中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 陈俊松, 杨渐, 蒋宏忱. 2020
- Junsong Chen, Jian Yang, Hongchen Jiang. 2020
- 湖泊硫循环微生物研究进展
- Research progress on microbes involved in lacustrine sulfur cycling
- 微生物学报, 60(6): 1177-1191
- Acta Microbiologica Sinica, 60(6): 1177-1191
-
文章历史
- 收稿日期:2020-02-08
- 修回日期:2020-04-08
- 网络出版日期:2020-04-26
湖泊是内陆水体的重要组成部分,约占内陆水体总面积的83%[1]。同时,湖泊也是地球表层系统中水、土、气等各个圈层相互作用的连接点[2],且对全球或区域内的物质元素循环具有重要影响[3]。在湖泊生态系统中,微生物是物质循环和能量转化的重要推动者,在维持湖泊生态系统平衡和驱动元素地球化学循环方面起到关键作用[4]。因此,了解微生物种群结构及其组成有助于揭示湖泊生态系统结构和功能[4]。
湖泊环境存在各种各样的有机和无机含硫化合物,比如黄铁矿(FeS2)和石膏(CaSO4)等矿物态含硫化合物,硫化氢和硫酸盐等无机含硫化合物,以及甲硫醇(methanthiol or methyl-mercaptan,MeSH)和二甲基三硫醚(dimethyltrisulfide,DMTS)等有机含硫化合物[5-6]。这些含硫化合物中硫的价态包括–2价到+6价。在湖泊中,微生物可将含硫化合物的硫元素由低价态氧化为更高的价态,同时也可将高价态硫还原为低价态的含硫化合物。例如:微生物可将S2O32–氧化为SO42–,也可将SO42–还原为HS–/S2–。同时,微生物还能将低价态的还原型硫化物(如S2O32–和HS–/S2–)氧化为单质硫(S0),单质硫可被进一步氧化为SO32–和SO42–或还原为HS–/S2–等。上述氧化还原过程构成了微生物驱动的硫元素循环。环境中驱动硫元素循环的微生物类群主要包括硫氧化菌(sulfur-oxidizing bacteria,SOB)和硫酸盐还原菌(sulfate-reducing bacteria,SRB)群[7]。SOB和SRB群落构成和分布受控于环境条件,且不同种类的SOB和SRB执行硫元素循环过程的途径和方式也有差异。因此,研究湖泊硫氧化和硫酸盐还原菌种群构成及其对环境条件的响应规律,对于了解湖泊硫循环过程机制具有重要意义[8]。本文综述湖泊中驱动无机含硫化合物氧化还原的微生物种群多样性及其响应环境变化的规律,并进而为认识湖泊生态系统硫循环过程提供理论参考。
2 湖泊微生物驱动的硫氧化过程 2.1 微生物驱动的无机硫氧化途径湖泊中微生物驱动的硫氧化过程包括无机硫氧化和有机硫氧化。由于湖泊中有机硫化物的存在形式多元化,目前关于微生物对有机硫化物的氧化/脱硫途径研究较少。而对湖泊中无机硫氧化过程的研究较为全面,可细分为硫化物氧化、单质硫氧化、亚硫酸盐氧化和硫代硫酸盐氧化四个过程。本文主要介绍微生物参与的无机硫氧化途径(图 1)。
2.1.1 硫化物氧化是将HS–/S2–转化为单质硫(S0)的过程:
几乎所有硫氧化菌(Sulfur-oxidizing bacteria, SOB)均可执行硫化物氧化。该过程依赖黄素细胞色素c硫化氢脱氢酶(Flavocytochrome c sulfide dehydrogenase,FSDH)和硫化物醌还原酶(Sulfide: quinone reductase,SQR)的催化[9]。SQR将硫化物氧化为单质硫,并通过辅基FAD将e–传递给醌,将醌还原为氢醌的膜蛋白[10]。前人研究表明,某些野生型菌株中缺少细胞色素c和sox基因,但可以氧化硫化物,表明SQR发挥主要作用[9]。目前已鉴定识别出6种SQR (Type-Ⅰ,Ⅱ,Ⅲ,Ⅳ,Ⅴ和Ⅵ),并且同一菌株中存在多种SQR[11]。SQR存在于许多光营养型和化能营养型细菌中,如Gallionella、Hydrogenophaga、Limnohabitans、Methylomonas、Nitrospira、Rhodoferax、Sulfuritalea、Chlorobium和Paracoccus等[12-13]。此外,在多种SOB中,FSDH是细胞周质中的可溶性蛋白或膜结合酶[14]。它主要由较大的黄素蛋白(FccB)和较小的黄素蛋白(FccA)组成,并存在于许多紫硫细菌和绿硫细菌细胞内[9]。大量研究显示FSDH和SQR适应不同的环境条件,例如,SQR适合于硫化物浓度较高的环境,而FSDH适合于硫化物浓度较低的环境[15]。
2.1.2 单质硫氧化包含反向异化亚硫酸盐还原酶(reverse dissimilatory sulfite reduction,rDsr)参与的Dsr途径和类杂二硫化物还原酶(heterodisulide reductases-like,Hdr)参与的Hdr途径:Dsr途径中低分子量的有机硫化物,如谷胱甘肽酰胺过硫化物,被认为是将硫从胞质或胞外硫沉积转移到胞质的载体分子。单质硫进入细胞内,被Cys-SSH硫中继系统运输。Cys-SSH系统包括Rhd、TusA与DsrEFH三种转运蛋白,它们通过Cys-S-将单质硫固定形成Cys-SS-,按顺序传递单质硫[16]。之后DsrC蛋白上2个保守并具有还原活性的CysA和CysB接收DsrEFH转运来的单质硫[17],并在蛋白复合体DsrMKJOP的催化下,将硫转化为三硫过氧化物,最后在DsrAB蛋白催化下形成SO32–并释放[18]。Dsr途径涉及到的基因包括rhd、tusA、dsrE2和dsrAB,前三者编码三种转运蛋白,dsrAB则编码硫化物转化的关键蛋白酶;已知SOB的Dsr途径至少存在tusA和dsrE2基因[18]。Hdr途径是近年来新发现的硫氧化途径,其中类杂二硫化物还原酶由基因簇hdrC1B1A-hdrC2B2编码[19],存在于Ectothiorhodospiraceae (外硫红螺旋菌科)的光营养型SOB[9]。Hdr途径与Dsr途径类似,都需利用硫转运蛋白Rhd、TusA、DsrEFH将硫转移到对应位点催化反应成SO32–[20-22]。不同于Dsr途径的是Hdr途径依赖Hdr复合体进行催化。目前,Hdr途径仍未得到实验证实。
2.1.3 硫代硫酸盐氧化途径:硫代硫酸盐氧化可以分为两种途径,一种是由Sox多酶复合体系参与的Paracoccus硫氧化途径(Paracoccus sulfur oxidation,PSO),直接将S2O32–转化为SO42–且不产生中间产物SO32–[23];另一种是连四硫途径(S4 intermeiate,S4I),在氧化S2O32–过程中形成S4O62–等中间产物[24]。PSO途径相关的基因是sox基因簇,包含SoxRSVWXYZ-ABCDEFGH共15个基因,它们编码了SoxXA、SoxYZ、SoxB和SoxCD四种关键蛋白。首先,异二聚体SoxYZ蛋白与S2O32–结合,在SoxXA蛋白的催化作用下将S2O32–转化为二硫化物,其中一个硫原子被SoxB催化为SO42–,另一个硫原子以SoxY蛋白上硫烷的形式存在,再被SoxCD复合体催化成硫砜,再进一步被SoxB转化为SO42–。在紫硫细菌中,S2O32–氧化过程会出现单质硫,主要原因是缺少SoxCD蛋白[25],但产生单质硫的机理仍不清楚。S4I途径是利用tsdA基因编码的TsdA蛋白酶催化S2O32–形成S4O62–并释放2e–。不同生物来源的TsdA酶表现出不同的催化偏向性,且不同生物体中e–受体也不同。另外,S4O62–与HS-在高pH环境中不稳定,因此S4I途径通常被认为发生在酸性环境。然而,在碱性苏打湖中却往往可以检测到连四硫代还原酶的转录,说明碱性环境中可能存在S4O62–,暗示碱性湖泊中存在新的S4O62–转化途径[26]。
2.1.4 亚硫酸盐的直接氧化和间接氧化途径:直接氧化依赖硫氧化还原酶(sulfur oxygenase/reductase,SOR)催化,通常在胞质膜外进行。该过程最具特征的酶是SorAB复合体[27]。缺少SorB亚基的Sor-A型钼蛋白被称为SorT蛋白,也可以参与亚硫酸盐氧化[28]。前人发现SO32–也可以被Sox系统作为底物氧化[29],与之相关的酶是SoxCD蛋白。SoxCD与SorAB类似,但二者参与的反应不同[30]:SoxCD介导6e–的反应,SorAB介导2e–的反应[15]。间接氧化发生在一些不产氧的紫硫细菌和绿硫细菌细胞质,主要通过腺苷酰硫酸(adenosine-5-phosphosulfate,APS)还原酶和ATP硫酸化酶催化。该过程中SO32–与腺苷酸单磷酸(AMP)在AprABM复合蛋白的作用下形成APS,随后APS在腺苷磷酸硫酰酶(sulfate adenylate transferase,Sat)催化下转化为SO42–和ATP。在AMP转化APS的过程会获得额外的能量,因此与直接氧化相比,间接氧化可能在自然环境更为有利[31]。
2.2 湖泊硫氧化菌种群构成目前已知的SOB主要分布在绿菌门(Chlorobi)、绿弯菌门(Chloroflexi)、厚壁菌门(Firmicutes)和变形菌门(Proteobacteria)[32]。湖泊中栖息着大量的SOB,且不同地球化学条件的湖泊中存在特殊的SOB(表 1)。例如,Kojima等[33]从淡水湖沉积物中分离出一种能氧化硫代硫酸盐、四硫酸盐和单质硫的新菌株Sulfurifustis variabilis skN76T,且其形态随环境温度发生变化。在咸水湖中,以硫代硫酸盐、硫化物和单质硫为底物的Sulfuriflexus mobilis和Thiomicrospira halophila也被分离出来[34-35]。Sorokin等从苏打湖中分离出Thioalkalimicrobium aerophilum、Thioalkalimicrobium sibericum、Thioalkalivibrio versutus、Thioalkalivibrio nitratis和Thioalkalivibrio denitrificans等纯菌株,并发现它们可以在pH > 10的培养基中正常生长[36-37]。因此,极端的湖泊环境中孕育着特殊的SOB,这些SOB是宝贵的生物资源。Yang等[38]通过检测soxB功能基因分析,发现青藏高原北部盐湖中的SOB主要属于Alphaproteobacteria纲和Betaproteobacteria纲。Watanabe等[39]利用aprA功能基因检测发现淡水湖泊SOB来自于Desulfovibrionales、Desulfobacteraceae、Desulfobulbaceae和Peptococcaceae。Kojima等[40]通过aprA、dsrA、sqr和soxB功能基因分析发现,湖泊SOB主要属于Betaproteobacteria,且与Sulfuritalea hydrogenivorans关系最为密切。Vavourakis等[26]
| Methods | Lake types | Identified major SOB groups | References |
| Cultivation | Fresh | Sulfurifustis variabilis | [33] |
| Cultivation | Saline | Sulfuriflexus mobilis, Thiomicrospira halophila | [34-35] |
| Cultivation | Soda | Thioalkalimicrobium aerophilum, Thioalkalimicrobium sibericum, Thioalkalivibrio versutus, Thioalkalivibrio nitratis, Thioalkalivibrio denitrificans | [36-37] |
| aprA function gene | Fresh | Desulfovibrionales, Desulfbactereae, Desulfobubulbaceae, Peptococcaceae | [39] |
| soxB function gene | Saline | Alphaproteobacteria, Betaproteobacteria | [38] |
| aprA, dsrA, sqr, soxB function genes | Saline | Betaproteobacteria, Sulfuritalea hydrogenivorans | [40] |
| Metagenomes and Metatranscriptomes | Soda | Thioalkalivibrio, Thiohalocapsa | [26] |
对西伯利亚苏打湖的宏基因组学研究表明其中的SOB群落主要属于Thioalkalivibrio属和Thiohalocapsa属。然而,硫氧化菌功能基因的参考序列有限,大部分OTU (operational taxonomic units)无法明确划分到某一属[39]。因此,仍需新技术去发掘未鉴定的SOB菌株。
2.3 影响湖泊硫氧化菌群落组成的环境因素湖泊中影响SOB群落组成的环境因素包括湖水氧化还原条件、盐度和pH等。(1)湖水氧化还原条件:湖泊中SOB群落组成随湖水氧化还原条件变化。例如,Sulfuritalea属SOB在滞水层的相对丰度较高,而Sulfurimonas和Sulfuricurvum的SOB则聚集在氧化层以下的区域[40];Thioalkalimicrobium (Gammaproteobacteria)、Rhodobaca和Roseinatronobacter (Alphaproteobacteria)在水体占主导优势,表层沉积物中Thioalkalivibrio相对丰度最高,然而在深层沉积物中Thiohalospira、Thioalkalibacter、Thioalkalispira (Gammaproteobacteria)和Sulfurimonas (Epsilonbactereota) SOB的相对丰度最高[25]。SOB群落组成随湖水氧化还原条件的变化可能与不同SOB的代谢底物差异有关[42]。Sulfuritalea可以氧化S2O32–和单质硫,而不能氧化硫化物[43]。Sulfurimonas和Sulfuricurvum SOB以氧化硫化物为主[41]。因此,硫化物的空间分布是SOB群落组成随湖水氧化还原条件变化的根本原因。(2)盐度:随着湖泊盐度的升高,微生物需要抵抗环境盐渗透压也随之升高,SOB的群落组成也由淡水种群向耐盐和嗜盐的菌群替变。如,本课题组发现淡水湖和微咸水湖中SOB以Betaproteobacteria占主导,高盐湖泊中则是以Alphaproteobacteria和Gammaproteobacteria为主[38]。Thioalkalimicrobium和Thioalkalivibrio是碱性苏打湖中主要的SOB种群,Thioalkalimicrobium在低盐的碱湖富集产物中占主导,而Thioalkalivibrio在高盐碱湖中占优势,且已分离的Thioalkalivibrio属菌株均能生存于4 mol/L Na+浓度的饱和碱盐水中[44]。(3) pH:SOB对pH的适应性非常广,根据其最适生长pH范围可分为嗜酸、嗜碱和中性SOB。碱性湖中,以嗜碱SOB为主,而酸性湖中,则由嗜酸性SOB主导。例如,Acidithiobacillus (Acidithiobacillia,原Gammaproteobacteria[44])和Sulfobacillus (Clostridia)在酸性湖泊中占优势[45];Thioalkalimicrobium和Thioalkalivibrio在各种碱水湖的表层沉积物中分布广泛[26, 47-48]。
2.4 湖泊硫氧化速率及影响因素湖泊中关于无机含硫化合物氧化速率的研究较少。Vavourakis等[26]进行模拟实验评估苏打湖中沉积物的S2O32–氧化速率,黑暗条件下沉积物表面以下0–2 cm区域S2O32–氧化速率约1.4 μmol S/(mL·h),2–4cm区域约0.5 μmol S/(mL·h),而光照环境中沉积物表面以下0–2 cm区域S2O32–氧化速率约0.5 μmol S/(mL·h)。硫代硫酸盐氧化速率与氧气浓度相关,沉积物浅表层氧气浓度高,促进硫氧化过程的进行。其次,部分还原性硫化物(大部分是FeS)被额外氧化,致使检测的氧化产物SO42–含量偏高[26]。光照对硫代硫酸盐氧化速率的影响可能与硫氧化菌的生理活性相关,有研究表明绿硫细菌适宜在低光强的环境中生长,光照条件抑制了沉积物中部分硫氧化菌的活性[49],进而使得硫代硫酸盐氧化速率降低。值得注意的是S2–/HS–离子会与湖泊水体或沉积物孔隙水中的Fe3+、Cu2+反应生成沉淀,影响其生化反应,干扰硫氧化速率的测定。因此,测定速率往往低于实际硫氧化速率。
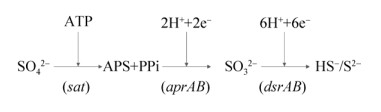
3 湖泊微生物驱动的硫酸盐还原过程 3.1 硫酸盐还原途径硫酸盐还原过程指SO42–转化为还原性硫化物或单质硫的过程。硫酸盐还原可分为同化硫酸盐还原和异化硫酸盐还原两种类型。其中,同化硫酸盐还原需要消耗能量,异化硫酸盐还原是一个产能过程,将硫酸盐作为呼吸链末端的电子受体,生成无机硫化物。硫酸盐还原过程本质是依赖微生物的多种蛋白催化和电子传递。参与微生物硫还原过程的酶主要包括ATP硫酸化酶、APS还原酶(APS reductase,Apr)和异化型亚硫酸盐还原酶(dissimilatory sulfite reductase,Dsr)。硫酸盐还原菌(sulfate reducing bacteria,SRB)参与了湖泊中的整个异化硫酸盐还原过程(图 2)。SO42–化学性质稳定,不易被还原。在异化硫酸盐还原过程中,SO42–首先转化为氧化性较强的腺嘌呤磷酰硫酸盐(APS,adenosine phosphosulphate)。该过程在ATP硫酸化酶的催化下消耗ATP进行。ATP硫酸化酶是一种含有钴和锌的金属蛋白,存在于同化和异化SO42–过程中,由Sat基因编码。然后APS还原为SO32–,SO32–继续还原为硫化物[50]。前者在APS还原酶催化下进行(由aprAB基因编码),后者由异化亚硫酸盐还原酶介导(由dsrAB基因编码,与其相关的蛋白还包括dsrCDE)[50]。异化亚硫酸盐还原酶是硫酸盐还原过程的关键蛋白之一,由α2β2结构组成,2个亚基分别被dsrA和dsrB基因编码表征。然而dsr基因在同一类群的硫酸盐还原菌中相当保守,而在不同类群的硫酸盐还原菌种群中存在较大差异[51]。因此,aprAB和dsrAB基因被广泛用于检测环境样品中的硫酸盐还原菌群落多样性。另外,基于转录组学的研究表明在Mono盐湖中dsrA和aprA基因的丰度随着湖泊深度增加[47];SRB群落多样性在缺氧的湖泊深处更加丰富[52],是因为大部分硫酸盐还原菌是严格厌氧的,只有少部分的脱硫弧菌属(Desulfovibrio)可以在微量氧的环境中生存。同时也有研究报道,SRB的dsrAB基因转录水平与环境SO42–浓度呈正相关[53]。
除dsrAB和aprAB基因外,还存在其他功能基因也与硫酸盐还原过程存在紧密联系。例如,编码醌相互作用膜结合氧化还原酶复合物的基因(qmoABC)被认为是Deltaproteobacteria纲SRB进行硫酸盐还原的必要条件。由qmoABC基因编码的QmoABC蛋白可以作为电子通道将电子从细胞膜传递给AprAB蛋白[54]。
3.2 湖泊硫酸盐还原菌种类构成近年来通过传统分离培养方法,大量的SRB菌株从不同类型湖泊中被分离和鉴定(表 2),如从高盐湖泊中分离的Desulfohalobium utahense[55]和Desulfovibrio oxyclinae[56]、淡水湖泊分离的Desulfosporosinus lacus[57]、寡营养湖泊分离的Desulfosporomusa polytropa[58]等。这些SRB纯菌株一定程度上扩增了我们对SRB生理特性的认识。然而,以上这些SRB纯菌株只能代谢乙酸盐等小分子有机物,且不能还原单质硫。目前,利用大分子有机硫化物和还原单质硫的SRB菌株仍很难分离获得。因此,传统纯培养技术对于我们认识SRB群落多样性仍具有局限性。通过检测环境中SRB硫酸盐还原过程中的关键酶基因(如dsrAB和aprBA)的分析方法,是认识环境SRB群落多样性和分布的最为有效的手段。如,Sorokin等对苏打湖中的dsrB基因研究表明苏打湖中SRB主要属于Desulfovibrionales和Desulfobacteriaceae[59]。Watanabe等检测淡水湖泊中的aprA基因发现SRB群落主要属于Desulfovibrionales、Desulfobulbaceae和Desulfobacteraceae[60]。Qin等[61]利用dsrB功能基因发现西藏盐湖中属于Halomonas和Acintobacter的SRB群落丰度最高。本课题组研究发现,在青藏高原盐湖中dsrB基因主要属于Desulfobacteraceae、Desulfobulbacea和Peptococcaceae[62]。
| Methods | Lake types | Major SRB groups | References |
| Cultivation | Fresh | Desulfosporosinus lacus | [57] |
| Cultivation | Soda | Desulfonatronospira thiodismutans | [45] |
| Cultivation | Saline | Desulfohalobium utahense, Desulfovibrio oxyclinae | [55] |
| aprA function gene | Fresh | Desulfovibrionales, Desulfobulbaceae, Desulfobacteraceae | [60] |
| dsrB function gene | Soda | Desulfovibrionales, Desulfobacteriaceae | [59] |
| dsrB function gene | Saline | Halomonas, Acintobacter | [61] |
| dsrB function gene | Saline | Desulfobacteraceae, Desulfobulbaceae, Peptococcaceae | [62] |
| Metagenomes and Metatranscriptomes | Soda | Deltaproteobacteria | [26] |
研究湖泊中SRB群落组成的方法还有宏基因组学和宏转录组学。相较于功能基因等方法,宏基因组学与宏转录组学可以为SRB菌株分离和生理表征提供新见解[63-64]。例如,Ng等基于宏基因组发现冰湖中SRB与绿硫细菌C-Ace硫化合物交换强烈[65]。Johnson等发现酸性盐湖中大量的基因与硫酸盐转化为APS以及随后从亚硫酸盐中产生硫化物有关[66]。Vavourakis等利用宏基因组学发现,苏打湖中所有编码完整硫酸盐还原途径(sat+aprAB+dsrAB)的物种都来源于Deltaproteobacteria;同时,作者还发现dsrB基因转录高度活跃,并与Desulfococcaceae中一个潜在的新属密切相关[26]。
3.3 影响湖泊硫酸盐还原菌群落组成的环境因素影响SRB群落组成的环境因素包括有机底物种类、氧气浓度、盐度和pH。(1)有机底物种类:不同的SRB菌属可利用的有机底物不同,导致其在湖泊中非均质分布。例如,在湖泊氧化带中Desulforegula相对丰度最高,而在氧化带10 m以下区域Desulfobacca、Desulfovibrio和Sulfurospirillum是优势菌属[41]。SRB群落的空间差异与菌属的代谢差异有关。例如,Desulforegula只能利用SO42–,不能在缺乏SO42–的环境中生长[67],Desulfobacca和Desulfovibrio可利用单质硫、S2O32–和SO32–等作为电子受体。(2)氧气浓度:传统研究认为SRB是严格厌氧菌,湖泊氧气浓度高的区域抑制了SRB的生长和多样性,但新的研究表明部分SRB菌属可以在有氧环境中生长,具有耐氧性[68-69]。例如,在富集培养中,Desulfovibrio的相对丰度随氧气浓度降低而升高,且耐氧上限为6.68 mg/L[70]。(3)盐度:与SOB群落类似,SRB群落组成在不同盐度湖泊中也存在差异。如淡水湖中Desulfobacca、Thermodesulfovibrio、Desulfurispora和Desulfosporosinus丰度较高[71]。高盐湖泊中Desulfofaba和Desulfotomaculum等耐盐嗜盐微生物占主导。(4) pH:pH是影响湖泊SRB群落组成的最重要环境因素之一。例如,在pH < 2的高盐湖泊中,SRB群落主要由Desulfomicrobium、Desulfosporosinus和Clostridium构成,且以Desulfomicrobium为主[46]。然而,在苏打湖(pH > 10)环境中,SRB群落主要由Desulfonatronovibrio、Desulfonatronospira、Desulfurivibrio和Dethiobacter构成[26]。
3.4 湖泊硫还原速率及影响因素前人研究部分湖泊的硫酸盐还原速率(sulfate reduction rate,SRR)发现,表层沉积物(0–2 cm) SRR最高,占总SRR的93%[72]。影响SRR的因素之一是SO42–浓度。SO42–浓度随沉积物深度增加而减少,SO42–含量下降,抑制了SRB的代谢活性,致使SRR降低[72]。中等盐度(30– 60 g/L)湖泊中SRR最高(2.2–31.2 μmol S/(kg·h));淡水湖(3–10 g/L)中SRR为0.1–1.3 μmol S/(kg·h);高盐湖(165–390 g/L)中SRR为0.4–3.9 μmol S/(kg·h)[48]。可以看出,淡水湖中SRR低于咸(盐)湖中SRR,但在高盐湖中SRR低于中等盐度湖泊SRR,表明盐度不是影响SRR的唯一因素。因此,湖泊中其他环境因素可能对SRR也有影响,例如氧气浓度、有机质含量、沉积物中矿物的内在特征。SRB大部分是厌氧菌,氧气含量高会抑制SRB的代谢活动,降低SRR。SRB以SO42–氧化有机物获取代谢所需的能量,有机物含量丰富的区域,SRB可利用的有机碳充足,利于硫酸盐还原过程的进行[73]。沉积物中矿物特征也可能影响周围功能微生物的群落组成。已有研究表明土壤中石膏会影响Syntrophobacteraceae的SRB菌群[74],推测湖泊中石膏可能对SRB群落和SRR产生影响。另外,矿物也可以间接影响生物群落组成,如碳酸盐、磷酸盐矿物对附近区域的pH具有缓冲作用[75],间接影响微生物的代谢活性,从而改变SRR。
4 硫循环与其他元素循环的耦合微生物在驱动硫化合物的氧化还原过程中,往往和其他元素(如碳、氮、磷等)循环相互耦合[44, 76-78]。许多SOB不仅参与湖泊的硫氧化,同时耦联硝酸盐还原、聚磷和固碳等功能。如苏打湖中分离的Thioalkalivibrio denitrificancs和Halomonas disederata菌体具有液泡结构,用于储存NO3–,且在硫氧化过程中将电子传递给NO3–,具有硝酸盐还原功能[35-36]。研究发现属于Beggiatoa、Thiomargarita与Magnetococcus的SOB与湖泊磷释放有关,即硫化物浓度较高的环境会造成它们释放磷酸[79]。Acidithiobacillus SOB细菌可以通过卡尔文循环途径固定CO2[80],在高盐碱湖中soxB和cbbL基因的克隆文库中Thioalkalivibrio的相对丰度重合较好,暗示Thioalkalivibrio SOB将硫氧化过程和固碳过程结合[81]。
硫酸盐还原过程与湖泊中磷和铁元素循环也存在紧密联系。研究表明,在SO42–浓度低的淡水湖中,SRB产生的单质硫及硫化氢会抑制铁还原菌的代谢,并与湖泊中的铁氧化物发生化学反应,生成二硫化亚铁,使得水体中铁循环趋向于硫化物介导的化学方式的铁还原(sulfide- mediated chemical iron reduction,SCIR);同时铁氧化物的溶解导致水体和沉积物中PO43–浓度显著增加[82]。因此,对湖泊富营养化的处理需要考虑湖泊硫循环的生物因素。另外,SRB与湖泊中的甲烷转化紧密相关。甲烷厌氧氧化古菌(anaerobic methanotrophic archaea,ANME)与SRB共生。在甲烷氧化的过程中,将SO42–作为最终电子受体,使得甲烷厌氧氧化过程与硫酸盐还原过程耦合[83-84]。SRB与产甲烷菌竞争乙酸盐和氢气等电子供体,影响湖泊碳循环过程。如:低硫酸盐水体中产甲烷过程是厌氧呼吸的主要形式;而在高硫酸盐水体中,硫酸盐还原菌代谢强烈,抑制产甲烷菌活性[85]。
5 总结和展望湖泊环境栖息着多种多样的硫氧化和硫酸盐还原微生物类群,它们介导着不同类型含硫化合物的相互转化。本文主要综述了硫氧化和硫酸盐还原微生物在湖泊生境中的硫代谢过程、群落分布特征与潜在活力及其影响因素。通过上述综述,可知不同湖泊中的硫氧化和硫酸盐还原微生物种群组成存在着差异,这些差异主要是由于不同微生物种群对不同湖泊环境因素(如有机质组分、代谢底物浓度、pH、氧气浓度、盐度)的偏好差异导致。同时,本文也涉及了湖泊中硫循环与碳氮磷铁等元素循环的耦合,表明驱动硫循环的微生物对其他元素生物地球化学循环也具有重要影响。然而,针对湖泊中微生物驱动的硫循环过程研究仍存在诸多不足,建议加强以下几个方面的研究:(1)利用高通量测序、宏基因组学和转录组学相结合探究极端环境湖泊(苏打湖、盐湖、冰下湖)中硫循环微生物的种类和代谢特征。Wright等[86]发现冰泉沉积物中硫相关的化能自养型Sulfurovum和Sulfuricurvum是主要的初级生产力形式。这表明极端环境中硫循环微生物执行了其他功能,许多新颖的硫循环微生物类群亟待发掘。(2)探究湖泊中矿物成分组成特征对硫循环微生物的影响。本课题组发现矿物成分对青藏高原湖泊沉积物微生物群落影响较其他环境因素更强烈[87]。这意味着区域性的矿物成分构成差异可能改变湖泊中的硫循环微生物群落构成。本课题正在进行的研究也发现不同含硫矿物(黄铁矿、磁黄铁矿、石膏)周围沉积物中微生物α多样性不同,说明矿物组分可能改变硫循环微生物群落多样性(未发表数据)。(3)需改良湖泊硫氧化速率测定的方法。S2–易与H+结合后挥发或与金属离子沉淀,造成测试数据的误差;同时,硫氧化过程的中间产物种类多,部分产物(如S4O62–)不稳定,氧化产物的定量失真,也会影响结果。(4)探究硫代谢途径和功能基因跟环境因子/空间因子的关联。部分硫循环代谢被认为发生某些特定环境,如S2O32–的四硫代谢途径被认为只存在于酸性湖泊。然而,新的研究显示在苏打湖中也存在S4O62–转化基因,表明可能存在特殊的途径或其他的环境因子影响该过程,这些潜在代谢途径还需进一步研究。
| [1] | Seitzinger S, Harrison JA, Bohlke JK, Bouwman A, Lowrance R, Peterson B, Tobias C, Dercht GV. Denitrification across landscapes and waterscapes:a synthesis. Ecological Applications, 2006, 16(6): 2064-2090. DOI:10.1890/1051-0761(2006)016[2064:DALAWA]2.0.CO;2 |
| [2] | Poulin BA, Ryan JN, Nagy KL, Stubbin A, Dittmar T, Orem W, Kranbbenhoft DP, Aiken GR. Spatial dependence of reduced sulfur in everglades dissolved organic matter controlled by sulfate enrichment. Environmental Science & Technology, 2017, 51(7): 3630-3639. |
| [3] |
Wu QL, Xing P, Li HB, Zeng J. Impacts of regime shift between phytoplankton and macrophyte on the microbial community structure and its carbon cycling in lakes. Microbiology China, 2013, 40(1): 87-97.
(in Chinese) 吴庆龙, 邢鹏, 李化炳, 曾巾. 草藻型稳态转换对湖泊微生物结构及其碳循环功能的影响. 微生物学通报, 2013, 40(1): 87-97. |
| [4] |
Luo JH, Tao Y, Xing P, Wu QL. Advance of metagenomics research for lake microbiomes. Journal of Lake Sciences, 2020, 32(1): 271-280.
(in Chinese) 罗建桦, 陶晔, 邢鹏, 吴庆龙. 湖泊微生物宏基因组学研究进展. 湖泊科学, 2020, 32(1): 271-280. |
| [5] | Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nature Reviews Microbiology, 2008, 6(6): 441-454. DOI:10.1038/nrmicro1892 |
| [6] |
Zhu JC, Wu YC, Yin HB. Temporal and spatial variations of sulfur speciations in the sediments of algae accumulation area in Lake Taihu. China Environmental Science, 2017, 37(12): 4690-4700.
(in Chinese) 朱瑾灿, 吴雨琛, 尹洪斌. 太湖蓝藻聚集区沉积物硫形态的时空变异特征. 中国环境科学, 2017, 37(12): 4690-4700. |
| [7] | Lens PNL, Kuenen JG. The biological sulfur cycle:novel opportunities for environmental biotechnology. Water Science and Technology, 2001, 44(8): 57-66. DOI:10.2166/wst.2001.0464 |
| [8] |
Jiang HL, Wu QL. Microbiological research on Chinese lake. Bulletin of Chinese Academy of Sciences, 2017, 2(32): 273-279.
(in Chinese) 江和龙, 吴庆龙. 中国湖泊卫生区组研究. 中国科学院院刊, 2017, 2(32): 273-279. |
| [9] | Frigaard N, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Advances in Microbial Physiology, 2008, 54: 103-200. DOI:10.1016/S0065-2911(08)00002-7 |
| [10] | Shahak Y, Hauska G. Sulfide oxidation from cyanobacteria to humans:sulfide-quinone oxidoreductase (SQR). Springer, Dordrecht, 2008, 27: 319-335. |
| [11] | Marcia M, Ermler U, Peng G, Michel H. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins:Structure, Function, and Bioinformatics, 2010, 78(5): 1073-1083. DOI:10.1002/prot.22665 |
| [12] | Pham VH, Yong J, Park S, Yoon D, Chung W, Rhee S. Molecular analysis of the diversity of the sulfide:quinone reductase (sqr) gene in sediment environments. Microbiology, 2008, 154(10): 3112-3121. DOI:10.1099/mic.0.2008/018580-0 |
| [13] | Luo J, Tan X, Liu K, Lin W. Survey of sulfur-oxidizing bacterial community in the Pearl River water using soxB, sqr, and dsrA as molecular biomarkers. 3 Biotech, 2018, 8(1): 73. |
| [14] | Kostanjevecki V, Brige A, Meyer TE, Cusanovich MA, Guisez Y, van Beeumen J. A membrane-bound flavocytochrome c-sulfide dehydrogenase from the purple phototrophic sulfur bacterium Ectothiorhodospira vacuolata. Journal of Bacteriology, 2000, 182(11): 3097-3103. DOI:10.1128/JB.182.11.3097-3103.2000 |
| [15] |
Liu Y, Jiang LJ, Shao ZZ. Advances in sulfur-oxidizing bacterial taxa and their sulfur oxidation pathways. Acta Microbiologica Sinica, 2018, 58(2): 191-201.
(in Chinese) 刘阳, 姜丽晶, 邵宗泽. 硫氧化细菌的种类及硫氧化途径的研究进展. 微生物学报, 2018, 58(2): 191-201. |
| [16] | Dahl C. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. lubmb Life, 2015, 67(4): 268-274. |
| [17] | Stockdreher Y, Venceslau SS, Josten M, Sahl HG, Pereira IA, Dahl C. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum:evidence for sulfur transfer from DsrEFH to DsrC. PLoS One, 2012, 7(7): e40785. DOI:10.1371/journal.pone.0040785 |
| [18] | Santos AA, Venceslau SS, Grein F, Leavitt WD, Dahl C, Johnston DT, Pereira IA. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science, 2015, 350(6267): 1541-1545. DOI:10.1126/science.aad3558 |
| [19] | Venceslau SS, Stockdreher Y, Dahl C, Pereira IA. The "bacterial heterodisulfide" DsrC is a key protein in dissimilatory sulfur metabolism. Biochimica Et Biophysica Acta (BBA) - Bioenergetics, 2014, 1837(7): 1148-1164. DOI:10.1016/j.bbabio.2014.03.007 |
| [20] | Bobadilla Fazzini RA, Cortés MP, Padilla L, Maturana D, Budinich M, Maass A, Parada P. Stoichiometric modeling of oxidation of reduced inorganic sulfur compounds (Riscs) in Acidithiobacillus thiooxidans. Biotechnology and Bioengineering, 2013, 110(8): 2242-2251. DOI:10.1002/bit.24875 |
| [21] | Guo X, Yin H, Liang Y, Hu Q, Zhou X, Xiao Y, Ma L, Zahng X, Qiu G, Liu X. Comparative genome analysis reveals metabolic versatility and environmental adaptations of Sulfobacillus thermosulfidooxidans strain ST. PLoS One, 2014, 9(6): e99417. DOI:10.1371/journal.pone.0099417 |
| [22] | Mangold S, Valdés J, Holmes DS, Dopson M. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Frontiers in Microbiology, 2011, 2: 17. |
| [23] | Ghosh W. Tetrathiobacter kashmirensis gen. nov., sp. nov., a novel mesophilic, neutrophilic, tetrathionate-oxidizing, facultatively chemolithotrophic betaproteobacterium isolated from soil from a temperate orchard in Jammu and Kashmir, India. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(5): 1779-1787. DOI:10.1099/ijs.0.63595-0 |
| [24] | Meyer B, Imhoff JF, Kuever J. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria-evolution of the Sox sulfur oxidation enzyme system. Environmental Microbiology, 2007, 9(12): 2957-2977. DOI:10.1111/j.1462-2920.2007.01407.x |
| [25] | Hensen D, Sperling D, Truper HG, Brune DC, Dahl C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Molecular Microbiol, 2006, 62(3): 794-810. DOI:10.1111/j.1365-2958.2006.05408.x |
| [26] | Vavourakis CD, Mehrshad M, Balkema C, van Hall R, Anei AS, Ghai R, Sorokin D, Muyzer G. Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake. BMC Biology, 2019, 17(1): 69. DOI:10.1186/s12915-019-0688-7 |
| [27] | Kappler U, Bailey S. Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and ac-type cytochrome subunit. Journal of Biological Chemistry, 2005, 280(26): 24999-25007. DOI:10.1074/jbc.M503237200 |
| [28] | Wilson JJ, Kappler U. Sulfite oxidation in Sinorhizobium meliloti. Biochimica Et Biophysica Acta (BBA) - Bioenergetics, 2009, 1787(12): 1516-1525. DOI:10.1016/j.bbabio.2009.07.005 |
| [29] | Dahl C. Sulfur metabolism in phototrophic bacteria//Hallenbeck PC. Modern topics in the phototrophic prokaryotes: metabolism, bioenergetics, and omics. Cham: Springer International Publishing, 2017: 27-66. |
| [30] | Quentmeier A, Kraft R, Kostka S, Klockenkämper R, Friedrich C. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Archives of Microbiology, 2000, 173(2): 117-125. DOI:10.1007/s002039900118 |
| [31] | Parey K, Demmer U, Warkentin E, Wynen N, Ermler U, Dahl C. Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS One, 2013, 8(9): e74707. DOI:10.1371/journal.pone.0074707 |
| [32] | Imhoff JF, Thiel V. Phylogeny and taxonomy of Chlorobiaceae. Photosynthesis Research, 2010, 104(2-3): 123-136. DOI:10.1007/s11120-009-9510-7 |
| [33] | Kojima H, Fukui M. Sulfuriflexus mobilis gen. nov., sp. nov., a sulfur-oxidizing bacterium isolated from a brackish lake sediment. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(9): 3515-3518. DOI:10.1099/ijsem.0.001227 |
| [34] | Shinohara A, Fukui M, Kojima H. Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov.. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(10): 3709-3713. |
| [35] | Sorokin DY. Thiomicrospira halophila sp. nov., a moderately halophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium from hypersaline lakes. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(10): 2375-2380. DOI:10.1099/ijs.0.64445-0 |
| [36] | Sorokin DY, Lysenko AM, Mityushina LL, Tourova TP, Jones BE, Rainey FA, Robertson LA, Kuenen GJ. Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp.nov., novel and Thioalkalivibrio denitrificancs sp.nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. International Journal of Systematic and Evolutionary Microbiology, 2001, 51(2): 565-580. DOI:10.1099/00207713-51-2-565 |
| [37] | Sorokin D. Oxidation of inorganic sulfur compounds by obligatory organotrophic bacteria. Mikrobiologiia, 2003, 72(6): 725-739. |
| [38] | Yang J, Jiang H, Dong H, Wu G, Hou W, Zhao W, Sun Y, Lai Z. Abundance and diversity of sulfur-oxidizing bacteria along a salinity gradient in four Qinghai-Tibetan Lakes, China. Geomicrobiology Journal, 2013, 30(9): 851-860. DOI:10.1080/01490451.2013.790921 |
| [39] | Watanabe T, Kojima H, Takano Y, Fukui M. Diversity of sulfur-cycle prokaryotes in freshwater lake sediments investigated using aprA as the functional marker gene. Systematic and Applied Microbiology, 2013, 36(6): 436-443. DOI:10.1016/j.syapm.2013.04.009 |
| [40] | Kojima H, Watanabe T, Iwata T, Fukui M. Identification of major planktonic sulfur oxidizers in stratified freshwater lake. PLoS One, 2014, 9(4): e93877. DOI:10.1371/journal.pone.0093877 |
| [41] | Berg JS, Jezequel D, Duverger A, Lamy D, Laberty-Robert C, Miot J. Microbial diversity involved in iron and cryptic sulfur cycling in the ferruginous, low-sulfate waters of Lake Pavin. PLoS One, 2019, 14(2): e212787. |
| [42] |
Wu SJ, Wang X, Ji QY, Wang MY, Zhao YP, Wang GX. Iron-sulfur distribution and its enviromental significance in three typical areas of western Lake Taihu. Journal of Lake Sciences, 2019, 31(4): 950-960.
(in Chinese) 吴松峻, 汪旋, 季秋忆, 王明钥, 赵艳萍, 王国祥. 太湖西岸典型区域沉积物的硫铁分布特征及环境意义. 湖泊科学, 2019, 31(4): 950-960. |
| [43] | Kojima H, Fukui M. Sulfuritalea hydrogenivorans gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(7): 1651-1655. DOI:10.1099/ijs.0.024968-0 |
| [44] | Sorokin DY, Kuenen JG, Muyzer G. The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Frontiers in Microbiology, 2011, 2: 44. |
| [45] | Williams KP, Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(8): 2901-2906. |
| [46] | Escudero L, Oetiker N, Gallardo K, Tebes-Cayo C, Guajardo M, Nuñez C, Davis-Belmar C, Pueyo J, Chong Díaz G, Demergasso C. A thiotrophic microbial community in an acidic brine lake in Northern Chile. Antonie van Leeuwenhoek, 2018, 111(8): 1403-1419. DOI:10.1007/s10482-018-1087-8 |
| [47] | Edwardson CF, Hollibaugh JT. Metatranscriptomic analysis of prokaryotic communities active in sulfur and arsenic cycling in Mono Lake, California, USA. The ISME journal, 2017, 11(10): 2195-2208. DOI:10.1038/ismej.2017.80 |
| [48] | Sorokin DY, Gorlenko VM, Namsaraev BB, Namsaraev ZB, Lysenko AM, Eshinimaev BT, Khmelenina VN, Trotsenko YA, Kuenen JG. Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia, 2004, 522(1): 235-248. |
| [49] | Diao M, Huisman J, Muyzer G. Spatio-temporal dynamics of sulfur bacteria during oxic-anoxic regime shifts in a seasonally stratified lake. FEMS Microbiology Ecology, 2018, 94(4). |
| [50] | Dorries M, Wohlbrand L, Kube M, Reinhardt R, Rabus R. Genome and catabolic subproteomes of the marine, nutritionally versatile, sulfate-reducing bacterium Desulfococcus multivorans DSM 2059. BMC Genomics, 2016, 17(1): 918. DOI:10.1186/s12864-016-3236-7 |
| [51] | Zhang Y, Zhen Y, Mi T, He H, Yu Z. Molecular characterization of sulfate-reducing bacteria community in surface sediments from the adjacent area of Changjiang Estuary. Journal of Ocean University of China, 2016, 15(1): 107-116. DOI:10.1007/s11802-016-2781-7 |
| [52] | Scholten J CM, Joye SB, Hollibaugh JT, Murrell JC. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microbial Ecology, 2005, 50(1): 29-39. DOI:10.1007/s00248-004-0085-8 |
| [53] | Neretin LN, Schippers A, Pernthaler A, Hamann K, Amann R, Jørgensen BB. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environmental Microbiology, 2003, 5(8): 660-671. DOI:10.1046/j.1462-2920.2003.00452.x |
| [54] | Pereira IAC, Ramos AR, Grein F, Marques MC, Silva SM, Venceslau SS. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Frontiers in Microbiology, 2011, 2: 69. |
| [55] | Jakobsen TF. Desulfohalobium utahense sp. nov., a moderately halophilic, sulfate-reducing bacterium isolated from Great Salt Lake. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(9): 2063-2069. DOI:10.1099/ijs.0.64323-0 |
| [56] | Krekeler D, Sigalevich P, Teske A, Cypionka H, Cohen Y. A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai), Desulfovibrio oxyclinae sp. nov.. Archives of Microbiology, 1997, 167(6): 369-375. DOI:10.1007/s002030050457 |
| [57] | Ramamoorthy S, Sass H, Langner H, Schumann P, Kroppenstedt RM, Spring S, Overmann J, Rosenzweig RF. Desulfosporosinus lacus sp. nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(12): 2729-2736. DOI:10.1099/ijs.0.63610-0 |
| [58] | Sass H, Overmann JR, R Tters H, Babenzien HD, Cypionka H. Desulfosporomusa polytropa gen. nov., sp. nov., a novel sulfate-reducing bacterium from sediments of an oligotrophic lake. Archives of Microbiology, 2004, 182(2-3): 204-211. |
| [59] | Foti M, Sorokin DY, Lomans B, Mussman M, Zacharova EE, Pimenov NV, Kuenen JG, Muyzer G. Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Applied and Environmental Microbiology, 2007, 73(7): 2093-2100. DOI:10.1128/AEM.02622-06 |
| [60] | Watanabe T, Kojima H, Fukui M. Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase. Scientific Reports, 2016, 6: 36262. DOI:10.1038/srep36262 |
| [61] | Qin H, Wang S, Feng K, He Z, Virta MPJ, Hou W, Dong H, Deng Y. Unraveling the diversity of sedimentary sulfate-reducing prokaryotes (SRP) across Tibetan saline lakes using epicPCR. Microbiome, 2019, 7(1): 71. DOI:10.1186/s40168-019-0688-4 |
| [62] |
Yang J, Jiang HC, Sun YJ, Wu G, Hou WG, Dong HL, Lai ZP. Abundance and diversity of sulfate-reducing bacteria in Qinghai-Tibetan Lakes. Journal of Salt Lake Research, 2013, 21(1): 7-13.
(in Chinese) 杨渐, 蒋宏忱, 孙永娟, 吴耿, 侯卫国, 董海良, 赖忠平. 青藏高原湖泊沉积物硫酸盐还原菌种群分布. 盐湖研究, 2013, 21(1): 7-13. |
| [63] | Yau S, Lauro FM, Williams TJ, Demaere MZ, Brown MV, Rich J, Gibson JAE, Cavicchioli R. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. The ISME Journal, 2013, 7(10): 1944-1961. DOI:10.1038/ismej.2013.69 |
| [64] | Edwardson CF, Hollibaugh JT. Composition and activity of microbial communities along the redox gradient of an alkaline, hypersaline, lake. Frontiers in Microbiology, 2018, 9: 14. DOI:10.3389/fmicb.2018.00014 |
| [65] | Ng C, Demaere MZ, Williams TJ, Lauro FM, Raftery M, Gibson JAE Andrews-Pfannkoch, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. The ISME Journal, 2010, 4(8): 1002-1019. DOI:10.1038/ismej.2010.28 |
| [66] | Johnson SS, Chevrette MG, Ehlmann BL, Benison KC. Insights from the metagenome of an acid salt lake:the role of biology in an extreme depositional environment. PLoS One, 2015, 10(4): e122869. |
| [67] | Rees GN, Patel BK. Desulforegula conservatrix gen. nov., sp. nov., a long-chain fatty acid-oxidizing, sulfate-reducing bacterium isolated from sediments of a freshwater lake. International Journal of Systematic and Evolutionary Microbiology, 2001, 51(5): 1911. DOI:10.1099/00207713-51-5-1911 |
| [68] | Ramel F, Amrani A, Pieulle L, Lamrabet O, Voordouw G, Seddiki N, Brèthes D, Company M, Dolla A, Brasseur G. Membrane-bound oxygen reductases of the anaerobic sulfate-reducing Desulfovibrio vulgaris Hildenborough:roles in oxygen defence and electron link with periplasmic hydrogen oxidation. Microbiology, 2013, 159(12): 2663-2673. |
| [69] | Ramel F, Brasseur G, Pieulle L, Valette O, Hirschler-Réa A, Fardeau ML, Dolla A. Growth of the obligate anaerobe Desulfovibrio vulgaris hildenborough under continuous low oxygen concentration sparging:Impact of the membrane-bound oxygen reductases. PLoS One, 2015, 10(4): e123455. |
| [70] |
Chen YW, Zhang CH. Characteristics of oxygen tolerance of sulfate-reducing bacteria in lakes and coastal waters under enrichment. Geological Journal of China Universities, 2019, 25(5): 705-713.
(in Chinese) 陈亚文, 张朝晖. 富集培养条件下湖泊和沿海海域水体硫酸盐还原菌的耐氧性特征. 高校地质学报, 2019, 25(5): 705-713. |
| [71] | Vuillemin A, Horn F, Friese A, Winkel M, Alawi M, Wagner D, Henny C, Orsi WD, Crowe SA, Kallmeyer J. Metabolic potential of microbial communities from ferruginous sediments. Environmental microbiology, 2018, 20(12): 4297-4313. DOI:10.1111/1462-2920.14343 |
| [72] | Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, Han S, Lanoil B, Oremland RS. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic- rich soda lakes:Mono and Searles Lakes, California. Applied and Environmental Microbiology, 2006, 72(10): 6514-6526. DOI:10.1128/AEM.01066-06 |
| [73] | Brodersen KE, Trevathan-Tackett SM, Nielsen DA, et al. Oxygen consumption and sulfate reduction in vegetated coastal habitats:Effects of physical disturbance. Frontiers in Marine Science, 2019, 6. |
| [74] | Liu P, Pommerenke B, Conrad R. Identification of Syntrophobacteraceae as major acetate-degrading sulfate reducing bacteria in Italian paddy soil. Environmental Microbiology, 2018, 20(1): 337-354. DOI:10.1111/1462-2920.14001 |
| [75] | Jones AA, Bennett PC. Mineral ecology:surface specific colonization and geochemical drivers of biofilm accumulation, composition, and phylogeny. Frontiers in Microbiology, 2017, 8: 491. |
| [76] | van de Velde S, Callebaut I, Gao Y, Meysman FJR. Impact of electrogenic sulfur oxidation on trace metal cycling in a coastal sediment. Chemical Geology, 2017, 452: 9-23. DOI:10.1016/j.chemgeo.2017.01.028 |
| [77] | Niu Z, Pan H, Guo X, Lu P, Feng J, Chen Y, Tou F, Liu M, Yang Y. Sulphate-reducing bacteria (SRB) in the Yangtze Estuary sediments:Abundance, distribution and implications for the bioavailibility of metals. Science of The Total Environment, 2018, 634: 296-304. DOI:10.1016/j.scitotenv.2018.03.345 |
| [78] | Zhao Y, Zhang Z, Wang G, Li X, Ma J, Chen S, Deng H, Annalisa OH. High sulfide production induced by algae decomposition and its potential stimulation to phosphorus mobility in sediment. Science of The Total Environment, 2019, 650: 163-172. DOI:10.1016/j.scitotenv.2018.09.010 |
| [79] | Rivas-Lamelo S, Benzerara K, Lefèvre CT, Monteil CL, Jézéquel D, Menguy N, Viollier E, Guyot F, Férard C, Poinsot M, Skouri-Panet F, Trcera N, Miot J, Duprat E. Magnetotactic bacteria as a new model for P sequestration in the ferruginous Lake Pavin. Geochemical Perspectives Letters, 2017: 35-41. |
| [80] | Esparza M, Cardenas JP, Bowien B, Jedlicki E, Holmes DS. Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans, carbon fixation in A. ferrooxidans. BMC Microbiology, 2010, 10: 229. DOI:10.1186/1471-2180-10-229 |
| [81] | Tourova TP, Slobodova NV, Bumazhkin BK, Kolganova TV, Muijzer G, Sorokin DY. Analysis of community composition of sulfur-oxidizing bacteria in hypersaline and soda lakes using soxB as a functional molecular marker. FEMS Microbiology Ecology, 2013, 84(2): 280-289. DOI:10.1111/1574-6941.12056 |
| [82] | Wu S, Zhao Y, Chen Y, Dong X, Wang M, Wang G. Sulfur cycling in freshwater sediments:A cryptic driving force of iron deposition and phosphorus mobilization. Science of The Total Environment, 2019, 657: 1294-1303. DOI:10.1016/j.scitotenv.2018.12.161 |
| [83] | Rotaru A, Thamdrup B. A new diet for methane oxidizers. Science, 2016, 351(6274): 658-659. DOI:10.1126/science.aaf0741 |
| [84] |
Wei SZ. Methanotrophs and their applications in enviroment treatment:A review. Chinese Journal of Applied Ecology, 2012, 23(8): 2309-2318.
(in Chinese) 魏素珍. 甲烷氧化菌及其在环境治理中的应用. 应用生态学报, 2012, 23(8): 2309-2318. |
| [85] | Kuivila KM, Murray JW, Devol AH, Novelli PC. Methane production, sulfate reduction and competition for substrates in the sediments of Lake Washington. Geochimica Et Cosmochimica Acta, 1989, 53(2): 409-416. DOI:10.1016/0016-7037(89)90392-X |
| [86] | Wright KE, Williamson C, Grasby SE, Spear JR, Templeton AS. Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit, Borup Fiord Pass. Frontiers in Microbiology, 2013, 4: 63. |
| [87] | Yang J, Jiang H, Sun X, Chen J, Xie Z, Dong H. Minerals play key roles in driving prokaryotic and fungal community in the surface sediments of the Qinghai-Tibetan lakes. FEMS Microbiology Ecology, 2020. |
 2020, Vol. 60
2020, Vol. 60