中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Xiaoxiao Dang, Lei Zhang, Wei Wang, Guangmei Wang, Runjin Liu, Zhihong Xie. 2020
- 党消消, 张蕾, 王伟, 王光美, 刘润进, 解志红. 2020
- Soil microbial community structure along the ecological succession in Yellow River Delta
- 黄河三角洲原生演替中土壤微生物群落结构分析
- Acta Microbiologica Sinica, 60(6): 1272-1283
- 微生物学报, 60(6): 1272-1283
-
文章历史
- 收稿日期:2020-01-07
- 修回日期:2020-04-11
- 网络出版日期:2020-04-26
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, Shandong Province, China;
4. School of Life Sciences, Yantai University, Yantai 264005, Shandong Province, China;
5. College of Plant Healthy and Medicine, Qingdao Agricultural University, Qingdao 266000, Shandong Province, China
2. 中国科学院大学, 北京 100049;
3. 中国科学院海洋大科学研究中心, 山东 青岛 266071;
4. 烟台大学生命科学学院, 山东 烟台 264005;
5. 青岛农业大学植物医学学院, 山东 青岛 266000
The Yellow River Delta is the second largest estuary delta in China, which is one of the areas with the highest biodiversity in the world[1]. The succession process occurs from ocean to terrestrial land in this region, along with a decrease in the soil salt content and an increase in the organic matter content. At present, few studies of the microbial characteristic associated with different successional process in the wetlands have been reported. This estuarine delta is naturally characterized by extensive coverage of primary salinization, and different types of halophytes grow along the natural saline gradient. Saline-alkali stress is one of the main abiotic factors that greatly restricts the survival, growth, and productivity of plants by affecting their physiological and development processes[1-2]. However, soil microorganisms can promote the growth and yield of the plants by increasing their tolerance to salinity and nutrient availability[3-5]. In turn, halophytes could improve the microenvironments of the soil, which promotes the survival of rhizosphere microorganisms[6].
Soil microorganism play critical roles in maintaining the ecological balance and have been regarded as sensitive indicators of the soil environments[7-8]. Microorganisms play important roles in biogeochemical cycling in the rhizosphere via nitrogen fixation, phosphorus uptake, and nutrient cycling[9-10]. The composition and abundance of soil bacteria are often related to soil chemistry and plant species[11-14]. Many studies have demonstrated that soil pH and salinity are the main factors that affect the composition of microbial communities[15-18]. In a global survey of bacterial communities from terrestrial environments, salinity has been reported to be the major environmental determinant of microbial community composition[19-20]. The structure of soil bacterial communities has also been shown to be associated with differences in soil pH[21-22]. In addition, plant species can influence the soil characteristics and microbial communities through the excretion of root exudates and oxygen into the rhizosphere[23-25]. However, the combined effects of plant species and soil properties on soil microorganisms are poorly understood.
The Yellow River Delta forms a natural saline-alkali gradient from the coastal to terrestrial land. Suaeda glauca (SG), Glycine soja and Phragmites australis (GP) are three typical plant communities along the succession region in the Yellow River Delta since 1990s. In this study, we compared the microbial community compositions and functional gene contents of rhizosphere microbial communities of Suaeda glauca (SG) and Glycine soja-Phragmites australis (GP). We compared halophyte rhizosphere microbial communities with the bulk soils of barren wetland in the coastal area of the Yellow River Delta. Therefore, the aim of this study was to investigate the combined effects of different vegetation covers and soil factors on both microbial communities and functional genes in Yellow River Delta rhizosphere soils.
1 Materials and Methods 1.1 Site description and samplingThree sampling sites were located in the Yellow River Delta (119.1°E, 37.7°N) in Dongying City, Shandong Province, China. The rhizosphere soils were collected as described previously with a few modifications[26]. A nested five-point sampling method was used. At each sampling site, surface sediments (0-5 cm) were sampled at five points, and the distance between each point was 5 m. Three subsamples were collected in each sampling site. After that, all the subsamples of each site were manually mixed to homogeneity and immediately transported to the laboratory. The soil samples were divided into two parts. One was stored at 4 ℃ to analyze its soil properties, while the other was stored at -80 ℃ until DNA extraction.
1.2 Soil physiochemical propertiesThe soil pH was measured as previously described using a pH meter (Mettler Toledo Instruments, Shanghai, China)[27]. The electrical conductivity (EC) was measured using a conducting meter (DDS-11A, Shanghai, China). The total organic carbon (TC), total nitrogen (TN), and total phosphorus (TP) were measured as previously described[28].
1.3 Metagenome sequencing and assemblyIllumina HiSeq-2000 sequencing of metagenomic DNA from the three samples generated 239489174 raw reads with a total of 24.19 billion bps. Sequences were cleaned and assembled using SeqPrep (https://github.com/jstjohn/SeqPrep), Sickle (https://github.com/najoshi/sickle) and SOAP[29]. Open reading frames (ORFs) were predicted using MetaGene software[30]. The ORFs with a length of ≥100 bp were selected and translated into amino acid sequences using the NCBI database. The predicted genes (with 95% sequence identity and 90% coverage) were clustered by CD-HIT software[31], and the longest sequences from each cluster were selected as representative sequences to build a non-redundant gene catalog. The reads were then mapped to the representative sequences with 95% identity using SOAP aligner[32], and the gene abundance in each sample was evaluated.
Representative sequences of the non-redundant gene catalog were aligned to the NR database using BLASTp for taxonomic annotations. The reads were also used for the taxonomic assessment by screening for SSU rRNAs using Metaxa2[33]. The non-redundant gene catalog was aligned against the Non-supervised Orthologous Groups (eggNOG) database using BLASTp for Clusters of Orthologous Groups (COG) annotation with an E-value cut-off of 1e-5[34]. The non-redundant gene catalog was then aligned against the Kyoto Encyclopedia of Genes and Genomes (KEGG) using BLASTp against the KEGG database with an E-value cutoff of 1e-5[35].
1.4 Accession numberThe raw sequencing data have been deposited into the NCBI Sequence Read Archive (SRA) database. The SRP accession number is SRP253127.
2 Results and Discussion 2.1 Soil propertiesSoil samples were taken from three sample sites, where the vegetation was Suaeda glauca (SG), Glycine soja-Phragmites australis (GP) and barren wetland (BW). The chemical composition of the soils is summarized in Table 1. All the soil samples were highly saline and had a high pH. The pH values are alkaline and decline along from the land to coast. The electrical conductivity (EC) values varied from 7.42 to 8.98. A previous study showed that the main component of soil salinity is sodium chloride, which exhibits a significant linear correlation with the EC in Yellow River Delta[36]. The total nitrogen and total carbon were highest in GP, which could have resulted from the improvement of soil nutrients by the legume Glycine soja. It has been estimated that soil microorganisms require a C:N ratio of approximately 25:1 to reach 40% of their growth efficiency[37]. The C:N ratio varied from 14.98 to 20.43 in the three samples, indicating that the microbial communities facilitated a high degree of organic nitrogen mineralization of the soil organic matter.
| Sample ID | Description | TN/% | TC/% | TP/% | C:N | pH | EC/(dsm-1) |
| SG | Suaeda glauca | 0.09±0.02 b | 1.79±0.07 b | 0.11±0.01 a | 20.43±2.4 a | 8.61±0.01 a | 7.42±0.09 b |
| GP | Glycine soja-Phragmites australis | 0.22±0.14 a | 3.31±0.20 a | 0.11±0.02 a | 14.98±1.9 a | 8.16±0.101 b | 8.98±0.59 a |
| BW | Barren wetland | 0.10±0.13 b | 1.76±0.14 b | 0.09±0.04 b | 17.86±1.6 a | 7.93±0.02 c | 7.52±0.01 b |
| Values are the means±standard deviation (SD; n=3). Values within a column followed by different lowercase letters indicate a statistically significant difference at P < 0.05. TN: total nitrogen; TC: total organic carbon; TP: total phosphorus; EC: electrical conductivity. | |||||||
2.2 Microbial taxonomic analysis at the phylum level
We first explored the microbial community composition by analyzing the reads using the ribosomal small subunit (SSU)[38]. The results demonstrated that the niche contains very high bacterial abundance (90%-93%), with a significantly lower proportion of Eukaryota (1.90%-2.88%) and Archaea (0.43%-3.56%) (Figure 1-A). The populations of Eukaryota and Archaea are found in low amounts in the three soil samples, which is consistent with the results of other environments[39-40]. The taxonomic analysis of SSUs of the bacterial fraction revealed that Proteobacteria was the most prevalent phylum represented in the metagenome, while other phyla, such as Acidobacteria, Chloroflexi, Actinobacteria, Bacteroidetes, Planctomycetes, and Gemmatimonadetes, were present as relatively minor contributors to the total bacterial abundance (Figure 1-A).

|
| Figure 1 The relative abundance of different species at the phylum level by taxonomic classification using the ribosomal small subunit (SSU) genes (A) and the relative reads number (B). BW: barren wetland soil; SG: Suaeda glauca rhizosphere soil; GP: Glycine soja-Phragmites australis rhizosphere soil |
We further analyzed the microbial community structure of the three samples by comparing the relative reads number. The results showed that there were 10 phyla in common that comprised a total of 90.32%, 85.05% and 85.95% in SG, GP, and BW, respectively. As shown in Figure 1-B, the dominant phyla in the three sample soils were Proteobacteria, Firmicutes, Actinobacteria and Chloroflexi. Proteobacteria was the most abundant phylum in all samples, accounting for a total of 71.5%, 61.4% and 55.5% of SG, GP and BW, respectively. Many studies have shown that Proteobacteria predominate in different soil types, which could indicate an involvement in the biogeochemical process in ecosystems[41-44]. The differences in distribution are probably related to the organic matter content in the soil, since the relative abundance of Proteobacteria can be influenced by nutrients availability[45].
The next most abundant phylum varied among the three samples, Bacteriodetes (5.87%) in SG, Actinobacteria (5.88%) in GP, and Gemmatimonadetes (7.89%) in BW, respectively. These results are consistent with those of previous studies, in which Bacteroidetes, Actinobacteria and Gemmatimonadetes were reported to be prevalent in the saline-alkaline sediment environment[46-48]. Bacteriodetes and Actinobacteria have been demonstrated to thrive in plant soils, while Gemmatimonadetes were enriched in barren soils in the Yellow River Delta[49]. The relative abundance of Bacteriodetes was decreased from 5.87% (SG) to 0.6% (GP), whereas that of Acidobacteria was increased from 0.50% (SG) to 5.88% (GP) and that of Nitrospirae was increased from 0.32% (SG) to 2.49% (GP). Since Acidobacteria serve as decomposers, they participate in the cycling of organic matter derived from plants, fungi and insects[50]. In this study, the relative abundance of Acidobacteria was higher in the rhizospheres of GP, which could be related to the higher contents of root exudates in that region.
2.3 Microbial taxonomic analysis at the genus levelThe richness of the bacterial community at the general level is illustrated in Figure 2. Twenty-four main genera (each with a relative abundance of > 1%) were detected in the three soil samples, and the main genera represented 15.83%-22.88%. The composition of bacterial communities differed substantially across different samples. Marinobacter (3.94%), Streptococcus (3.92%), Alcanivorax (2.83%), Gemmatimonas (1.73%), Pseudomonas (1.25%), Steroidobacter (1.09%), Thioalkalivibrio (1.07%), and Fluviicola (1.03%) were the dominant genera in SG. Sinorhizobium (10.82%), Nitrospira (2.32%), Pseudolabrys (2.25%), Pyrinomonas (1.87%), Bradyrhizobium (1.68%), Streptococcus (1.49%), Lysobacter (1.40%), and Rhodoplanes (1.06%) were the dominant genera in GP. Gemmatimonas (6.81%), Kiloniella (1.51%), Streptococcus (1.38%), Alcanivorax (1.38%), Fodinicurvata (1.37%), Rhodovibrio (1.25%), Azospirillum (1.14%), and Thioalkalivibrio (1.00%) were the dominant genera in BW. Among the top 24 genera, the dominant genera were composed of Sinorhizobium, Gemmatimonas, Marinobacter and Streptococcus, many species of which have been reported to have beneficial effects for C or N cycling in the soil[48]. In contrast with those in the BW and SG, the main microbes in the communities of GP at later succession stages included the genera of Sinorhizobium (10.82%), Nitrospira (2.32%), and Bradyrhizobium (1.68%). These species have been demonstrated to play important roles in nitrogen cycle[51]. The legume Glycine soja appears to facilitate the expansion of the Sinorhizobium species in GP, since this microorganism is a nitrogen-fixing mutualistic symbiont of the legume. The relative lower population of Bradyrhizobium could be attributed to the influence of saline soil. Previous studies have identified that Sinorhizobium as the dominant species in alkaline-saline soil, whereas Bradyrhizobium was found to be dominant in neutral to acidic soil[52-54]. Nitrospira plays a role in nitrification, which is also exist in YRD agriculture soils in previous studies[55-56]. In concert, these results suggest that different vegetation covers and saline soils have an important impact on the bacterial communities.

|
| Figure 2 Bacterial community structure of each sampling point at the genus level |
2.4 Distribution of putative metabolic genes of microbial communities
To better understand the functional potential of the microorganisms represented in the three sampling sites, the metabolic functions of the microbial communities in SG, GP and BW were investigated by aligning to the eggNOG databases. The results demonstrated that the number of total predicted functional genes of SG and GP was 25.2% and 71.2% lower than those of the BW, respectively (Figure 3-A). In addition to the function of unknown genes, the main functional gene classes in the three samples were amino acid metabolism, energy production and conversion, and inorganic ion transport and metabolism genes (Figure 3-A).
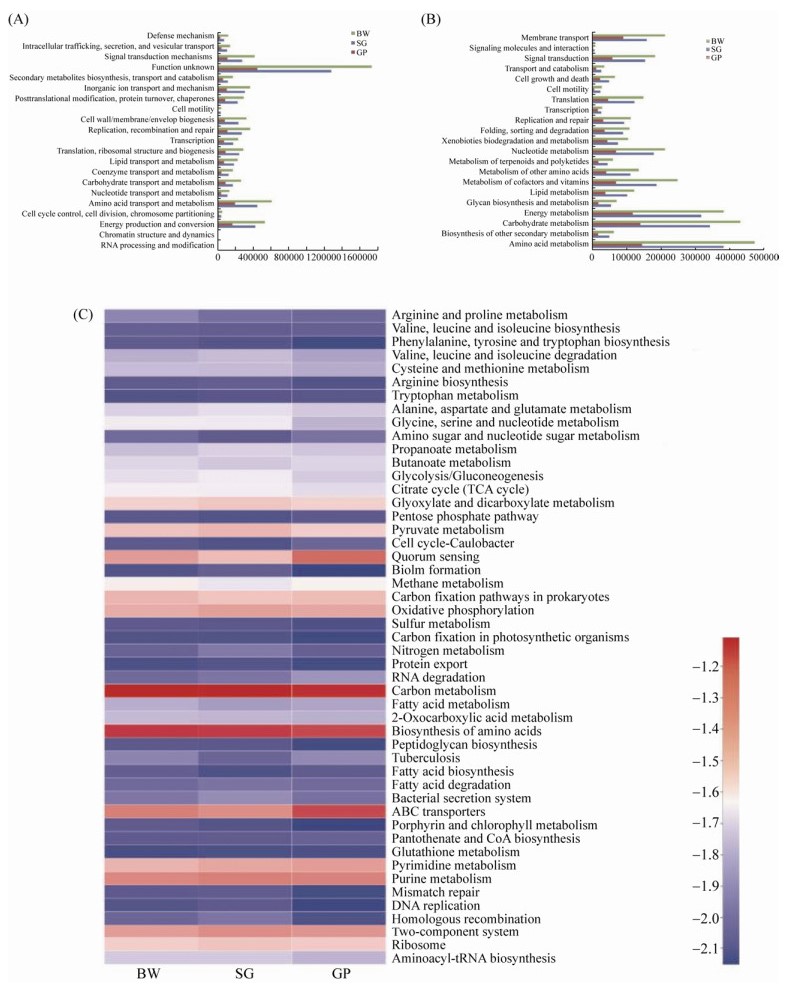
|
| Figure 3 COG (A) and KEGG analysis (B) of the predicted functional genes in the three samples. Heatmap showing the differences of the three investigated communities based on the KEGG level 3 (C) |
Furthermore, the predicted genes were aligned to the KO (Kyoto Encyclopedia of Genes and Genomes Orthology) database. The results demonstrated that the total number of predicted functional genes of SG and GP was 19.8% and 68.0% lower than that of the BW, respectively (Figure 3-B). Many genes were attributed to three categories, namely amino acid transport and metabolism, carbohydrate metabolism, and energy metabolism (Figure 3-B). Moreover, we compared the functional genes at KEGG level 3 using a heatmap analysis. As shown in Figure 3-C, a higher number of sequences in quorum sensing and ABC transporters was found in GP compared with those of SG and BW. The quorum sensing genes were potentially implicated in regulating the legume-rhizobia nodulation symbiosis process[57]. The ABC transporters genes could be related to the high EC values of the soil sample in GP, since the ABC transporter proteins have been demonstrated to play a role in osmotolerance[46, 58]. In summary, our data show that the vegetation types and soil characteristics play important roles in the structure of bacterial communities.
We further compared functional genes that participate in the nitrogen cycling in the three samples. As shown in Figure 4, the ATP-dependent glutamine synthetase-encoding gene glnA had the highest abundance in each sample. The next abundant genes were gltB and gltD, which encode glutamate-oxoglutarate amidotransferase and can cooperate with glnA in indirect ammonia fixation. The abundance of N fixation gene (nifH) in SG and GP was higher than that of BW. And more dissimilatory N reduction gene (nasA, nrfA, and nirADK) was detected in SG than that of GP and BW. For the ammonification process, the abundance of gdh of SG was lower than those of GP and BW. This observation could be related to C:N ratio in the soil, as the ammonification process can be influenced by nitrogen availability[59]. The mere presence of gene does not necessarily mean the fuction, and metatransciptomic analysis and cultivation of targeted functional guilds are warranted for further study.
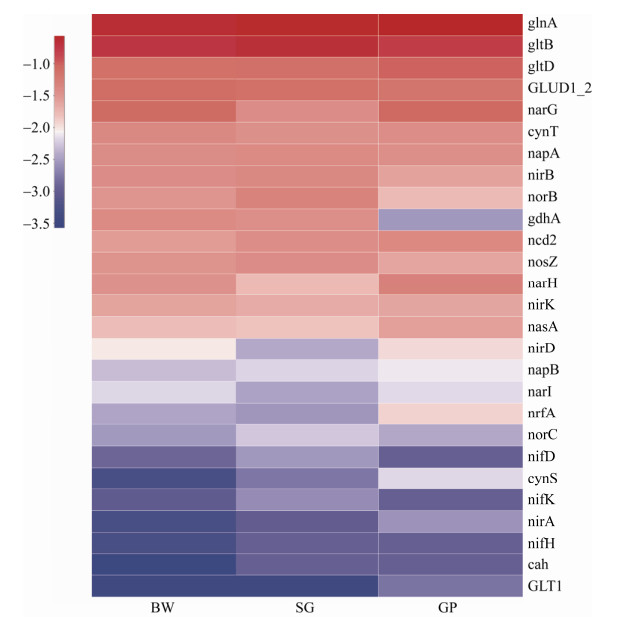
|
| Figure 4 Heatmap analysis of genes potentially involved in nitrogen cycle in the three samples |
| [1] | Cui BS, Yang QC, Yang ZF, Zhang KJ. Evaluating the ecological performance of wetland restoration in the Yellow River Delta, China. Ecological Engineering, 2009, 35(7): 1090-1103. DOI:10.1016/j.ecoleng.2009.03.022 |
| [2] | Fang H, Liu G, Kearney M. Georelational analysis of soil type, soil salt content, landform, and land use in the Yellow River Delta, China. Environmental Management, 2005, 35(1): 72-83. DOI:10.1007/s00267-004-3066-2 |
| [3] | Komaresofla BR, Alikhani HA, Etesami H, Khoshkholgh-Sima NA. Improved growth and salinity tolerance of the halophyte Salicornia sp. by co-inoculation with endophytic and rhizosphere bacteria. Applied Soil Ecology, 2019, 138: 160-170. DOI:10.1016/j.apsoil.2019.02.022 |
| [4] | Shahzad R, Khan AL, Bilal S, Muhammad W, Kang SM, Lee IJ. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environmental and Experimental Botany, 2017, 136: 68-77. DOI:10.1016/j.envexpbot.2017.01.010 |
| [5] | Zhao S, Zhou N, Zhao ZY, Zhang K, Wu GH, Tian CY. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Current Microbiology, 2016, 73(4): 574-581. DOI:10.1007/s00284-016-1096-7 |
| [6] | Munroe JW, Soto G, Virginio Filho EDM, Fulthorpe R, Isaac ME. Soil microbial and nutrient properties in the rhizosphere of coffee under agroforestry management. Applied Soil Ecology, 2015, 93: 40-46. DOI:10.1016/j.apsoil.2015.04.003 |
| [7] | Raiesi F, Salek-Gilani S. The potential activity of soil extracellular enzymes as an indicator for ecological restoration of rangeland soils after agricultural abandonment. Applied Soil Ecology, 2018, 126: 140-147. DOI:10.1016/j.apsoil.2018.02.022 |
| [8] | Khan AG. Rote of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. Journal of Trace Elements in Medicine and Biology, 2005, 18(4): 355-364. DOI:10.1016/j.jtemb.2005.02.006 |
| [9] | Zhang D, Wang C, Li X, Yang X, Zhao L, Liu L, Zhu C, Li R. Linking plant ecological stoichiometry with soil nutrient and bacterial communities in apple orchards. Applied Soil Ecology, 2018, 126: 1-10. DOI:10.1016/j.apsoil.2017.12.017 |
| [10] | Revillini D, Gehring CA, Johnson NC. The role of locally adapted mycorrhizas and rhizobacteria in plant-soil feedback systems. Functional Ecology, 2016, 30(7): 1086-1098. DOI:10.1111/1365-2435.12668 |
| [11] | Jesus EdC, Marsh TL, Tiedje JM, Moreira FMdeS. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME Journal, 2009, 3(9): 1004-1011. DOI:10.1038/ismej.2009.47 |
| [12] | Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Applied and Environmental Microbiology, 2006, 72(7): 4522-4531. DOI:10.1128/AEM.00149-06 |
| [13] | Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology, 2009, 68(1): 1-13. |
| [14] | Tian XY, Zhang CS. Illumina-based analysis of endophytic and rhizosphere bacterial diversity of the coastal halophyte Messerschmidia sibirica. Frontiers in Microbiology, 2017, 8: 2288. DOI:10.3389/fmicb.2017.02288 |
| [15] | Wang ZY, Xin YZ, Gao DM, Li FM, Morgan J, Xing BS. Microbial community characteristics in a degraded wetland of the Yellow River Delta. Pedosphere, 2010, 20(4): 466-478. DOI:10.1016/S1002-0160(10)60036-7 |
| [16] | Yuan BC, Li ZZ, Liu H, Gao M, Zhang YY. Microbial biomass and activity in salt affected soils under and conditions. Applied Soil Ecology, 2007, 35(2): 319-328. DOI:10.1016/j.apsoil.2006.07.004 |
| [17] | Tripathi S, Kumari S, Chakraborty A, Gupta A, Chakrabarti K, Bandyapadhyay BK. Microbial biomass and its activities in salt-affected coastal soils. Biology and Fertility of Soils, 2006, 42(3): 273-277. DOI:10.1007/s00374-005-0037-6 |
| [18] | Rietz DN, Haynes RJ. Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biology and Biochemistry, 2003, 35(6): 845-854. DOI:10.1016/S0038-0717(03)00125-1 |
| [19] | Lozupone CA, Knight R. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(27): 11436-11440. DOI:10.1073/pnas.0611525104 |
| [20] | Campbell BJ, Kirchman DL. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME Journal, 2013, 7(1): 210-220. DOI:10.1038/ismej.2012.93 |
| [21] | Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 2009, 75(15): 5111-5120. DOI:10.1128/AEM.00335-09 |
| [22] | Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME Journal, 2010, 4(10): 1340-1351. DOI:10.1038/ismej.2010.58 |
| [23] | Dinesh R, Srinivasan V, Hamza S, Parthasarathy VA, Aipe KC. Physico-chemical, biochemical and microbial properties of the rhizospheric soils of tree species used as supports for black pepper cultivation in the humid tropics. Geoderma, 2010, 158(3/4): 252-258. |
| [24] | Cao D, Shi F, Koike T, Lu Z, Sun J. Halophyte plant communities affecting enzyme activity and microbes in saline soils of the Yellow River Delta in China. Clean Soil Air Water, 2014, 42(10): 1433-1440. DOI:10.1002/clen.201300007 |
| [25] | Stringlis IA, Yu K, Feussner K, de Jonge R, van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(22): E5213-E5222. DOI:10.1073/pnas.1722335115 |
| [26] | Chaudhary DR, Gautam RK, Yousuf B, Mishra A, Jha B. Nutrients, microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Applied Soil Ecology, 2015, 91: 16-26. DOI:10.1016/j.apsoil.2015.02.003 |
| [27] | Jing C, Xu Z, Zou P, Tang Q, Li Y, You X, Zhang C. Coastal halophytes alter properties and microbial community structure of the saline soils in the Yellow River Delta, China. Applied Soil Ecology, 2019, 134: 1-7. DOI:10.1016/j.apsoil.2018.10.009 |
| [28] | Walkley A, Black IA. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Science, 1934, 37(1): 29-38. DOI:10.1097/00010694-193401000-00003 |
| [29] | Xie YL, Xie YL, Wu GX, Tang JB, Luo RB, Patterson J, Liu SL, Huang WH, He GZ, Gu SC, Li SK, Zhou X, Lam TW, Li YR, Xu X, Wong GKS, Wang J. SOAP denovo-Trans:de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics, 2014, 30(12): 1660-1666. DOI:10.1093/bioinformatics/btu077 |
| [30] | Noguchi H, Park J, Takagi T. MetaGene:prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Research, 2006, 34(19): 5623-5630. DOI:10.1093/nar/gkl723 |
| [31] | Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT:accelerated for clustering the next-generation sequencing data. Bioinformatics, 2012, 28(23): 3150-3152. DOI:10.1093/bioinformatics/bts565 |
| [32] | Li R, Li Y, Kristiansen K, Wang J. SOAP:short oligonucleotide alignment program. Bioinformatics, 2008, 24(5): 713-714. DOI:10.1093/bioinformatics/btn025 |
| [33] | Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. Metaxa2:improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 2015, 15(6): 1403-1414. DOI:10.1111/1755-0998.12399 |
| [34] | Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database:a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1): 33-36. |
| [35] | Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C, Wei L. KOBAS 2.0:a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 2011, 39: W316-W322. DOI:10.1093/nar/gkr483 |
| [36] | Wang YL, Gong P. Soil salinity measurements on the Yellow River Delta. Journal of Nanjing University (Natural Sciences), 2006, 42(6): 602-610. |
| [37] | Chapin FS, Pamela AM, Harold AM, Chapin MC. Principles of terrestrial ecosystem ecology. New York: Springer-Verlag, 2002. |
| [38] | Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjoling A, Nilsson RH. Metaxa2 Diversity Tools:Easing microbial community analysis with Metaxa2. Ecological Informatics, 2016, 33: 45-50. DOI:10.1016/j.ecoinf.2016.04.004 |
| [39] | Wong FKY, Lacap DC, Lau MCY, Aitchison JC, Cowan DA, Pointing SB. Hypolithic microbial community of quartz pavement in the high-altitude tundra of central Tibet. Microbial Ecology, 2010, 60(4): 730-739. DOI:10.1007/s00248-010-9653-2 |
| [40] | Vikram S, Guerrero LD, Makhalanyane TP, Le PT, Seely M, Cowan DA. Metagenomic analysis provides insights into functional capacity in a hyperarid desert soil niche community. Environmental Microbiology, 2016, 18(6): 1875-1888. DOI:10.1111/1462-2920.13088 |
| [41] | Chen Y, Jiang Y, Huang H, Mou L, Ru J, Zhao J, Xiao S. Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Science of the Total Environment, 2018, 637: 1400-1412. |
| [42] | Yu Y, Wang H, Liu J, Wang Q, Shen T, Guo W, Wang R. Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River Estuary. European Journal Soil Biology, 2012, 4(6): 12-21. |
| [43] | Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N. A global atlas of the dominant bacteria found in soil. Science, 2018, 359(6373): 320-325. DOI:10.1126/science.aap9516 |
| [44] | Hu Y, Wang L, Tang Y, Li Y, Chen J, Xi X. Variability in soil microbial community and activity between coastal and riparian wetlands in the Yangtze River estuary-potential impacts on carbon sequestration. Soil Biology and Biochemistry, 2014, 70: 221-228. DOI:10.1016/j.soilbio.2013.12.025 |
| [45] | Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biology and Biochemistry, 2010, 42(6): 896-903. DOI:10.1016/j.soilbio.2010.02.003 |
| [46] | Ahmed V, Verma MK, Gupta S, Mandhan V, Chauhan NS. Metagenomic profiling of soil microbes to mine salt stress tolerance genes. Frontiers in Microbiology, 2018, 9: 159. DOI:10.3389/fmicb.2018.00159 |
| [47] | Canfora L, Bacci G, Pinzari F, Lo Papa G, Dazzi C, Benedetti A. Salinity and bacterial diversity:To what extent does the concentration of salt affect the bacterial community in a saline soil?. PLoS One, 2014, 9(9): e106662. DOI:10.1371/journal.pone.0106662 |
| [48] | Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K. Gemmatimonas aurantiaca gen. nov., sp nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(4): 1155-1163. DOI:10.1099/ijs.0.02520-0 |
| [49] | Wei GS, Li MC, Shi WC, Tian RM, Chang CY, Wang ZR, Wang NX, Zhao GX, Gao Z. Similar drivers but different effects lead to distinct ecological patterns of soil bacterial and archaeal communities. Soil Biology and Biochemistry, 2020, 144: 107759. DOI:10.1016/j.soilbio.2020.107759 |
| [50] | Ward NL, Challacombe JF, Janssen PH, Bernard H, Coutinho PM, Martin W, Gary X, Haft DH, Michelle S, Jonathan B. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Applied and Environmental Microbiology, 2009, 75(7): 2046-2056. DOI:10.1128/AEM.02294-08 |
| [51] | Beeckman F, Motte H, Beeckman T. Nitrification in agricultural soils:impact, actors and mitigation. Current Opinion in Biotechnology, 2018, 50: 166-173. DOI:10.1016/j.copbio.2018.01.014 |
| [52] | Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(22): 8629-8634. DOI:10.1073/pnas.1120436109 |
| [53] | Man CX, Wang H, Chen WF, Sui XH, Wang ET, Chen WX. Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil, 2008, 310: 77-87. DOI:10.1007/s11104-008-9631-3 |
| [54] | Zhang YM, Li Y, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the north China plain. Applied and Environmental Microbiology, 2011, 77(18): 6331-6342. DOI:10.1128/AEM.00542-11 |
| [55] | Pjevac P, Schauberger C, Poghosyan L, Herbold CW, Van Kessel MA, Daebeler A, Steinberger M, Jetten MS, Lücker S, Wagner M. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.3389/fmicb.2017.01508 |
| [56] | Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [57] | Gonzalez JE, Marketon MM. Quorum sensing in nitrogen-fixing rhizobia. Microbiology and Molecular Biology Reviews, 2003, 67(4): 574-592. DOI:10.1128/MMBR.67.4.574-592.2003 |
| [58] | Takami H, Takaki Y, Uchiyama I. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Research, 2002, 30(18): 3927-3935. DOI:10.1093/nar/gkf526 |
| [59] | Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, Jorgensen BB, Jetten MSM. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(18): 6478-6483. DOI:10.1073/pnas.0502088102 |
 2020, Vol. 60
2020, Vol. 60