中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 孙小溪, 蒋宏忱. 2020
- Xiaoxi Sun, Hongchen Jiang. 2020
- 湖泊微生物硝化过程研究进展
- Research progress in microbial nitrification in lakes
- 微生物学报, 60(6): 1148-1161
- Acta Microbiologica Sinica, 60(6): 1148-1161
-
文章历史
- 收稿日期:2020-02-11
- 修回日期:2020-03-31
- 网络出版日期:2020-04-09
硝化作用是指微生物将NH4+氧化为NO3–的过程,这一过程在很大程度上决定了湖泊无机氮库的形态分布,并且影响着湖泊初级生产力,在影响湖泊氮循环和控制温室气体N2O释放方面扮演着关键角色[1-3]。历史上,我们对于硝化作用的认知一直在不断更新。20世纪以来,学术界普遍认为只有细菌(AOB)能够实现氨氧化;直到2005年,有学者在研究海洋[4]和土壤[5]的微生物基因组时发现古菌中存在类似的amoA基因,并成功分离得到第一株氨氧化古菌(AOA) Nitrosopumilis maritimus[6],从而证明古菌也具有氨氧化功能;于2015年,完全氨氧化(comammox)微生物被发现可以独立实现从氨到硝酸盐的氧化过程[7-8],极大地改变了我们对于硝化微生物的认识;于2019年,AOA Nitrosocosmicus oleophilus被发现在酸性条件下可表达一氧化氮还原酶P450 NOR。经过计算,当pH为5.5时,N. oleophilus产生的N2O中有超过50%来源于此生物途径[9],这一发现颠覆了AOA只能通过非生物途径产生N2O的固有认识。在湖泊生态系统中,硝化作用为反硝化作用源源不断地提供底物,二者作为紧密联系的有机整体,在降低湖泊氮载荷、减轻湖泊富营养化方面发挥着关键作用[10-12]。因此,研究硝化作用有助于我们全面认识湖泊氮循环的规律和生态意义。本文将综述湖泊生境中不同类型的硝化过程及相关的微生物群落组成,并综合微生物代谢机理分析硝化过程的影响因素,总结现有研究的不足,展望未来研究的方向。
1 湖泊中的分步硝化过程硝化作用可以通过分步实现,主要包含氨氧化(反应式1)和亚硝酸盐氧化(反应式2)两个反应步骤。

|
公式(1) |

|
公式(2) |
氨氧化是分步硝化作用的第一步同时也是限速步骤。由于这一步骤依赖氨单加氧酶(AMO)来完成,因此执行该步骤的微生物群落通常用编码AMO的amoA基因来鉴定。大多数AOB属于严格好氧的β变形菌纲和γ变形菌纲,例如Nitrosomonas、Nitrosospira和Nitrosococcus属[13-15]。AOA根据系统发育关系可分为以下属:Nitrosopumilus、Nitrosotalea、Nitrosocaldus以及Nitrososphaera[16]。湖泊生境中的AOB主要为Nitrosospira[14]和Nitrosococcus,后者主要分布于盐湖中[17-18];AOA主要为Nitrosotalea和Nitrosopumilus中的低盐度分支[19]。在青藏高原盐湖中,Yang等(2013)发现AOA主要为Nitrososphaera和Nitrosopumilus以及一个“低盐度分支”,与世界其他湖泊AOA群落组成有着显著差异[20],暗示了盐湖生境中AOA分布具有地理差异的特殊性。AOA与AOB具有不同的环境偏好和生态位,受到湖泊生境中一系列环境因子的控制,例如氨浓度、pH、氧浓度、盐度、硫化物浓度以及磷酸盐浓度等,但本质上与二者生理生化机制的差异有关(表 1)[21-22]。AOA具有极高的底物亲和力(半饱和常数K=133 nmol/L)和极低的底物临界值(≤10 nmol/L),分别是AOB的200倍和1/100,二者数量级的差异使得AOA在底物极度匮乏的环境中占绝对优势[21]。从分子机制上看,AOA在进化过程中获得的V-ATPase十分关键,该酶可以帮助AOA产生ATP和质子驱动力,进而适应酸性、高盐、高压等逆境[23-24]。具体到湖泊环境中,大多数情况下,AOA在种群丰度上均高于AOB[13, 25-28]。但也存在相反的情形,根据前人报道,在青藏高原盐湖沉积物[13]、极度富营养化的湖泊[12]以及用于水产养殖的太湖东部湾[25]等环境中,AOB的丰度高于AOA。目前鲜有学者建立起湖泊生境中AOA、AOB群落特征与pH的联系,可能与大部分研究湖泊pH梯度较小且偏碱性的特点有关[12, 20, 29]。在pH范围4.8–7.7的丹麦湖泊沉积物中AOA相对丰度被发现与pH呈负相关,暗示了AOA对低pH的偏好性[26]。纯培养研究显示,pH降低会引起氨的质子化,从而降低底物的生物可利用性,使得耐受寡营养的AOA生存下来,而AOB几乎不能在pH低于6.5的环境中进行氨氧化[30-33]。另外,AOA通常能够在高硫化物的条件下生存并表达氨氧化活性,而AOB的生长则受到抑制,这可能与AOB中含铜AMO的合成受到抑制有关[34-35]。湖泊溶解氧含量也是影响氨氧化速率和相关微生物的关键因素。纯菌动力学研究表明,AOA对氧气的亲和力远远高于AOB,引起了二者对氧气的适应机制存在本质上的差异,使得AOA在低氧环境中具有竞争优势[21, 36]。例如,在盐湖环境中,随着湖水深度增加至氧跃层,溶解氧浓度大幅降低,氨氧化速率的峰值也出现在溶解氧浓度较低的深层湖水和季节性低氧区,并且与AOA丰度具有很好的相关性而与AOB丰度不相关,暗示着AOA在低氧区对于氨氧化过程的实质性贡献[37]。综上,AOA往往在寡营养、低pH、低氧和含硫的湖泊环境中比AOB更占优势。
| Parameter | AOA | AOB |
| Substrate affinity | High | Low |
| Substrate threshold | Low | High |
| Ammonia | Low | High |
| pH | From acidic to alkaline | Near neutral pH |
| Oxygen | Low | High |
| Sulfide | Presence | Absence |
| Phoaphate | Low | High |
盐度也是影响AOA与AOB丰度和多样性的一个重要因素,但根据已有研究尚不能得出一致的规律。有研究显示,高盐环境中AOB的丰度和活性较低而AOA多样性与氨氧化活力较高[38-41]。因此,AOA被认为在极端环境(如高盐湖泊)中更有竞争力,而AOB的生长则受到限制[21, 42]。也有研究描述了不同的现象。如本课题组研究发现,青藏高原盐湖沉积物中AOB在基因组和转录组层面的功能基因丰度均高于AOA[13]。从能量角度看,高盐环境下微生物需要产生大量的能量来抵御盐度压力,这种反应的热力学特征一定程度上决定了其生存的盐度上限[43]。本课题组前期研究揭示了青藏高原不同盐度湖泊AOA的群落组成和丰度规律,在盐度高达160 g/L的超盐湖中检测到了古菌amoA基因[20],却并未在这些湖泊水体中检测到可观的氨氧化速率(未发表数据)。考虑到青藏高原湖泊受到极强的太阳辐射,影响着硝化微生物的生态位分布与活力[44-45],光抑制和盐度抑制共同作用是否导致氨氧化群落生态位下移(比如在沉积物中表现出较高的活力)?还需要更多深入的研究来揭示。
1.2 亚硝酸盐氧化数十年来,亚硝酸盐氧化过程受到的关注程度远不及氨氧化过程。造成这一现象的原因一方面可能是由于氨氧化被认为是硝化作用的限速步骤,另一方面可能与亚硝酸盐氧化菌(NOB)难以分离培养的特性有关[46]。实际上,亚硝酸盐的消耗途径直接决定了氮是以亚硝酸盐、硝酸盐以及铵盐的形式保存在湖泊生境中还是以一氧化氮、氧化亚氮或氮气形式逸散到大气中,是影响湖泊氮收支的一个关键步骤。
根据系统发育NOB可以分为Nitrobacter、Nitrotoga、Nitrococcus、Nitrospira、Nitrospina、Nitrolancea与“Candidatus Nitromaritima” 7个属,属于Proteobacteria、Chloroflexi、Nitrospirae以及Nitrospinae 4个门[47]。湖泊河流等淡水环境中的NOB主要为Nitrobacter、Nitrotoga、Nitrospira lineage Ⅰ和Ⅱ;盐湖等高盐环境中NOB则广泛分布于除Nitrolancea之外的6个属中。
亚硝酸盐氧化通过亚硝酸盐氧化还原酶(NXR)实现,该酶通过将2个电子传递到呼吸链中来完成一次反应。NXR与细胞质膜联系紧密,由NxrA、NxrB与NxrC 3个亚基组成[48-49]。负责与底物结合的NxrA亚基分为胞质型和胞浆型[48],前者分布于Nitrospira[48]、Nitrospina[50]和“Candidatus Nitromaritima”[51],后者分布于Nitrobacter、Nitrolancea和Nitrococcus中[46, 52]。胞质型的NXR能够在氧化亚硝酸盐的同时从水中获得氢离子,从而获得质子动力势(proton motive force,PMF)并作为细胞的能量来源[48]。具有胞浆型NXR的NOB在反应中产生质子却不能贡献PMF,且需要消耗额外的能量用于硝酸盐与亚硝酸盐的膜内外传递[47]。鉴于亚硝酸盐氧化产能较低,这种微小的区别很可能成为这类NOB的生理瓶颈,使其在特定的环境中处于劣势。NOB具有丰富的代谢机制,参与了湖泊中多种元素循环。例如:在缺氧条件下,NOB能够耦合甲酸盐氧化和硝酸盐还原并保持活性;以氢气为能量来源,还能够固定水体无机碳[53-55]。值得注意的是,NOB与氨氧化菌群常存在着共生和交叉饲喂的现象:首先,NOB氧化环境中的尿素生成氨和CO2,缺乏尿素降解酶的AOA/AOB从NOB获得氨,并将其氧化为亚硝酸根,随后NOB获得亚硝酸根并将其氧化,实现双方生长的促进和硝化速率的提升[54, 56]。
NOB群落结构与功能及其硝化速率取决于亚硝酸根浓度、无机碳浓度、温度以及盐度等环境因子。生理差异(胞浆型和胞质型NXR,以下简称为胞浆型NOB和胞质型NOB)使不同NOB响应环境变化的机制和程度不同。NOB对底物亲和力的差异[47],使不同类型NOB对底物浓度的响应不一:底物亲和力高的胞质型NOB(如Nitrospira lineage Ⅰ、Nitrospina)更加适应亚硝酸盐浓度低的环境,如淡水湖泊;而底物亲和力较低的胞浆型NOB (如Nitrobacter)则更加适应局部的或者季节性亚硝酸盐聚集的环境[47, 57-58]。无机碳的缺乏能够很大程度上抑制Nitrobacter的生长与活力;但随着二氧化碳浓度的增加,Nitrobacter的生长和活力又得到恢复,这种现象可能与碳限制条件下该菌的碳浓缩机制有关[59]。温度也是影响NOB不同种群分布的重要因素,Nitrobacter、Nitrospira与Nitrotoga生长温度范围分别为17–28 ℃、10–28 ℃和10–17 ℃,温度适应范围的差异对解释不同NOB的季节性分布具有一定的指示作用[60]。
在盐湖极端环境中,氮循环微生物群落的生态位分布、组成和活力均受到盐度的影响,尤其是产能较低的微生物[43, 61]。前人几乎没有在海水盐度以上的环境中观察到亚硝酸盐氧化的生物过程,可能是由于NOB的生理生化特点使其难以在高盐环境中生存。在高盐环境中,微生物需要大量的能量来抵御盐度压力,而亚硝酸盐氧化过程产能(74 kJ/mol N)远小于硝化过程(275 kJ/mol N)和反硝化过程(800 kJ/mol N);此外NOB需要大量能量来完成自养固碳,约百倍于亚硝氧化过程的产能[43, 62-63]。然而,AOA (高达160 g/L)和γ-AOB (40–94 g/L)却能够在高盐环境中生存并保持氨氧化活性[20, 43, 61]。理论上,两步硝化反应热力学上的差异会引起某些高盐、超盐湖泊中亚硝酸根离子的积累。然而,本课题组长期观测的青藏高原北部盐湖中并未检测到亚硝的积累[13, 20, 64],这暗示着亚硝通过其他途径被消耗,如反硝化过程和厌氧氨氧化过程[65]。如此一来,盐湖中两步硝化过程的平衡可能被打破,氨氧化可能与其他微生物建立了新的耦合机制,值得深入探讨。
2 湖泊中的完全氨氧化过程完全氨氧化作用(comammox)由Nitrospira lineage Ⅱ实现。Comammox Nitrospira曾经一直被认为是NOB的一员,该微生物体内包含氨氧化(AMO、HAO)和亚硝酸盐氧化作用(NXR)的所有酶。Comammox的氨单加氧酶在系统发育上与已知的AMO有着明显差异[7],据此可以将其分为Clade A和Clade B两支[8]。其中Clade A可以进一步分为Clade A.1和Clade A.2[66]。根据目前研究,湖泊环境中的Comammox主要为Clade A.1,而Clade A.2和Clade B的丰度较低[66-67]。
与分步硝化作用相比,Comammox最大的优势体现在能量方面,即利用同等的底物产能更多[7],Comammox反应途径决定了它生成ATP的效率更高[68]。与NOB Nitrospira相比,Comammox Nitrospira实现氨氧化与羟胺氧化的机制不同,虽然无法同化亚硝酸盐、不能直接利用外部亚硝酸盐作为氮源,却拥有更多样的尿素转运蛋白和内稳态基因,因此能够独自实现从尿素降解到完全氨氧化的过程[69]。与氨氧化菌相比,Comammox的氨氧化和羟胺氧化基因与β-AOB最为相近,暗示了它们在进化上可能存在关联[7-8, 69]。另外,Comammox的基因组包含一种促进微生物低氧浓度下生长的基因,使其在微氧条件下具有明显的竞争优势[69-70]。与AOA类似,Comammox缺少参与一氧化氮还原的基因,不能完成大部分AOB具备的通过一氧化氮还原酶(NOR)产生N2O的反应(表 2),即硝化反硝化代谢过程。目前仅观察到Comammox通过非生物途径生成N2O,生成量与AOA相当,远低于AOB[71]。因此,研究Comammox这一“绿色硝化菌”的分布规律及影响因素将对全球气候变化有一定的指示作用。
为了更好地了解这种新近发现的氮循环途径的生态学意义,学者们对比传统硝化菌,研究了Comammox分布规律、硝化过程中的相对贡献及其影响因素。基因组数据显示,Comammox广泛分布于各种湖泊生境中(包括盐湖),往往与AOA和AOB共存[8, 66-67, 72-73]。目前在大范围湖泊生境中定量Comammox相对贡献的研究仍然欠缺。在武汉东湖与安徽巢湖沉积物样品中,Comammox功能基因丰度高于AOA和AOB[72-73];Xia等(2018)综合宏基因组分析也得出了一致的结论,即淡水生态系统中Comammox群落的相对丰度在所有氨氧化功能群中占据较高的比例(52.3%±37.5%),暗示该过程对湖泊硝化作用不可忽视的贡献[66]。影响AOA和AOB生态位分异的主要因素为氨亲和力的差异和pH,而Comammox需要与氨氧化菌竞争氨来维持生长。因此,有学者推测氨浓度以及亲和力差异是影响它们分布的一个重要因素[8, 74]。动力学研究显示,Comammox N. inopinata对氨的亲和力高于AOB和大部分AOA,能够在底物浓度极低(< 5 μmol/L)的条件下竞争中胜出[74-76]。此外,氧气浓度也是十分重要的影响因素,基于基因组的研究表明Comammox拥有特殊的低氧基因,使其具有更高的氧亲和力;前人在湖泊低氧环境中检测到Comammox丰度更高的现象,暗示着Comammox比AOA和AOB更适应低氧气浓度的环境[69-70, 72]。值得注意的是,数量上的优势并不一定等同于生化活力的高效性[77]。因此,Comammox在湖泊生境中的生态意义还需要深入探究。
3 湖泊硝化过程对温室气体N2O的贡献作为温室气体之一,N2O对全球变暖效应的贡献约占6%。百年尺度上单分子N2O的增温潜势(global warming potential)为CO2的298倍[78-79]。除温室效应外,N2O还能够在平流层进一步被氧化为NO,是导致臭氧层破坏的主要原因之一[80]。N2O作为一种反应副产物或中间产物,全球N2O排放量的70%来源于微生物参与的硝化和反硝化过程[81-82]。
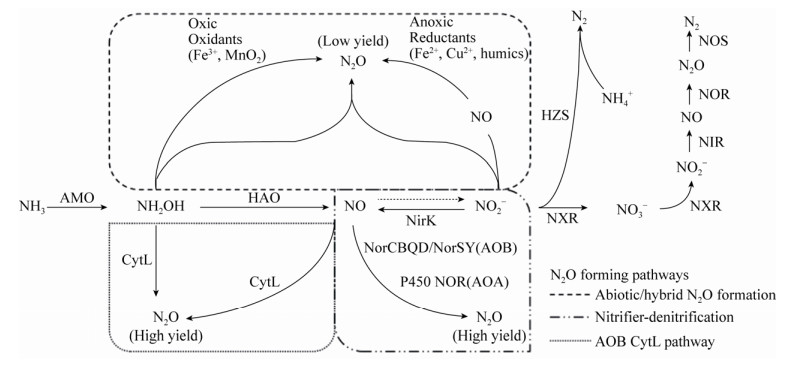
关于湖泊反硝化过程产N2O机理和贡献的研究已经相对成熟,而硝化过程则往往被忽视。实际上,AOA、AOB和Comammox均能产生N2O气体,但其产生机制与产量却差别迥异。N2O主要通过以下三种途径产生(表 2,图 1):1)非生物途径,也称杂交生成N2O。在好氧条件下,AOA、AOB和Comammox氨氧化的中间产物NH2OH与NO2–化学反应生成N2O[83-84]。2)硝化菌反硝化途径。在低氧条件下,AOA和AOB通过亚硝酸盐还原酶NirK将NO2–还原为NO,实现反应的第一步;实现第二步反应的酶有所差异:AOB通过一氧化氮还原酶NOR产生N2O;AOA中没有编码这种酶的基因norB,而是在低pH下表达的P450 NOR催化下生成N2O[9, 85-86]。3) AOB独有的CytL蛋白催化途径。在厌氧条件下,CytL氧化NH2OH生成N2O;或在NH2OH存在的条件下,结合NO并将其还原为N2O[87]。尽管部分Comammox也含有类似CytL蛋白(相似度小于55%),却并没有发现Comammox通过该途径生成N2O[88]。纯培养和微宇宙实验一致显示,AOA和Comammox贡献的N2O产量低于AOB,AOA的N2O/NO2–比值为0.04%–0.07%,AOB则为0.095%–0.270%[84, 89-91]。综合来看,与生物成因相比,非生物成因的N2O占比极低,不到3%;且受pH影响很大,pH越低,化学生成N2O的速率越高[92]。综合来看,硝化过程产生的N2O主要来自于AOB,但目前的研究结果集中在纯培养实验层面,在实际湖泊生境中硝化过程对温室气体N2O的贡献如何,如何定量区分硝化过程和反硝化过程的N2O生成比例是当前湖泊硝化过程研究的前沿问题。Zhang等(2015)提出了N2O排放的三通道模型,并通过同位素成对标记和来源区分法解析了N2O的来源,结果表明除了反硝化过程,自养硝化和异养硝化过程也是重要的N2O产生途径,甚至在某些陆地生态系统中超过反硝化作用,主导了N2O的产生[93-94]。将该技术应用于湖泊生境有助于全面认识硝化过程对温室气体N2O的贡献,深刻理解硝化过程在全球气候变化中扮演的角色。
| Gene | AOA | AOB | Comammox |
| nirK | +/– | +/– | +/– |
| norCBQDSY | – | +/– | – |
| p450nor | +/– | – | – |
| cytL | – | +/– | +/– |
| +/– denotes gene detected in several but not all organisms, - denotes not detected. | |||
|
| 图 1 氨氧化过程中N2O形成的生物和非生物途径 Figure 1 Biotic and abiotic pathways involved in N2O production derived from ammonia oxidation. Solid and dashed arrows depict confirmed and proposed reaction pathways, respectively. The enzymes or redox involved in the reaction are noted next to the arrow. AOB and Comammox contain AMO and HAO, while AOA does not contain HAO. The enzyme that catalyzes the oxidation of nitric oxide to nitrite is unknown. AMO: ammonia monooxygenase; HAO: hydroxylamine oxidoreductase; NirK: copper-containing nitrite reductase; NorCBQD/NorSY: nitric oxide reductase in AOB; P450 NOR: cytochrome P450 nitric oxide reductase in AOA; CytL: periplasmic tetraheme cyt. c P460 protein; NXR: nitrite oxidoreductase; NIR: nitrite reductase; NOR: nitric oxide reductase; NOS: nitrous oxide reductase; HZS: hydrazine synthase. |
4 总结和展望
尽管湖泊中硝化菌群的生理生态特征各不相同,既分工合作又相互竞争,但它们最终作为一个整体完成了湖泊的硝化过程。湖泊生境作为氮循环的热点区域,氮转化速率整体上高于河口和海洋等水生系统(表 3)[1, 95]。总体来说,环境条件控制着硝化过程发生的位置和速率。硝化过程很大程度上依赖于氧气和氨的生物可利用性,还受到pH、硫化物浓度、温度、盐度以及光等环境因子的限制[95]。一般来说,底物充足的富营养湖泊比寡营养湖泊的硝化速率要高,这与AOB的丰度和活力密切相关[12, 26]。湖泊水体硝化速率的峰值往往出现在深层,受到氧气浓度、光抑制作用和浮游生物竞争机制的综合影响[1, 37, 96]。水体酸化会显著降低硝化速率,在pH小于3的湖泊中硝化作用被完全抑制[97-98];大多数化能无机自养型硝化菌在中性偏微碱性环境中(pH 7.0–8.5)的生长和硝化活力最佳[99];极端的碱性条件并不会完全抑制沉积物中的硝化作用,如Nitrobacter alkalicus能够在pH高达10的苏打湖沉积物中生存[100],这可能与沉积物中粘土矿物的缓冲作用有关[101-102],而这种机制也一定程度上解释了沉积物(105–107 cells/cm3)比湖泊水体(101–104 cells/cm3)的硝化菌群和硝化速率高出几个数量级的现象(表 3)[95, 99, 103]。
综合湖泊硝化过程与其驱动微生物群落的研究发现,微生物参与的硝化过程很大程度上决定了湖泊无机氮库的赋存形式和分布规律,在湖泊内部氮循环和温室气体N2O的释放方面起着不可忽视的作用,对区域乃至全球气候变化有着深远的影响。AOA、AOB、Comammox等硝化微生物新代谢途径的发现大大拓宽了传统观念中硝化作用的范围,加深了我们对于硝化作用响应气候变化的理解。但通过总结前人的研究和本课题组相关数据可以发现,仍有一些值得深入探究的科学问题,例如:(1)本课题组前期研究观察到青藏高原盐湖中氨氧化速率和氨氧化微生物群落分布规律不一致的现象,考虑到青藏高原湖泊受到极强的太阳辐射,光和盐度共同抑制作用将如何影响着硝化微生物的生态位分布与活力[44-45],值得进一步探究;(2)亚硝酸盐氧化菌(0–35 g/L)和氨氧化菌(0–160 g/L)在盐度适应范围方面存在差异[43, 61-62],这种差异是否会引起盐湖中两步硝化过程的失衡?氨氧化微生物是否与其他微生物建立了新的耦合机制(如厌氧氨氧化、反硝化与氨氧化过程的耦合)? (3)定量区分硝化过程和反硝化过程的N2O生成比例是当前湖泊硝化过程研究的前沿问题,应用同位素成对标记技术和来源区分计算方法将有助于全面认识硝化过程在全球气候变化中的贡献。这些问题目前尚无定论,但解决上述问题无疑可以帮助我们更好地理解各个环境中的氮循环过程及其环境效应。
| Ecosystem | Lake | River | Coastal marine | Deep ocean | |||||||
| Sed | Water | Sed | Water | Sed | Water | Sed | Water | ||||
| Rates | 2–20 | 0–1 | 3–23 | 0–4 | 0.2–7.0 | 0.002–0.200 | 0.003–0.100 | 0.001–0.010 | |||
| Sed denotes Sediment, coastal marine and deep ocean are divided according to maximum depth of less then 100 m and above 100 m. Units for sediment nitrification rates are mmol N/(m2·d), for water nitrification rates are mmol N/(L·d). | |||||||||||
| [1] | Small GE, Bullerjahn GS, Sterner RW, Beall BFN, Brovold S, Finlay JC, McKay RML, Mukherjee M. Rates and controls of nitrification in a large oligotrophic lake. Limnology and Oceanography, 2013, 58(1): 276-286. DOI:10.4319/lo.2013.58.1.0276 |
| [2] | Howarth RW, Marino R. Nitrogen as the limiting nutrient for eutrophication in coastal marine ecosystems:evolving views over three decades. Limnology and Oceanography, 2006, 51(1): 364-376. |
| [3] | Prosser JI, Hink L, Gubry-Rangin C, Nicol GW. Nitrous oxide production by ammonia oxidizers:Physiological diversity, niche differentiation and potential mitigation strategies. Global Change Biology, 2020, 26(1): 103-118. DOI:10.1111/gcb.14877 |
| [4] | Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science, 2004, 304(5667): 66-74. DOI:10.1126/science.1093857 |
| [5] | Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environmental Microbiology, 2005, 7(12): 1985-1995. DOI:10.1111/j.1462-2920.2005.00906.x |
| [6] | Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature, 2005, 437(7058): 543-546. DOI:10.1038/nature03911 |
| [7] | van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, den Camp HJMO, Kartal B, Jetten MSM, Lücker S. Complete nitrification by a single microorganism. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [8] | Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, Von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [9] | Jung MY, Gwak JH, Rohe L, Giesemann A, Kim JG, Well R, Madsen EL, Herbold CW, Wagner M, Rhee SK. Indications for enzymatic denitrification to N2O at low pH in an ammonia-oxidizing archaeon. The ISME Journal, 2019, 13(10): 2633-2638. DOI:10.1038/s41396-019-0460-6 |
| [10] | Hampel JJ, McCarthy MJ, Gardner WS, Zhang L, Xu H, Zhu GW, Newell SE. Nitrification and ammonium dynamics in Taihu Lake, China:seasonal competition for ammonium between nitrifiers and cyanobacteria. Biogeosciences, 2018, 15(3): 733-748. DOI:10.5194/bg-15-733-2018 |
| [11] | Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environmental Microbiology, 2008, 10(11): 3002-3016. DOI:10.1111/j.1462-2920.2008.01764.x |
| [12] | Hou J, Song CL, Cao XY, Zhou YY. Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Research, 2013, 47(7): 2285-2296. DOI:10.1016/j.watres.2013.01.042 |
| [13] | Jiang HC, Dong HL, Yu BS, Lv G, Deng SC, Berzins N, Dai MH. Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, Northwestern China. Geomicrobiology Journal, 2009, 26(3): 199-211. DOI:10.1080/01490450902744004 |
| [14] | Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria:a model for molecular microbial ecology. Annual Reviews of Microbiology, 2001, 55: 485-529. DOI:10.1146/annurev.micro.55.1.485 |
| [15] | Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA. Evolutionary relationships among ammonia-and nitrite-oxidizing bacteria. Journal of Bacteriology, 1994, 176(21): 6623-6630. DOI:10.1128/JB.176.21.6623-6630.1994 |
| [16] | Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. amoA-moAk B, Loy A, Wagner M. lechl S, Gröngröft A, Richter A, Overmann J, Reinhof amoA genes from soils of four different geographic regions. Environmental Microbiology, 2012, 14(2): 525-539. DOI:10.1111/j.1462-2920.2011.02666.x |
| [17] | Koops HP, Pommerening-Röser A. Nitrosococcus//Winogradsky. Bergey's Manual of Systematics of Archaea and Bacteria. New York: John Wiley & Sons, 2015. |
| [18] | Campbell MA, Chain PSG, Dang HY, El Sheikh AF, Norton JM, Ward NL, Ward BB, Klotz MG. Nitrosococcus watsonii sp. nov., a new species of marine obligate ammonia-oxidizing bacteria that is not omnipresent in the world's oceans:calls to validate the names 'Nitrosococcus halophilus' and 'Nitrosomonas mobilis'. FEMS Microbiology Ecology, 2011, 76(1): 39-48. DOI:10.1111/j.1574-6941.2010.01027.x |
| [19] | Cao HL, Auguet JC, Gu JD. Global ecological pattern of ammonia-oxidizing archaea. PLoS One, 2013, 8(2): e52853. DOI:10.1371/journal.pone.0052853 |
| [20] | Yang J, Jiang HC, Dong HL, Wang HY, Wu G, Hou WG, Liu WG, Zhang CL, Sun YJ, Lai ZP. amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Frontiers in Microbiology, 2013, 4: 329. |
| [21] | Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature, 2009, 461(7266): 976-979. DOI:10.1038/nature08465 |
| [22] | Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiology Reviews, 2009, 33(5): 855-869. DOI:10.1111/j.1574-6976.2009.00179.x |
| [23] | Gubry-Rangin C, Kratsch C, Williams TA, McHardy AC, Embley TM, Prosser JI, Macqueen DJ. Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(30): 9370-9375. DOI:10.1073/pnas.1419329112 |
| [24] | Wang BZ, Qin W, Ren Y, Zhou X, Jung MY, Han P, Eloe-Fadrosh EA, Li M, Zheng Y, Lu L, Yan X, Ji JB, Liu Y, Liu LM, Heiner C, Hall R, Martens-Habbena W, Herbold CW, Rhee SK, Bartlett DH, Huang L, Ingalls AE, Wagner M, Stahl DA, Jia ZJ. Expansion of Thaumarchaeota habitat range is correlated with horizontal transfer of ATPase operons. The ISME Journal, 2019, 13(12): 3067-3079. DOI:10.1038/s41396-019-0493-x |
| [25] | Wu YC, Xiang Y, Wang JJ, Zhong JC, He JZ, Wu QL. Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environmental Microbiology Reports, 2010, 2(4): 569-576. DOI:10.1111/j.1758-2229.2010.00146.x |
| [26] | Herrmann M, Saunders AM, Schramm A. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Applied and Environmental Microbiology, 2009, 75(10): 3127-3136. DOI:10.1128/AEM.02806-08 |
| [27] | Auguet JC, Triadó-Margarit X, Nomokonova N, Camarero L, Casamayor EO. Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. The ISME Journal, 2012, 6(9): 1786-1797. DOI:10.1038/ismej.2012.33 |
| [28] | Bollmann A, Bullerjahn GS, McKay RM. Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. PLoS One, 2014, 9(5): e97068. DOI:10.1371/journal.pone.0097068 |
| [29] | Liu B, Li YM, Zhang JP, Zhou XH, Wu CD. Abundance and diversity of ammonia-oxidizing microorganisms in the sediments of Jinshan Lake. Current microbiology, 2014, 69(5): 751-757. DOI:10.1007/s00284-014-0646-0 |
| [30] | Burton SAQ, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Applied and Environmental Microbiology, 2001, 67(7): 2952-2957. DOI:10.1128/AEM.67.7.2952-2957.2001 |
| [31] | Zhang LM, Hu HW, Shen JP, He JZ. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. The ISME Journal, 2012, 6(5): 1032-1045. DOI:10.1038/ismej.2011.168 |
| [32] | Yao HY, Gao YM, Nicol GW, Campbell CD, Prosser JI, Zhang LM, Han WY, Singh BK. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Applied and Environmental Microbiology, 2011, 77(13): 4618-4625. DOI:10.1128/AEM.00136-11 |
| [33] | He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di HJ. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environmental Microbiology, 2007, 9(9): 2364-2374. DOI:10.1111/j.1462-2920.2007.01358.x |
| [34] | Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(6): 2134-2139. DOI:10.1073/pnas.0708857105 |
| [35] | Sears K, Alleman JE, Barnard JL, Oleszkiewicz JA. Impacts of reduced sulfur components on active and resting ammonia oxidizers. Journal of Industrial Microbiology and Biotechnology, 2004, 31(8): 369-378. DOI:10.1007/s10295-004-0157-2 |
| [36] | Martens-Habbena W, Stahl DA. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Methods in Enzymology, 2011, 496: 465-487. DOI:10.1016/B978-0-12-386489-5.00019-1 |
| [37] | Carini SA, Joye SB. Nitrification in Mono Lake, California:Activity and community composition during contrasting hydrological regimes. Limnology and Oceanography, 2008, 53(6): 2546-2557. DOI:10.4319/lo.2008.53.6.2546 |
| [38] | Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. The ISME Journal, 2007, 1(7): 660-662. DOI:10.1038/ismej.2007.79 |
| [39] | Zhang Y, Chen LJ, Dai TJ, Tian JP, Wen DH. The influence of salinity on the abundance, transcriptional activity, and diversity of AOA and AOB in an estuarine sediment:a microcosm study. Applied Microbiology and Biotechnology, 2015, 99(22): 9825-9833. DOI:10.1007/s00253-015-6804-x |
| [40] | Gao J, Hou LJ, Zheng YL, Liu M, Yin GY, Yu CD, Gao DZ. Shifts in the community dynamics and activity of ammonia-oxidizing prokaryotes along the Yangtze Estuarine salinity gradient. Journal of Geophysical Research:Biogeosciences, 2018, 123(11): 3458-3469. DOI:10.1029/2017JG004182 |
| [41] | Hu AY, Yao TD, Jiao NZ, Liu YQ, Yang Z, Liu XB. Community structures of ammonia-oxidising archaea and bacteria in high-altitude lakes on the Tibetan Plateau. Freshwater Biology, 2010, 55(11): 2375-2390. DOI:10.1111/j.1365-2427.2010.02454.x |
| [42] | Valentine DL. Opinion:Adaptations to energy stress dictate the ecology and evolution of the archaea. Nature Reviews Microbiology, 2007, 5(4): 316-323. DOI:10.1038/nrmicro1619 |
| [43] | Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environmental Microbiology, 2011, 13(8): 1908-1923. DOI:10.1111/j.1462-2920.2010.02365.x |
| [44] | Zhang SB, Xia XH, Li SL, Zhang LW, Wang GQ, Li MS, Shi YN, Chen NW. Ammonia oxidizers in high-elevation rivers of the Qinghai-Tibet Plateau display distinctive distribution patterns. Applied and Environmental Microbiology, 2019, 85(22). DOI:10.1128/AEM.01701-19 |
| [45] | Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiology Letters, 2012, 327(1): 41-46. DOI:10.1111/j.1574-6968.2011.02457.x |
| [46] | Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy E, Wagner M, Daims H. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environmental Microbiology, 2014, 16(10): 3055-3071. DOI:10.1111/1462-2920.12300 |
| [47] | Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends in Microbiology, 2016, 24(9): 699-712. DOI:10.1016/j.tim.2016.05.004 |
| [48] | Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damsté JSS, Spieck E, Le Paslier D, Daims H. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(30): 13479-13484. DOI:10.1073/pnas.1003860107 |
| [49] |
Wang ZJ, Wang S, Liu YY, Feng K, Deng Y. The applications of metagenomics in the detection of environmental microbes involving in nitrogen cycle. Biotechnology Bulletin, 2018, 34(1): 1-14.
(in Chinese) 王朱珺, 王尚, 刘洋荧, 冯凯, 邓晔. 宏基因组技术在氮循环功能微生物分子检测研究中的应用. 生物技术通报, 2018, 34(1): 1-14. |
| [50] | Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Frontiers in microbiology, 2013, 4: 27. |
| [51] | Ngugi DK, Blom J, Stepanauskas R, Stingl U. Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. The ISME Journal, 2016, 10(6): 1383-1399. DOI:10.1038/ismej.2015.214 |
| [52] | Sorokin DY, Lücker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WIC, Damsté JSS, Le Paslier D, Muyzer G, Wagner M, Van Loosdrecht MCM, Daims H. Nitrification expanded:discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. The ISME Journal, 2012, 6(12): 2245-2256. DOI:10.1038/ismej.2012.70 |
| [53] | Koch H, Galushko A, Albertsen M, Schintlmeister A, Gruber-Dorninger C, Lücker S, Pelletier E, Le Paslier D, Spieck E, Richter A, Nielsen PH, Wagner M, Daims H. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science, 2014, 345(6200): 1052-1054. DOI:10.1126/science.1256985 |
| [54] | Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen PH, Wagner M, Daims H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(36): 11371-11376. DOI:10.1073/pnas.1506533112 |
| [55] | Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, Mathyer ME, Hallam SJ, Lopez-Garcia P, Takaki Y, Nunoura T, Woyke T, Herndl GJ, Stepanauskas R. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science, 2017, 358(6366): 1046-1051. DOI:10.1126/science.aan8260 |
| [56] | Cai MW, Ng SK, Lim CK, Lu HY, Jia YY, Lee PKH. Physiological and metagenomic characterizations of the synergistic relationships between ammonia- and nitrite-oxidizing bacteria in freshwater nitrification. Frontiers in microbiology, 2018, 9: 280. DOI:10.3389/fmicb.2018.00280 |
| [57] | Nowka B, Daims H, Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria:nitrite availability as a key factor in niche differentiation. Applied and Environmental Microbiology, 2015, 81(2): 745-753. DOI:10.1128/AEM.02734-14 |
| [58] | Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environmental Microbiology, 2006, 8(8): 1487-1495. DOI:10.1111/j.1462-2920.2006.01033.x |
| [59] | Kim YM, Winkler MKH, van Loosdrecht MCM, Chandran K. The effect of inorganic carbon limitation on nitrite oxidizing bacteria. Proceeding of Water Environment Federation, 2012, 2012(15). DOI:10.2175/193864712811725834 |
| [60] | Alawi M, Off S, Kaya M, Spieck E. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environmental Microbiology Reports, 2009, 1(3): 184-190. DOI:10.1111/j.1758-2229.2009.00029.x |
| [61] | Gonzalez-Silva BM, Jonassen KR, Bakke I, Østgaard K, Vadstein O. Nitrification at different salinities:Biofilm community composition and physiological plasticity. Water Research, 2016, 95: 48-58. DOI:10.1016/j.watres.2016.02.050 |
| [62] | Oren A. The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations:implications for the functioning of salt lake ecosystems. Hydrobiologia, 2001, 466(1/3): 61-72. DOI:10.1023/A:1014557116838 |
| [63] | Zhang Y, Qin W, Hou L, Zakem EJ, Wan XH, Zhao ZH, Liu L, Hunt KA, Jiao NZ, Kao SJ, Tang K, Xie XB, Shen JM, Li YF, Chen MM, Dai XF, Liu C, Deng WC, Dai MH, Ingalls AE, Stahl DA, Herndl GJ. Nitrifier adaptation to low energy flux controls inventory of reduced nitrogen in the dark ocean. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(9): 4823-4830. DOI:10.1073/pnas.1912367117 |
| [64] | Yang J, Jiang HC, Wu G, Hou WG, Sun YJ, Lai ZP, Dong HL. Co-occurrence of nitrite-dependent anaerobic methane oxidizing and anaerobic ammonia oxidizing bacteria in two Qinghai-Tibetan saline lakes. Frontiers of Earth Science, 2012, 6(4): 383-391. |
| [65] | Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MMM. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(17): 7104-7109. DOI:10.1073/pnas.0611081104 |
| [66] | Xia F, Wang JG, Zhu T, Zou B, Rhee SK, Quan ZX. Ubiquity and diversity of complete ammonia oxidizers (comammox). Applied and Environmental Microbiology, 2018, 84(24): e01390-18. |
| [67] | Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, Steinberger M, Jetten MSM, Lücker S, Wagner M, Daims H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.3389/fmicb.2017.01508 |
| [68] | Costa E, Pérez J, Kreft JU. Why is metabolic labour divided in nitrification?. Trends in Microbiology, 2006, 14(5): 213-219. DOI:10.1016/j.tim.2006.03.006 |
| [69] | Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. The ISME Journal, 2018, 12(7): 1779-1793. DOI:10.1038/s41396-018-0083-3 |
| [70] | Camejo PY, Santo Domingo J, McMahon KD, Noguera DR. Genome-enabled insights into the ecophysiology of the comammox bacterium "Candidatus Nitrospira nitrosa". MSystems, 2017, 2(5): e00059-17. |
| [71] | Kits KD, Jung MY, Vierheilig J, Pjevac P, Sedlacek CJ, Liu SR, Herbold C, Stein LY, Richter A, Wissel H, Brüggemann N, Wagner M, Daims H. Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata. Nature Communications, 2019, 10(1): 1836. DOI:10.1038/s41467-019-09790-x |
| [72] | Shi Y, Jiang YY, Wang SY, Wang XM, Zhu GB. Biogeographic distribution of comammox bacteria in diverse terrestrial habitats. Science of The Total Environment, 2020, 717: 137257. DOI:10.1016/j.scitotenv.2020.137257 |
| [73] | Xu Y, Lu J, Wang Y, Liu G, Wan X, Hua Y, Zhu D, Zhao J. Diversity and abundance of comammox bacteria in the sediments of an urban lake. Journal of Applied Microbiology, 2020. DOI:10.1111/jam.14593 |
| [74] | Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [75] | Kuypers MMM. Microbiology:a fight for scraps of ammonia. Nature, 2017, 549(7671): 162-163. DOI:10.1038/549162a |
| [76] |
Shi GS, Bai L, Zhou LG. Research advances of complete ammonia oxidizers and its nitrification. Journal of Jilin Jianzhu University, 2018, 35(5): 57-63.
(in Chinese) 史国帅, 白莉, 周立光. 完全氨氧化菌及其硝化作用的研究进展. 吉林建筑大学学报, 2018, 35(5): 57-63. DOI:10.3969/j.issn.1009-0185.2018.05.011 |
| [77] | Di HJ, Cameron KC, Shen JP, Winefield C, O'callaghan M, Bowatte S, He JZ. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nature Geoscience, 2009, 2(9): 621-624. DOI:10.1038/ngeo613 |
| [78] | W MO. The state of the greenhouse gases in the atmosphere based on global observations through 2012. Vienna: WMO Greenhouse Gas Bulletin, 2013. |
| [79] | Neubauer SC, Megonigal JP. Moving beyond global warming potentials to quantify the climatic role of ecosystems. Ecosystems, 2015, 18(6): 1000-1013. DOI:10.1007/s10021-015-9879-4 |
| [80] | Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O):the dominant ozone-depleting substance emitted in the 21st century. Science, 2009, 326(5949): 123-125. DOI:10.1126/science.1176985 |
| [81] | Maavara T, Lauerwald R, Laruelle GG, Akbarzadeh Z, Bouskill NJ, Van Cappellen P, Regnier P. Nitrous oxide emissions from inland waters:Are IPCC estimates too high?. Global Change Biology, 2019, 25(2): 473-488. DOI:10.1111/gcb.14504 |
| [82] | IPCC. The physical science basis. Contribution of working group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Cambridge, New York: Cambridge University Press, 2007. |
| [83] | Liu SR, Han P, Hink L, Prosser JI, Wagner M, Bruggemann N. Abiotic conversion of extracellular NH2OH contributes to N2O emission during ammonia oxidation. Environmental Science & Technology, 2017, 51(22): 13122-13132. |
| [84] | Stieglmeier M, Mooshammer M, Kitzler B, Wanek W, Zechmeister-Boltenstern S, Richter A, Schleper C. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. The ISME Journal, 2014, 8(5): 1135-1146. DOI:10.1038/ismej.2013.220 |
| [85] | Caranto JD, Lancaster KM. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(31): 8217-8222. DOI:10.1073/pnas.1704504114 |
| [86] | Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR, Yang Y, Orsi WD, Moran DM, Saito MA. Genomic and proteomic characterization of "Candidatus Nitrosopelagicus brevis":an ammonia-oxidizing archaeon from the open ocean. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(4): 1173-1178. DOI:10.1073/pnas.1416223112 |
| [87] | Caranto JD, Vilbert AC, Lancaster KM. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(51): 14704-14709. DOI:10.1073/pnas.1611051113 |
| [88] | Koch H, van Kessel MAHJ, Lücker S. Complete nitrification:insights into the ecophysiology of comammox Nitrospira. Applied Microbiology and Biotechnology, 2019, 103(1): 177-189. DOI:10.1007/s00253-018-9486-3 |
| [89] | Jung MY, Well R, Min D, Giesemann A, Park SJ, Kim JG, Kim SJ, Rhee SK. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. The ISME Journal, 2014, 8(5): 1115-1125. DOI:10.1038/ismej.2013.205 |
| [90] | Hink L, Nicol GW, Prosser JI. Archaea produce lower yields of N2O than bacteria during aerobic ammonia oxidation in soil. Environmental Microbiology, 2017, 19(12): 4829-4837. DOI:10.1111/1462-2920.13282 |
| [91] | Hink L, Gubry-Rangin C, Nicol GW, Prosser JI. The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. The ISME Journal, 2018, 12(4): 1084-1093. DOI:10.1038/s41396-017-0025-5 |
| [92] | Su QX, Domingo-Felez C, Jensen MM, Smets BF. Abiotic nitrous oxide (N2O) production is strongly pH dependent, but contributes little to overall N2O emissions in biological nitrogen removal systems. Environmental Science & Technology, 2019, 53(7): 3508-3516. |
| [93] | Zhang JB, Müller C, Cai ZC. Heterotrophic nitrification of organic N and its contribution to nitrous oxide emissions in soils. Soil Biology and Biochemistry, 2015, 84: 199-209. DOI:10.1016/j.soilbio.2015.02.028 |
| [94] | Zhang Y, Zhao W, Cai Z, Müller C, Zhang J. Heterotrophic nitrification is responsible for large rates of N2O emission from subtropical acid forest soil in China. European Journal of Soil Science, 2018, 69(4): 646-654. DOI:10.1111/ejss.12557 |
| [95] | Canfield DE, Kristensen E, Thamdrup B. Aquatic Geomicrobiology. Vol. 48. Amsterdam: Academic Press, 2005. |
| [96] | Bellinger BJ, Jicha TM, Lehto LP, Seifert-Monson LR, Bolgrien DW, Starry MA, Angradi TR, Pearson MS, Elonen C, Hill BH. Sediment nitrification and denitrification in a Lake Superior estuary. Journal of Great Lakes Research, 2014, 40(2): 392-403. DOI:10.1016/j.jglr.2014.03.012 |
| [97] | Beman JM, Chow CE, King AL, Feng YY, Fuhrman JA, Andersson A, Bates NR, Popp BN, Hutchins DA. Global declines in oceanic nitrification rates as a consequence of ocean acidification. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(1): 208-213. DOI:10.1073/pnas.1011053108 |
| [98] | Jeschke C, Falagán C, Knoller K, Schultze M, Koschorreck M. No nitrification in lakes below pH 3. Environmental Science & Technology, 2013, 47(24): 14018-14023. |
| [99] | Verstraete W, Focht DD. Biochemical ecology of nitrification and denitrification//Alexander M. Advances in Microbial Ecology.Boston, MA: Springer, 1977. |
| [100] | Sorokin DY, Muyzer G, Brinkhoff T, Kuenen JG, Jetten MS. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Archives of Microbiology, 1998, 170(5): 345-352. DOI:10.1007/s002030050652 |
| [101] | Ransom B, Bennett RH, Baerwald R, Hulbert MH, Burkett PJ. In situ conditions and interactions between microbes and minerals in fine-grained marine sediments:A TEM microfabric perspective. American Mineralogist, 1999, 84(1/2): 183-192. |
| [102] | Oren A. Halobacterium sodomense sp. nov., a Dead Sea halobacterium with an extremely high magnesium requirement. International Journal of Systematic Bacteriology, 1983, 33(2): 381-386. DOI:10.1099/00207713-33-2-381 |
| [103] | Pauer JJ, Auer MT. Nitrification in the water column and sediment of a hypereutrophic lake and adjoining river system. Water Research, 2000, 34(4): 1247-1254. DOI:10.1016/S0043-1354(99)00258-4 |
 2020, Vol. 60
2020, Vol. 60