.3390/ijerph15112482 |
| [3] | Johnson DE, Weinberg L, Ciarkowski J, West P, Colwell RR. Wound infection caused by Kanagawa-negative Vibrio parahaemolyticus. Journal of Clinical Microbiology, 1984, 20(4): 811-812. DOI:10.1128/JCM.20.4.811-812.1984 |
| [4] | Lee TE, Chen TI, Yang YT, Ko TP, Huang YT, Huang JY, Huang MF, Lin SJ, Chen CY, Lin SS, Lightner DV, Wang HC, Wang AHJ, Wang HC, Hor LI, Lo CF. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(34): 10798-10803. DOI:10.1073/pnas.1503129112 |
| [5] | Vezzulli L, Brettar I, Pezzati E, Reid PC, Colwell RR, Höfle MG, Pruzzo C. Long-term effects of ocean warming on the prokaryotic community:evidence from the vibrios. The ISME Journal, 2012, 6(1): 21-30. DOI:10.1038/ismej.2011.89 |
| [6] | Lee KK, Liu PC, Huang CY. Vibrio parahaemolyticus infectious for both humans and edible mollusk abalone. Microbes and Infection, 2003, 5(6): 481-485. DOI:10.1016/S1286-4579(03)00065-0 |
| [7] | Martinez-Urtaza J, Bowers JC, Trinanes J, DePaola A. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Research International, 2010, 43(7): 1780-1790. DOI:10.1016/j.foodres.2010.04.001 |
| [8] | Iwamoto Y, Suzuki Y, Kurita A, Watanabe Y, Shimizu T, Ohgami H, Yanagihara Y. Rapid and sensitive PCR detection of Vibrio trachuri pathogenic to Japanese horse mackerel (Trachurus japonicus). Microbiology and Immunology, 1995, 39(12): 1003-1006. DOI:10.1111/j.1348-0421.1995.tb03290.x |
| [9] | Deepanjali A, Kumar HS, Karunasagar I, Karunasagar I. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Applied and Environmental Microbiology, 2005, 71(7): 3575-3580. DOI:10.1128/AEM.71.7.3575-3580.2005 |
| [10] | Velazquez-Roman J, León-Sicairos N, Flores-Villaseñor H, Villafaña-Rauda S, Canizalez-Roman A. Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Applied and Environmental Microbiology, 2012, 78(6): 1794-1803. DOI:10.1128/AEM.06953-11 |
| [11] | Okuda J, IshibashI M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. Journal of Clinical Microbiology, 1997, 35(12): 3150-3155. DOI:10.1128/JCM.35.12.3150-3155.1997 |
| [12] | Wong HC, Liu SH, Wang TK, Lee CL, Chiou CS, Liu DP, Nishibuchi M, Lee BK. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Applied and Environmental Microbiology, 2000, 66(9): 3981-3986. DOI:10.1128/AEM.66.9.3981-3986.2000 |
| [13] | Ansaruzzaman M, Marcelino L, Deen JL, Bhuiyan NA, Wang XY, Safa A, Sultana M, Chowdhury A, Nair GB, Sack DA, von Seidlein L, Puri MK, Ali M, Chaignat CL, Clemens JD, Barreto A. Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique:spread of the pandemic into the African continent. Journal of Clinical Microbiology, 2005, 43(6): 2559-2562. DOI:10.1128/JCM.43.6.2559-2562.2005 |
| [14] | Nishibuchi M, Fasano A, Russell RG, Kaper JB. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infection and Immunity, 1992, 60(9): 3539-3545. DOI:10.1128/IAI.60.9.3539-3545.1992 |
| [15] | Raimondi F, Kao JPY, Fiorentini C, Fabbri A, Donelli G, Gasparini N, Rubino A, Fasano A. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infection and Immunity, 2000, 68(6): 3180-3185. DOI:10.1128/IAI.68.6.3180-3185.2000 |
| [16] | Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infection and Immunity, 2010, 78(4): 1772-1780. DOI:10.1128/IAI.01051-09 |
| [17] | Matsuda S, Okada N, Kodama T, Honda T, Iida T. A cytotoxic type III secretion effector of Vibrio parahaemolyticus targets vacuolar H+-ATPase subunit c and ruptures host cell lysosomes. PLoS Pathogens, 2012, 8(7): e1002803. DOI:10.1371/journal.ppat.1002803 |
| [18] | Yu Y, Yang H, Li J, Zhang PP, Wu BB, Zhu BL, Zhang Y, Fang WH. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Archives of Microbiology, 2012, 194(10): 827-835. DOI:10.1007/s00203-012-0816-z |
| [19] | Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One, 2013, 8(4): e61086. DOI:10.1371/journal.pone.0061086 |
| [20] | Broberg CA, Calder TJ, Orth K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes and Infection, 2011, 13(12/13): 992-1001. |
| [21] | Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infection and Immunity, 2004, 72(11): 6659-6665. DOI:10.1128/IAI.72.11.6659-6665.2004 |
| [22] | Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(34): 12497-12502. DOI:10.1073/pnas.0802773105 |
| [23] | Jones JL, Lüdeke CHM, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. Journal of Clinical Microbiology, 2012, 50(7): 2343-2352. DOI:10.1128/JCM.00196-12 |
| [24] | Boyd EF, Cohen ALV, Naughton LM, Ussery DW, Binnewies TT, Stine OC, Parent MA. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiology, 2008, 8: 110. DOI:10.1186/1471-2180-8-110 |
| [25] | Dor S, Herman G, Updegraff BL. Vibrio parahaemolyticus Type VI Secretion System 1 is activated in marine conditions to target bacteria, and is differentially regulated from System 2. PLos One, 2013, 8(4): e61086. DOI:10.1371/journal.pone.0061086 |
| [26] | Ceccarelli D, Hasan NA, Huq A, Colwell RR. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Frontiers in Cellular and Infection Microbiology, 2013, 3: 97. |
| [27] | Troisfontaines P. Type III Secretion:More Systems Than You Think. Physiology, 2005, 20(5): 326-339. DOI:10.1152/physiol.00011.2005 |
| [28] | Yang J, Tauschek M, Robins-Browne RM. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends in Microbiology, 2011, 19(3): 128-135. DOI:10.1016/j.tim.2010.12.001 |
| [29] | Hovey AK, Frank DW. Analyses of the DNA-binding and ranscriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. Journal of Bacteriology, 1995, 177(15): 4427-4436. DOI:10.1128/JB.177.15.4427-4436.1995 |
| [30] | Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiology and Molecular Biology Reviews, 1998, 62(2): 379-433. |
| [31] | Liu AC, Thomas NA. Transcriptional profiling of Vibrio parahaemolyticus exsA reveals a complex activation network for type III secretion. Frontiers in Microbiology, 2015, 6: 1089. |
| [32] | Kodama T, Yamazaki C, Park KS, Akeda Y, Iida T, Honda T. Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiology Letters, 2010, 311(1): 10-17. DOI:10.1111/j.1574-6968.2010.02066.x |
| [33] | Zhou XH, Konkel ME, Call DR. Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence, 2010, 1(4): 260-272. DOI:10.4161/viru.1.4.12318 |
| [34] | Nydam SD, Shah DH, Call DR. Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Frontiers in Cellular and Infection Microbiology, 2014, 4(1): 47-61. |
| [35] | Erwin DP, Nydam SD, Call DR. Vibrio parahaemolyticus ExsE is requisite for initial adhesion and subsequent type III secretion system 1-dependent autophagy in HeLa cells. Microbiology, 2012, 158(9): 2303-2314. DOI:10.1099/mic.0.059931-0 |
| [36] | Williams SG, Manning PA. Transcription of the Vibrio cholerae haemolysin gene, hlyA, and cloning of a positive regulatory locus, hlyU. Molecular Microbiology, 1991, 5(8): 2031-2038. DOI:10.1111/j.1365-2958.1991.tb00825.x |
| [37] | Mukherjee D, Pal A, Chakravarty D, Chakrabarti P. Identification of the target DNA sequence and characterization of DNA binding features of HlyU, and suggestion of a redox switch for hlyA expression in the human pathogen Vibrio cholerae from in silico studies. Nucleic Acids Research, 2015, 43(3): 1407-1417. |
| [38] | Liu MQ, Alice AF, Naka H, Crosa JH. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infection and Immunity, 2007, 75(7): 3282-3289. DOI:10.1128/IAI.00045-07 |
| [39] | Li L, Mou XY, Nelson DR. HlyU is a positive regulator of hemolysin expression in Vibrio anguillarum. Journal of Bacteriology, 2011, 193(18): 4779-4789. DOI:10.1128/JB.01033-10 |
| [40] | Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes & Development, 2007, 21(12): 1456-1471. DOI:10.1101/gad.1543107 |
| [41] | Getz LJ, Thomas NA. The transcriptional regulator HlyU positively regulates expression of exsA, leading to type III secretion system 1 activation in Vibrio parahaemolyticus. Journal of Bacteriology, 2018, 200(15): e00653-17. DOI:10.1128/JB.00653-17 |
| [42] | Zhou XH, Shah DH, Konkel ME, Call DR. Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Molecular Microbiology, 2008, 69(3): 747-764. DOI:10.1111/j.1365-2958.2008.06326.x |
| [43] | Crawford JA, Krukonis ES, Dirita VJ. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Molecular Microbiology, 2003, 47(5): 1459-1473. DOI:10.1046/j.1365-2958.2003.03398.x |
| [44] | Dirita VJ, Parsot C, Jander G, Mekalanos JJ. Regulatory cascade controls virulence in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America, 1991, 88(12): 5403-5407. DOI:10.1073/pnas.88.12.5403 |
| [45] | Hung DT, Mekalanos JJ. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(8): 3028-3033. DOI:10.1073/pnas.0409559102 |
| [46] | Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(18): 10220-10224. DOI:10.1073/pnas.170219997 |
| [47] | Valeru SP, Wai SN, Saeed A, Sandström G, Abd H. ToxR of Vibrio cholerae affects biofilm, rugosity and survival with Acanthamoeba castellanii. BMC Research Notes, 2012, 5: 33. DOI:10.1186/1756-0500-5-33 |
| [48] | Khouadja S, Suffredini E, Baccouche B. Occurrence of virulence genes among Vibrio cholerae and Vibrio parahaemolyticus strains from treated wastewaters. Environmental Monitoring & Assessment, 2014, 186(10): 6935-6945. DOI:10.1007/s10661-014-3900-9 |
| [49] | Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infection and Immunity, 2012, 80(5): 1834-1845. DOI:10.1128/IAI.06284-11 |
| [50] | Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel zur Wiesch P, Zhou XH, Davis BM, Waldor MK. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(22): 6283-6288. DOI:10.1073/pnas.1601718113 |
| [51] | Gode-Potratz CJ, Chodur DM, McCarter LL. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. Journal of Bacteriology, 2010, 192(22): 6025-6038. DOI:10.1128/JB.00654-10 |
| [52] | Osei-Adjei G, Gao H, Zhang Y, Zhang LY, Yang WH, Yang HY, Yin Z, Huang XX, Zhang YQ, Zhou DS. Regulatory actions of ToxR and CalR on their own genes and type III secretion system 1 in Vibrio parahaemolyticus. Oncotarget, 2017, 8(39): 65809-65822. DOI:10.18632/oncotarget.19498 |
| [53] | Yanagihara I, Nakahira K, Yamane T, Kaieda S, Mayanagi K, Hamada D, Fukui T, Ohnishi K, Kajiyama S, Shimizu T, Sato M, Ikegami T, Ikeguchi M, Honda T, Hashimoto H. Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin. Journal of Biological Chemistry, 2010, 285(21): 16267-16274. DOI:10.1074/jbc.M109.074526 |
| [54] | Okuda J, Nishibuchi M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct haemolysin gene. Molecular Microbiology, 1998, 30(3): 499-511. DOI:10.1046/j.1365-2958.1998.01072.x |
| [55] | Lin Z, Kumagai K, Baba K, Mekalanos JJ, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. Journal of Bacteriology, 1993, 175(12): 3844-3855. DOI:10.1128/JB.175.12.3844-3855.1993 |
| [56] | Zhang YQ, Zhang Y, Gao H, Zhang LY, Yin Z, Huang XX, Zhou DS, Yang HY, Yang WH, Wang L. Vibrio parahaemolyticus CalR down regulates the thermostable direct hemolysin (TDH) gene transcription and thereby inhibits hemolytic activity. Gene, 2017, 613: 39-44. DOI:10.1016/j.gene.2017.03.001 |
| [57] | Zhang YQ, Gao H, Osei-adjei G, Zhang Y, Yang WH, Yang HY, Yin Z, Huang XX, Zhou DS. Transcriptional regulation of the type VI secretion system 1 genes by quorum sensing and ToxR in Vibrio parahaemolyticus. Frontiers in Microbiology, 2017, 8: 2005. DOI:10.3389/fmicb.2017.02005 |
| [58] | Okada R, Matsuda S, Iida T. Vibrio parahaemolyticus VtrA is a membrane-bound regulator and is activated via oligomerization. PLoS One, 2017, 12(11): e0187846. DOI:10.1371/journal.pone.0187846 |
| [59] | Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, Park KS, Cantarelli VV, Dryselius R, Iida T, Honda T. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One, 2010, 5(1): e8678. DOI:10.1371/journal.pone.0008678 |
| [60] | Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, Orth K. Bile salt receptor complex activates a pathogenic type III secretion system. eLife, 2016, 5: e15718. DOI:10.7554/eLife.15718 |
| [61] | Rivera-Cancel G, Orth K. Biochemical basis for activation of virulence genes by bile salts in Vibrio parahaemolyticus. Gut Microbes, 2017, 8(4): 366-373. DOI:10.1080/19490976.2017.1287655 |
| [62] | Shi MT, Li N, Xue YY, Zhong ZT, Yang MH. The 58th cysteine of TcpP is essential for Vibrio cholerae virulence factor production and pathogenesis. Frontiers in Microbiology, 2020, 11: 118. DOI:10.3389/fmicb.2020.00118 |
| [63] | Givskov M, Rasmussen TB, Ren DV, Balaban N. Bacterial cell-to-cell communication (quorum sensing)//Balaban M. Control of Biofilm Infections by Signal Manipulation. Berlin, Heidelberg: Springer, 2007: 13-38. 10.1007/7142_2007_007 |
| [64] | Srivastava D, Waters CM. A tangled web:regulatory connections between quorum sensing and cyclic Di-GMP. Journal of Bacteriology, 2012, 194(17): 4485-4493. DOI:10.1128/JB.00379-12 |
| [65] | Zhang YQ, Qiu YF, Tan YF, Guo ZB, Yang RF, Zhou DS. Transcriptional regulation of opaR, qrr2-4and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS One, 2012, 7(4): e34622. DOI:10.1371/journal.pone.0034622 |
| [66] | Sun FJ, Zhang YQ, Wang L, Yan XJ, Tan YF, Guo ZB, Qiu JF, Yang RF, Xia PY, Zhou DS. Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus. PLoS One, 2012, 7(9): e44210. DOI:10.1371/journal.pone.0044210 |
| [67] | Wang L, Ling Y, Jiang HW, Qiu YF, Qiu JF, Chen HP, Yang RF, Zhou DS. AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. International Journal of Food Microbiology, 2013, 160(3): 245-251. DOI:10.1016/j.ijfoodmicro.2012.11.004 |
| [68] | Zhang YQ, Hu LH, Qiu Y, Osei-Adjei G, Tang H, Zhang Y, Zhang R, Sheng XM, Xu SG, Yang WH, Yang HY, Yin Z, Yang RF, Huang XX, Zhou DS. QsvR integrates into quorum sensing circuit to control Vibrio parahaemolyticus virulence. Environmental Microbiology, 2019, 21(3): 1054-1067. DOI:10.1111/1462-2920.14524 |
| [69] | Gode-Potratz CJ, McCarter LL. Quorum sensing and silencing in Vibrio parahaemolyticus. Journal of Bacteriology, 2011, 193(16): 4224-4237. DOI:10.1128/JB.00432-11 |
| [70] | Wang L, Zhou DS, Mao PY, Zhang YQ, Hou J, Hu Y, Li J, Hou SJ, Yang RF, Wang RH, Qiu JF. Cell density-and quorum sensing-dependent expression of type VI secretion system 2 in Vibrio parahaemolyticus. PLoS One, 2013, 8(8): e73363. DOI:10.1371/journal.pone.0073363 |
| [71] | Wu CQ, Zhang T, Zhang WW, Shi MT, Tu F, Yu A, Li MM, Yang MH. Two DsbA proteins are important for Vibrio parahaemolyticus pathogenesis. Frontiers in Microbiology, 2019, 10: 1103. DOI:10.3389/fmicb.2019.01103 |
 2020, Vol. 60
2020, Vol. 60