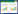中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张英杰, 廖子亚, 赵百锁. 2020
- Zhang Yingjie, Liao Ziya, Zhao Baisuo. 2020
- 嗜盐菌中甘氨酸甜菜碱的合成途径及其生物学功能
- Glycine betaine: biosynthesis and biological function in halophilic microorganisms
- 微生物学报, 60(6): 1074-1089
- Acta Microbiologica Sinica, 60(6): 1074-1089
-
文章历史
- 收稿日期:2019-11-16
- 修回日期:2020-01-09
- 网络出版日期:2020-01-21
2. 重庆市辅仁中学校, 重庆 400066
2. Chongqing Furen High School, Chongqing 400066, China
在嗜盐菌长期的盐适应或短期的盐胁迫过程中,甘氨酸甜菜碱(Glycine Betaine,又称三甲基甘氨酸,N, N, N-trimethylglycine,C5H11NO2)发挥着十分重要的作用[1-4]。甘氨酸甜菜碱属于甘氨酸的一种衍生物(图 1),具有3个甲基和双电荷性质且电荷在分子内分布呈中性;它分子量小(Mw=117.15),是一种季铵型生物碱,具有亲水性和亲脂性、易溶于水,是一种极为重要的相容性溶质。在高盐条件下,该物质可以被嗜盐菌从外界环境中吸收或通过生物合成来维持胞内的高渗透压,以适应细胞外高盐或盐胁迫环境。同时,它能稳定细胞结构,保持酶、蛋白质复合物的活性和细胞膜的完整性,来抵御外部环境中盐碱、高温或冷冻条件导致的迫害[3, 5]。此外,在低盐条件下,该物质也可以被嗜盐菌作为碳源或氮源利用[6]。

|
| 图 1 甘氨酸甜菜碱的结构图 Figure 1 Structure of glycine betaine. |
迄今为止,学术界已发表多篇有影响力的有关嗜盐菌盐适应机制(“相容性溶质机制”和“内盐机制”)及其在生物技术中的应用等综述[1-5, 7-9]。虽然相容性溶质的重要成员之一甘氨酸甜菜碱(以下简称为“甜菜碱”)的2种合成基因及途径的报道尚有一些,但是其合成途径在不同嗜盐菌中的分布特点及潜在应用却鲜有报道。基于目前微生物基因组数据库中已公开的嗜盐菌标准菌株的全基因组序列,本综述将对嗜盐菌(特指微生物适宜生长的盐度在0.5 mol/L以上的纯培养标准菌株)中合成甜菜碱2种途径的分布特点,嗜盐菌之间的亲缘关系及其在现代生物技术的应用,以及本领域内潜在的前沿性研究热点等方面进行述评。希望为后续的研究工作提供新思路,以激发更多学者在该研究领域内的兴趣并取得丰硕的科学研究成果。
1 嗜盐菌中甜菜碱的合成途径大多数嗜盐菌常常从外界环境摄取外源性甜菜碱来适应长期的高盐度环境和应对短期的盐胁迫[2, 5, 9]。但是,若环境中缺乏甜菜碱时,一些嗜盐菌可以通过生物合成来应对极端条件,避免细胞死亡从而得以生存繁殖[7, 10-11]。因此,甜菜碱的生物合成在有些嗜盐菌中成为必不可少的环节且发挥着很重要的作用。
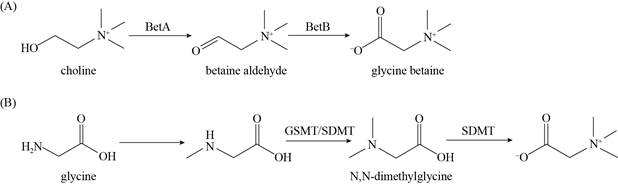
据报道[2, 4, 11],嗜盐菌通过2种途径来合成甜菜碱:①胆碱氧化合成途径:即以胆碱为底物,利用转运蛋白运输到细胞内的胆碱首先被胆碱脱氢酶(choline dehydrogenase,BetA,betA编码)氧化为甜菜碱乙醛,然后被甜菜碱乙醛脱氢酶(betaine aldehyde dehydrogenase,BetB,betB编码)再氧化为甜菜碱(图 2-A)。采用胆碱途径合成甘氨酸甜菜碱的嗜盐细菌来源种类比较多,共有75株(图 3,表 1)。其中,代表性菌株伸长盐单胞菌Halomonas elongate DSM3043 (革兰氏阴性嗜盐细菌的模式菌株)和达坂喜盐芽孢杆菌Halobacillus dabanensis D-8T (革兰氏阳性嗜盐细菌的模式菌株)的甜菜碱胆碱合成途径被广泛报道[12-14]。②甘氨酸甲基化途径:即以甘氨酸为底物,S-腺苷- L-甲硫氨酸(S-adenosyl-L-methionine,AdoMet)作为甲基供体,通过3次连续的N-甲基化,从甘氨酸依次合成肌氨酸、二甲基甘氨酸和甜菜碱。催化该过程的为2个S-腺苷甲硫氨酸依赖性甲基转移酶(甘氨酸肌氨酸甲基转移酶glycine sarcosine N-methyltransferase,GSMT,gsmA编码;肌氨酸二甲基甘氨酸甲基转移酶sarcosine dimethylglycine N-methyltransferase,SDMT,sdmA编码) (图 2-B)。采用甘氨酸甲基化途径合成甘氨酸甜菜碱的嗜盐菌有14株(图 4,表 2)。深入研究的嗜盐细菌有3株,分别为嗜盐放线多孢菌Actinopolyspora halophila ATCC27976T、盐绿需盐红螺菌Ectothiorhodospira halochloris ATCC35916T (现为Halorhodospira halochloris AT)、盐生隐杆藻Aphanothece halophytica strain 7418[11, 15-17]。2003年,Waditee等首次分离并实验证明了嗜盐蓝细菌A. halophytica strain 7418中GSMT和SDMT参与甜菜碱合成的三个甲基化步骤[17]。此外,嗜盐古菌葡萄牙甲烷嗜盐菌Methanohalophilus portucalensis FDF1T合成的MpGSMT催化3步甲基化反应的第1步和第2步[10]。MpGSMT可以分为MpGMT和MpSMT,分别催化甘氨酸甲基化为肌氨酸,肌氨酸甲基化为二甲基甘氨酸。MpSDMT催化甲基化反应的第2步和第3步,MpSDMT可以分为MpSMT和MpDMT,MpDMT催化二甲基甘氨酸甲基化为甜菜碱。研究还发现,基因gsmt和sdmt在排列上有一段重复序列,可能编码相同的酶蛋白序列[10]。嗜盐古菌中存在甜菜碱从头合成途径是令人惊喜的发现,打破了以往仅仅以“内盐机制”为主的观点,也为研究嗜盐古菌中“相容性溶质机制”提供了新视角。最新研究发现[6, 18],伍氏醋酸杆菌Acetobacterium woodii及需盐色盐杆菌Chromohalobacter salexigens DSM 3043T在甘氨酸甲基化合成途径中,产物甜菜碱反馈抑制作用于GSMT上,因此在甘氨酸从头合成途径中,可能发挥关键酶作用的酶为GSMT。
|
| 图 2 嗜盐菌中甘氨酸甜菜碱的2种合成途径 Figure 2 Two synthetic pathways of glycine betaine in halophilic bacteria. A: choline oxidation pathway; B: glycine methylation pathway. |

|
| 图 3 基于16S rRNA基因序列的胆碱氧化合成途径嗜盐细菌的亲缘图 Figure 3 Phylogenetic relationship based on 16S rRNA gene sequence of halophilic bacteria that synthesize glycine betaine through the choline pathway. The scale bar indicates the phylogenetic distance. |
| Species | Accession number | [Na+] opt./(mol/L) | [Na+] range/(mol/L) | pH opt. | pH range |
| Actinopolyspora halophila DSM 43834T | AQUI01000002 | 2.56–3.42 | 1.71–5.13 | ND | ND |
| Aestuariibacter salexigens DSM 15300T | AY207502 | 0.34–1.03 | 0.17–1.71 | 7.0–8.0 | ND |
| Afifella marina DSM 2698T | FMVW01000007 | 0.17–0.85 | 0–0.68 | 6.9–7.1 | ND |
| Alcanivorax borkumensis DSM 11573T | AM286690 | 0.51–1.71 | 0.17–2.14 | ND | ND |
| Alcanivorax dieselolei DSM 16502T | CP003466 | 0.51–1.28 | 0.17–2.56 | ND | ND |
| Alcanivorax pacificus W11-5T | AJGP01000039 | 0.51–0.85 | 0.08–2.05 | ND | ND |
| Alkalicoccus saliphilus DSM 15402T | AJ493660 | 2.74 | 0.17–4.27 | 9.0 | 6.5–10.0 |
| Alkalicoccus halolimnae BZ-SZ-XJ29T | KX618877 | 1.42–2.10 | 0.74–4.15 | 7.5 | 6.0–10.5 |
| Arhodomonas aquaeolei ATCC 49307T | M26631 | 2.56 | 1.03–3.42 | 7.0 | 6.0–8.0 |
| Bacillus aidingensis DSM 18341T | DQ504377 | 2.05 | 1.37–5.64 | 7.2 | 6.0–9.5 |
| Bacillus atrophaeus DSM7264T | AB021181 | 1.20 | ND | 5.6–5.7 | ND |
| Bacillus chagannorensis DSM 18086T | AM492159 | 1.20 | 0.51–3.42 | 8.5 | 5.8–10.0 |
| Bacillus selenitireducens DSM 15326T | ABHZ01000019 | 0.41–1.03 | 0.34–3.76 | 8.5–10.0 | ND |
| Bacillus urumqiensis BZ-SZ-XJ18T | KM066107 | 1.80 | 0.22–4.32 | 8.5–9.5 | 6.5–10.0 |
| Candidatus Chlorothrix halophila | AY395567 | 1.71 | 0.85–2.05 | 7.8–8.4 | ND |
| Chromohalobacter salexigens DSM 3043T | CP000285 | 1.28–1.71 | 0.15–4.27 | 7.5 | 5.0–10.0 |
| Desulfohalobium retbaense DSM 5692T | CP001734 | 1.71 | 0–4.10 | 6.5–7.0 | 5.5–8.0 |
| Desulfovermiculus halophilus DSM 18834T | DQ139408 | 1.37–1.71 | 0.51–3.93 | 7.2 | 6.4–8.2 |
| Desulfovibrio bastinii DSM 16055T | AY359868 | 0.68 | 0.17–2.05 | 5.8–6.2 | 5.2–7.4 |
| Desulfovibrio gracilis DSM 16080T | FUYC01000042 | 0.85–1.03 | 0.34–2.05 | 6.8–7.2 | 5.4–8.4 |
| Desulfovibrio oxyclinae DSM 11498T | U33316 | 0.85–1.71 | 0.43–3.85 | 7.0–7.5 | ND |
| Desulfovibrio vietnamensis DSM 10520T | X93994 | 0.85 | 0–1.71 | 7.5 | 5.0–10.0 |
| Ectothiorhodospira haloalkaliphila ATCC 51935T | KK214998 | 0.85 | 0.43–2.56 | 8.5–10 | ND |
| Enterovibrio calviensis DSM 14347T | AF118021 | 0.43–0.60 | 0.26–2.05 | ND | ND |
| Flexistipes sinusarabici DSM 4947T | CP002858 | 0.51 | 0.51–3.08 | ND | 6.0–8.0 |
| Fodinicurvata fenggangensis DSM 21160T | JMLV01000013 | 0.85 | 0.26—3.42 | 7.5 | 6.5–8.5 |
| Gracilibacillus lacisalsi DSM 19029T | DQ664540 | 0.85–1.20 | 0.17–3.08 | 7.5–8.0 | 5.5–10 |
| Hahella ganghwensis DSM 17046T | AQXX01000025 | 0.68–1.03 | 0.17–1.71 | 7.0–8.0 | 5.0–10.0 |
| Halalkalibacillus halophilus DSM 18494T | AB264529 | 1.71–2.56 | 0.85–4.27 | 8.5–9.0 | 5.5–10.0 |
| Halobacillus halophilus DSM 2266T | HE717023 | 0.51–0.85 | 0–2.56 | 7.8 | 7.0–9.0 |
| Halobacillus kuroshimensis DSM 18393T | AB195680 | 1.03 | 0.08–4.27 | 7.5–8.0 | 5.5–10.0 |
| Haloglycomyces albus DSM 45210T | AZUQ01000001 | 1.37–2.05 | 0.51–3.08 | 7.0–7.5 | 5.0–9.0 |
| Halomonas anticariensis FP35T | ASTJ01000004 | 1.28 | 0.08–2.56 | ND | 6.0–9.0 |
| Halomonas elongata DSM 2581T | FN869568 | 0.60–1.37 | 0–5.47 | ND | 5.0–9.0 |
| Halomonas halocynthiae DSM 14573T | AJ417388 | 0.51–1.03 | 0.08–2.56 | 7.5–8.5 | 5.0–11.0 |
| Halomonas halodenitrificans DSM 735T | L04942 | 0.30–2.0 | 0–5.5 | ND | ND |
| Halomonas ilicicola DSM 19980T | EU218533 | 1.71 | 0.34–3.0 | 6.5 | 6.0–9.0 |
| Halomonas jeotgali HwaT | AMQY01000015 | 1.71 | 0.85–4.27 | 7.0–8.0 | 5.0–10.0 |
| Halomonas lutea DSM 23508T | ARKK01000003 | 0.85–1.71 | 0.17–3.42 | 7.5 | 5.0–9.0 |
| Halomonas urumqiensis BZ-SZ-XJ27T | KM066108 | 1.42 | 0.22–4.32 | 8.0–8.5 | 6.0–10.0 |
| Halomonas zhanjiangensis DSM 21076T | FJ429198 | 0.51–0.85 | 0.17–3.42 | 7.5 | 6.0–10.5 |
| Halonatronum saccharophilum DSM 13868T | AZYG01000049 | 1.20–2.05 | 0.51–2.91 | 8.0–8.5 | 7.7–10.3 |
| Idiomarina loihiensis L2TRT | AE017340 | 1.28–1.71 | ND | ND | ND |
| Kushneria aurantia A10T | AM941746 | 1.71 | 0.85–3.0 | 7.0–8.0 | 5.5–8.5 |
| Kushneria indalinina CG2.1T | AJ427627 | 1.28–1.71 | 0.51–4.27 | 7.2 | 5.0–9.0 |
| Lentibacillus juripiscarius JCM 12147T | AB127980 | 1.71 | 0.51–5.13 | 7.0 | 5.0–9.0 |
| Limimaricola hongkongensis DSM 17492T | APGJ01000007 | ND | 0.34–2.39 | ND | 5.0–10.0 |
| Litorimicrobium taeanense DSM 22007T | jgi.1055387 | 0.34–0.68 | 0.17–1.03 | 6.5–7.5 | 6.0–8.0 |
| Marinobacter algicola DG893T | ABCP01000031 | 0.51–1.03 | 0.17–2.05 | 7.5 | 5.0–10.0 |
| Marinobacter daepoensis DSM 16072T | AY517633 | 0.34–1.03 | 0–3.08 | 7.0–8.0 | 5.5–ND |
| Marinobacter hydrocarbonoclasticus ATCC 49840T | FO203363 | 0.60 | 0.08–3.5 | 7.0–7.5 | 6.0–9.5 |
| Marinobacterium halophilum DSM 17586T | AY563030 | ND | 0.51–2.05 | ND | 5.3–9.3 |
| Marinobacterium lutimaris DSM 22012T | jgi.1107934 | 0.34–0.85 | 0.17–1.71 | 6.5–7.5 | 6.0–8.0 |
| Marinomonas ushuaiensis DSM 15871T | AJ627909 | 1.03 | 0.17–ND | 8.0 | 7.0–12.5 |
| Methylophaga aminisulfidivorans KCTC 12909T | AFIG01000001 | 0.51 | 0.26–1.54 | 6.8–7.0 | 6.0–8.0 |
| Nocardiopsis chromatogenes YIM 90109T | AY619715 | 0.85–1.37 | 0–3.08 | 7.2 | 6.0–9.0 |
| Nocardiopsis gilva YIM 90087T | AY619712 | 0.85–1.37 | 0–3.08 | 7.2 | 6.0–9.0 |
| Nocardiopsis salina YIM 90010T | AY373031 | 1.71 | 0.51–3.42 | 7.2 | 6.0–9.0 |
| Oceanicella actignis DSM 22673T | JQ864435 | 0.34–0.85 | 0.17–1.54 | 7.5–8.0 | 5.5–9.5 |
| Pontibacillus marinus BH030004T | AVPF01000156 | 0.34–0.85 | 0.17–1.54 | 7.0–7.5 | 6.0–9.0 |
| Rubidibacter lacunae KORDI 51-2T | ASSJ01000063 | 0.85 | 0.34–1.20 | ND | ND |
| Ruegeria lacuscaerulensis ITI-1157T | ACNX01000031 | 0.60 | 0–1.20 | 7.5 | ND |
| Saccharomonospora halophila 8T | AICX01000084 | 1.71 | 1.71–5.13 | ND | ND |
| Saccharomonospora paurometabolica YIM 90007T | AF540959 | 1.71 | ND | ND | ND |
| Salinibacter ruber DSM 13855T | CP000159 | 2.56–5.13 | 2.56–ND | 6.5–8.0 | 6.0–8.5 |
| Salinicoccus albus DSM 19776T | EF177692 | 1.71 | 0.17–5.13 | 8.5 | 6.0–10.0 |
| Salinicoccus kunmingensis DSM 17847T | DQ837380 | 1.36–1.71 | 0.08–4.27 | 8.0 | 6.0–10.0 |
| Salinimonas chungwhensis DSM 16280T | AY553295 | 0.34–0.85 | 0–2.56 | 7.0–8.0 | ND |
| Salipiger mucosus DSM 16094T | ARRM01000007 | 1.54–1.71 | 0.08–3.42 | ND | 6.0–10.0 |
| Salsuginibacillus kocurii DSM 18087T | AM492160 | 1.71 | 0.51–3.42 | 8.5 | 5.8–10.0 |
| Sedimentitalea nanhaiensis DSM 24252T | AXBG01000021 | 0.17–0.68 | 0.10–1.03 | 7.0–8.5 | 6.0–9.3 |
| Streptomyces zinciresistens K42T | AGBF01000432 | 0.68 | 0.68–1.20 | 7.5 | 5.0–11.0 |
| Thalassospira profundimaris WP0211T | AY186195 | 0.51–0.68 | 0.34–1.37 | ND | ND |
| Thalassospira xiamenensis DSM 17429T | CP004388 | 0.34–0.68 | 0.08–1.71 | ND | ND |
| Thioalkalivibrio paradoxus ARh 1T | AF151432 | 0.50–1.00 | 0.30–4.30 | 10.2 | ND |
| ND: Data not shown in the published paper. | |||||

|
| 图 4 基于16S rRNA基因序列的甘氨酸甲基化合成途径嗜盐菌的亲缘图 Figure 4 Phylogenetic relationship based on 16S rRNA gene sequence of halophiles that synthesize glycine betaine through glycine methylation pathway. The scale bar indicates the phylogenetic distance. Numbers at the branch nodes are bootstrap values for 1000 replicates. |
| Species | Accession number | [Na+] opt./(mol/L) | [Na+] range/(mol/L) | pH opt. | pH range |
| Actinopolyspora halophila DSM 43834T | AQUI01000002 | 2.56–3.42 | 1.71–5.13 | ND | ND |
| Aphanothece halophytica ATCC 43922T | AJ000721 | 1.00–4.00 | 0–5.0 | ND | ND |
| Arhodomonas aquaeolei DSM 8974 | M26631 | 2.56 | 1.03–3.42 | 6.5–7.5 | 6.0–8.0 |
| Candidatus Chlorothrix halophile | AY395567 | 1.71 | 0.85–2.05 | 7.8–8.4 | ND |
| Desulfonatronospira thiodismutans ASO3-1T | ACJN01000005 | 1.71 | 0–3.08 | 9.0 | 7.0–10.0 |
| Ectothiorhodospira haloalkaliphila ATCC 51935T | KK214998 | 0.85–1.03 | 0.43–2.56 | 8.5–10.0 | ND |
| Fodinicurvata fenggangensis DSM 21160T | JMLV01000013 | 0.85 | 0.26–3.42 | 7.5 | 6.5–8.5 |
| Halorhodospira halochloris AT | AP017372 | 2.40–4.62 | 1.54–5.13 | 8.1–9.1 | ND |
| Halorhodospira halophila SL1T | CP000544 | 1.88–3.76 | 1.54–5.13 | 7.4–7.9 | ND |
| Methanohalophilus portucalensis FDF1T | JWTK01000010 | 0.51–2.56 | ND | 6.5–7.5 | ND |
| Natranaerobius thermophilus JW/NM-WN-LFT | CP001034 | 3.90 | 3.10–4.90 | 9.5 | 8.3–10.6 |
| Rubidibacter lacunae KORDI 51-2T | ASSJ01000063 | 0.85 | 0.34–1.20 | ND | ND |
| Saccharomonospora paurometabolica YIM 90007T | AF540959 | 1.71 | ND | ND | ND |
| Thioalkalivibrio paradoxus ARh 1T | AF151432 | 0.50–1.00 | 0.30–4.30 | 10.2 | ND |
| ND: Data not shown in the published paper. | |||||
2 甜菜碱来源于多种类型的嗜盐菌
截止笔者撰写时,从JGI-IMG (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi)和EZBioCloud (https://www.ezbiocloud.net)数据库中共获得了134株含有甜菜碱生物合成基因的嗜盐菌标准菌株。其中,75株嗜盐细菌(占总数~56.0%,表 1)和53株嗜盐古菌(~39.6%,表 3)含有胆碱氧化途径所需的2个基因;13株嗜盐细菌(~9.7%)和1株嗜盐古菌(~0.7%)拥有甘氨酸甲基化途径所需的2个基因(表 2)。令人兴奋的是,我们发现有8株嗜盐细菌同时携带甜菜碱2种生物合成途径所需的基因(表 4),说明他们的盐适应调控机制可能存在独特性和复杂性,此类科学问题在以往的报道中还没有提及。因此,共存的2种甜菜碱合成途径在这8株嗜盐细菌中究竟是如何发挥作用,是否存在优先合成机制等还需要更深入的研究。本课题组正在从生理生化、分子遗传和组学工具(基因组、转录组和蛋白组)等方面进行全面的、系统的和深入的研究,期望不久的将来揭开2种甜菜碱生物合成共存途径的科学面纱。
| Species | Accession number | [Na+] opt./(mol/L) | [Na+] range/(mol/L) | pH opt. | pH range |
| Haloferax alexandrinus JCM 10717T | AB037474 | 4.27 | 1.71–5.30 | 7.2 | 5.5–7.5 |
| Haloferax denitrificans S1T | AOLP01000013 | 2.00–3.00 | 1.50–4.50 | 6.7 | 6.0–8.0 |
| Haloferax elongans ATCC BAA-1513T | DQ860977 | 2.60–3.40 | 1.70–5.10 | 7.4 | 7.0–9.0 |
| Haloferax gibbonsii ATCC 33959T | AOLJ01000012 | 2.56–4.27 | 1.71–5.30 | 6.5–7.0 | 5.5–8.0 |
| Haloferax larsenii JCM 13917T | AOLI01000021 | 2.20–3.40 | 1.00–4.80 | 6.5–7.0 | 6.0–8.5 |
| Haloferax lucentense DSM 14919T | AOLH01000027 | 4.27 | 1.71–5.13 | 7.5 | 5.0–9.0 |
| Haloferax mediterranei R-4T | CP001868 | 2.90 | 2.05–4.27 | 6.5 | ND |
| Haloferax mucosum PA12T | AOLN01000016 | 2.60–3.40 | 1.70–5.10 | 7.4 | 6.0–10.0 |
| Haloferax prahovense TL6T | AB258305 | 3.50 | 2.50–5.20 | 7.0–7.5 | 6.0–8.5 |
| Haloferax sulfurifontis M6T | AOLM01000015 | ND | 1.03–ND | 6.4–6.8 | 4.5–9.0 |
| Haloferax volcanii DS2T | CP001956 | 1.70–2.50 | ND | ND | ND |
| Halobaculum gomorrense DSM 9297T | L37444 | 1.50–2.50 | 1.00–ND | 6.0–7.0 | 5.5–8.0 |
| Halogeometricum borinquense DSM 11551T | ABTX01000001 | 3.42–4.27 | 1.37–ND | ND | ND |
| Halogeometricum pallidum JCM 14848T | AOIV01000004 | 3.70 | 1.70–4.30 | 6.5 | 5.0–8.5 |
| Haloplanus natans DSM 17983T | KE386573 | 3.00 | 2.60–4.30 | 7.0 | 6.5–8.0 |
| Halorubrum aidingense JCM 13560T | DQ355813 | 2.60 | 1.70–4.30 | 7.5 | 7.0–9.0 |
| Halorubrum arcis AJ201T | DQ355793 | 3.40–3.90 | 2.20–5.20 | 7.5 | 6.0–8.5 |
| Halorubrum californiense SF3-213T | EF139654 | 3.50–4.50 | 2.50–5.00 | 7.3 | 6.8–8.5 |
| Halorubrum coriense Ch2T | L00922 | ND | 1.50–5.30 | ND | ND |
| Halorubrum ezzemoulense DSM 17463T | DQ118426 | 3.42 | 2.56–4.27 | 7.0–7.5 | 6.5–9.0 |
| Halorubrum kocurii JCM 14978T | AOJH01000060 | 3.42 | 2.50–ND | 7.5 | 6.0–9. |
| Halorubrum lipolyticum DSM 21995T | DQ355814 | 2.60 | 1.70–4.80 | 7.5 | 7.0–9.0 |
| Halorubrum litoreum JCM 13561T | AOJF01000057 | 3.42 | 2.00–5.10 | 7.0–7.5 | 6.5–9.0 |
| Halorubrum saccharovorum M6T | U17364 | 3.50–4.50 | 1.50–5.20 | ND | ND |
| Halorubrum distributum JCM 9100T | D63572 | 4.27 | 1.50–5.30 | ND | ND |
| Halorubrum sodomense DSM 3755T | jgi.1055354 | 1.70–2.50 | 0.50–4.30 | ND | ND |
| Halorubrum tebenquichense DSM 14210T | AOJD01000002 | ND | 2.50–5.20 | ND | 7.0–10.0 |
| Halorubrum terrestre JCM 10247T | AB090169 | 4.27 | 2.56–5.13 | 7.5 | 5.0–9.0 |
| Halopiger xanaduensis SH-6T | CP002839 | 4.30 | 2.50–5.00 | 7.5–8.0 | 6.0–11.0 |
| Halobiforma lacisalsi AJ5T | CP019285 | 2.60–4.30 | 1.70–ND | 7.5 | 6.5–9.0 |
| Haloterrigena limicola JCM 13563T | DQ367241 | 3.10 | 1.70–5.10 | 7.0 | 6.5–9.0 |
| Haloterrigena saccharevitans AB14T | LWLN01000002 | 3.00–3.40 | 1.70–ND | 7.5 | 6.5–8.5 |
| Haloterrigena salina JCM 13891T | AM942763 | 3.40 | 2.50–5.00 | 7.0–8.0 | 6.0–9.0 |
| Halovivax asiaticus JCM 14624T | AOIQ01000016 | 3.40 | 2.50–4.30 | 7.0–7.5 | 6.0–9.0 |
| Halovivax ruber XH-70T | CP003050 | 3.40 | 2.50–4.30 | 7.0–7.5 | 6.0–9.0 |
| Natrinema altunense JCM 12890T | JNCS01000019 | 3.00–4.30 | 1.70–ND | 7.0–7.7 | 6.0–8.0 |
| Natrinema gari JCM 14663T | AB289741 | 2.60–3.40 | 1.70–5.1 | 6.0–6.5 | 5.5–8.5 |
| Natrinema pallidum DSM 3751T | AJ002949 | 3.40–4.30 | 1.70–ND | 7.2–7.6 | 6.0–8.4 |
| Natrinema pellirubrum DSM 15462T | AGIN01000009 | 3.40–4.30 | 2.00–ND | 7.2–7.8 | 6.0–8.6 |
| Natrinema versiforme JCM 10478T | AB023426 | 3.40–4.30 | 1.50–ND | 6.5–7.0 | 6.0–8.0 |
| Natronococcus jeotgali DSM 18795T | EF077631 | 3.93–4.27 | 1.28–5.13 | 7.5 | 7.0–9.5 |
| Natronorubrum tibetense GA33T | KB913017 | 3.40 | 2.00–5.10 | 9.0 | 8.5–11.0 |
| Natronococcus occultus SP4T | CP003929 | 2.80 | 1.40–5.20 | 9.5 | 8.5–11.0 |
| Haladaptatus paucihalophilus DX253T | AQXI01000001 | 2.60–3.10 | 0.80–5.10 | 6.0–6.5 | 5.0–7.5 |
| Halalkalicoccus jeotgali B3T | CP002062 | 2.56 | 1.71–5.13 | 7.0 | 6.5–9.0 |
| Haloarcula amylolytica JCM 13557T | DQ826512 | 2.90–3.20 | 2.00–5.10 | 7.0–7.5 | 6.5–9.0 |
| Haloarcula argentinensis DSM 12282T | EF645681 | 2.50–3.00 | 2.00–4.50 | ND | ND |
| Haloarcula japonica DSM 6131T | AB355986 | 3.42 | 2.56–5.13 | 7.0–7.5 | 6.0–8.0 |
| Halococcus saccharolyticus DSM 5350T | AOMD01000030 | 4.27 | 2.56–5.3 | 7.2 | 6.0–8.0 |
| Halococcus salifodinae DSM 8989T | AOME01000075 | 3.42–4.27 | 2.56–ND | ND | 6.8–9.5 |
| Halosimplex carlsbadense JCM 11222T | HQ263562 | 4.27 | 3.42–5.13 | ND | 7.0–8.0 |
| Natronomonas moolapensis DSM 18674T | HF582854 | 3.10–3.42 | 2.39–6.15 | 7.0–7.5 | 5.5–8.5 |
| Salinigranum rubrum GX10T | GU951431 | 3.10 | 1.40–4.80 | 7.0 | 5.0–9.0 |
| ND: Data not shown in the published paper. | |||||
| Species | Accession number | [Na+] opt./(mol/L) | [Na+] range/(mol/L) | pH opt. | pH range |
| Actinopolyspora halophila DSM 43834T | AQUI01000002 | 2.56–3.42 | 1.71–5.13 | ND | ND |
| Saccharomonospora paurometabolica YIM 90007T | AF540959 | 1.71 | ND | ND | ND |
| Arhodomonas aquaeolei DSM 8974 | M26631 | 2.56 | 1.03–3.42 | 6.5–7.5 | 6.0–8.0 |
| Ectothiorhodospira haloalkaliphila ATCC 51935T | KK214998 | 0.85–1.03 | 0.43–2.56 | 8.5–10 | ND |
| Fodinicurvata fenggangensis DSM 21160T | JMLV01000013 | 0.85 | 0.26–3.42 | 7.5 | 6.5–8.5 |
| Thioalkalivibrio paradoxus ARh 1T | AF151432 | 0.50–1.00 | 0.30–4.30 | 10.2 | ND |
| Candidatus Chlorothrix halophile | AY395567 | 1.71 | 0.85–2.05 | 7.8–8.4 | ND |
| Rubidibacter lacunae KORDI 51-2T | ASSJ01000063 | 0.85 | 0.34–1.20 | ND | ND |
| ND: Data not shown in the published paper. | |||||
采用胆碱氧化途径合成甜菜碱的嗜盐细菌标准菌株在门分类水平上,依次为变形菌门(Proteobacteria,占总数~61.3%)、厚壁菌门(Firmicutes,~22.7%)、放线菌门(Actinobacteria,~10.7%),其余则不足2%,分布在脱铁杆菌门(Deferribacteres)、绿弯菌门(Chloroflexi)、蓝细菌门(Cyanobacteria)和Rhodothermaeota (原属于Bacteroidetes[19])。在纲的水平上,依次为γ-变形菌纲(γ-Proteobacteria,~40.0%)、α-变形菌纲(α-Proteobacteria,~13.3%)、芽孢杆菌纲(Bacilli,~20.0%)、δ-变形菌纲(δ-Proteobacteria,~8.0%)和放线菌纲(Actinobacteria,~10.0%)。此外,梭菌纲(Clostridia)、脱铁杆菌纲(Deferribacteres)、绿球菌纲(Chroobacteria)和绿弯菌纲(Chloroflexia)等所占比例都小于2% (图 3和表 1)。其中,本课题组测序分析了变形菌门中的Halomonas urumqiensis BZ-SZ-XJ27T,以及厚壁菌门中的Bacillus urumqiensis BZ-SZ-XJ18T和Alkalicoccus saliphilus DSM 15402T全基因组序列[20-23]。系统在嗜盐细菌适应极端环境中发育分析表明,这些嗜盐细菌在亲缘关系上相距甚远(图 3),但是它们都编码胆碱氧化途径所需的胆碱脱氢酶(BetA)和甜菜碱乙醛脱氢酶(BetB),由此可见胆碱氧化途径以及所合成的甜菜碱的作用举足轻重。
而采用胆碱氧化途径合成甜菜碱的嗜盐古菌都属于广古菌门(Euryarchaeota)中的盐杆菌纲(Halobacteria) (图 5和表 3)。在科的水平上,仅有1株菌属于Halorubraceae,其余菌株依次被归类为盐富饶菌科(Haloferacaceae,占总数~52.8%)、盐细菌科(Halobacteriaceae,~17.0%)和钠白菌科(Natrialbaceae,~28.3%)。合成甜菜碱的嗜盐古菌多数被归类于盐富饶菌科,这类菌的嗜盐浓度一般都高于嗜盐细菌,对Na+的适应浓度普遍大于4.0 mol/L。一般而言,嗜盐古菌采用“内盐机制”来适应高盐环境[24-25]。但是,我们也发现有多种类型的嗜盐古菌携带甜菜碱胆碱氧化合成所需基因,暗示着一些嗜盐古菌也有可能应用“相容性溶质机制”来抵御外界的高盐条件。以上假说还需要更多的理论及实验数据来验证。究竟哪一种机制为主还是二者相辅相成,需要进一步阐明。

|
| 图 5 基于16S rRNA基因序列的胆碱氧化合成途径嗜盐古菌的亲缘图 Figure 5 Phylogenetic relationship based on 16S rRNA gene sequence of halophilic archaea that synthesize glycine betaine through choline pathway. The scale bar indicates the phylogenetic distance. |
相对于胆碱氧化途径,嗜盐菌中的甘氨酸甲基化途径的报道还比较少[10-11, 15-17, 26-27],数据库分析表明,仅有14株嗜盐菌含有甘氨酸甲基化途径所需的基因(图 4和表 2)。从门分类水平上,依次为变形菌门(占总数~50.0%)、放线菌门(~14.3%)、蓝细菌门(~14.3%),而厚壁菌门和绿弯菌门数量上较少,均占~7%。在纲分类水平上,多数属于γ-变形菌纲(~35.7%)和放线菌纲(~14.3%),一部分属于α-变形菌纲(~11.7%)。而δ-变形菌纲、绿弯菌纲、梭菌纲和绿球菌纲所占数量较少(~7.1%)。梭菌纲中嗜热盐碱厌氧菌Natranaerobius thermophilus JWNM-WN-LFT是一株厌氧嗜盐、嗜碱和嗜热的多嗜极细菌,本研究组采用蛋白组学iTRAQ技术、RNA-Seq技术和数字PCR技术,在不同极端条件下检测该菌的抗逆特性,发现了甜菜碱甲基化途径相关基因的表达。实验表明,GSMT和SDMT从低盐碱热条件下(3.0 mol/L Na+、pH 8.8和42 ℃)到高盐碱热条件下(4.0 mol/L Na+、pH 9.8和52 ℃),GSMT的基因上调1.75倍,SDMT的基因上调2.02倍(数据未发表)。此外,仅发现1株嗜盐古菌(葡萄牙甲烷嗜盐菌,Methanohalophilus portucalensis FDF1T)采用了甘氨酸甲基化途径合成甜菜碱[10]。值得关注的是,采用甘氨酸甲基化途径的嗜盐细菌和嗜盐古菌都编码合成甘氨酸肌醇甲基转移酶和肌氨酸二甲基甘氨酸甲基转移酶,但两种酶在嗜盐细菌和嗜盐古菌中结构和功能是否存在差异,还需要后续的实验工作来揭示其可能存在的深刻含义。
此外,有8株嗜盐细菌同时存在以胆碱或甘氨酸为底物的两种生物合成途径(表 4),包括变形菌:水油非红单胞菌Arhodomonas aquaeolei DSM 8974T、喜盐碱外硫红螺菌Ectothiorhodospira haloalkaliphila ATCC 51935T、Fodinicurvata fenggangensis DSM 21160T和Thioalkalivibrio paradoxus ARh 1T,放线菌:嗜盐放线多孢菌Actinopolyspora halophila DSM 43834T、寡食糖单孢菌Saccharomonospora paurometabolica YIM 90007T,绿弯菌:Candidatus Chlorothrix halophileT,和蓝细菌Rubidibacter lacunae KORDI 51-2T。8株嗜盐细菌对于Na+的最适浓度范围为0.50–2.56 mol/L,生长浓度范围为0–4.30 mol/L。这些嗜盐菌存在两种合成途径,但两种合成途径如何相互协调发挥作用,是否存在某种诱导因素使嗜盐菌仅选择其中一种机制合成甜菜碱,这是目前相关领域的研究中没有提及的,也是值得相关学者进行深入研究的很好的切入点。值得关注的是,采用甘氨酸从头合成甜菜碱是相对较为耗能的策略机制,因为甲基化反应是自然界中高耗能反应之一,每转移一个S-腺苷-L-甲硫氨酸供体需要细胞消耗12个ATP[15]。因此甘氨酸甲基化从头合成途径在自然界中较为罕见,但仍有部分嗜盐菌采用此种途径,其中的生物学功能还需要进一步探索。
3 甜菜碱在盐适应及盐胁迫中起重要的作用嗜盐菌中甜菜碱的生物学功能被报道了一些[2, 4, 9, 11, 28-29],但对采用不同合成途径的嗜盐菌中甜菜碱基因的作用是否存在差异鲜有报道。由于嗜盐菌不是分类学中的专有名词,不同门属之间的嗜盐菌的生理特性、携带的基因结构及其功能上存在着较大的差异。因此本文选取胆碱途径和甘氨酸途径中较为代表性的菌株进行探讨,希望对甜菜碱的盐适应机理进行系统的阐述。
作为革兰氏阴性的中度嗜盐细菌的模式菌株,伸长盐单胞菌(Halomonas elongata)中胆碱合成途径已被深入研究[12-13, 30-31]。研究表明,在2.0 mol/L NaCl高盐条件下,H. elongata DSM3043细胞吸收胆碱的速率比0.5 mol/L NaCl低盐条件下快4倍。此外,在2.0 mol/L NaCl条件下,该菌株从环境中吸收胆碱合成甜菜碱的菌株传代数是合成途径受阻菌株的2倍。作为革兰氏阳性中度嗜盐细菌的模式菌株,达坂喜盐芽孢杆菌(Halobacillus dabanensis D-8T)在1.0 mol/L和2.0 mol/L NaCl条件下,培养基中添加胆碱的OD600值比未处理的菌株高6倍[14]。胆碱虽然不是相容性渗透保护物质,但对于能进行生物合成甜菜碱的嗜盐菌十分重要。嗜盐古菌的盐适应机理已经得到了广泛的研究,普遍被认可的嗜盐机理为“内盐机制”,而盐杆菌纲作为研究较为透彻的嗜盐古菌,其盐适应机制已经得到了广泛的研究,早在1970s就有报道嗜盐古菌的“内盐机制”[32-33],即在高盐条件下嗜盐古菌通过吸收钾离子来增加细胞内的渗透压,以抵御细胞外盐环境或响应盐冲击所致的胁迫环境[24-25, 34]。但是,迄今为止尚未发现关于嗜盐古菌中存在胆碱氧化途径合成甜菜碱的报道,对于嗜盐古菌中甜菜碱的报道仅仅涉及转运部分,尚无报道涉及嗜盐古菌采用胆碱氧化途径合成甜菜碱[2, 29, 34]。此外,尚无本文所述的53株携带胆碱氧化途径相关基因的嗜盐古菌的报道,那么在这53株古菌中究竟还是以一种机制为主还是两者相辅相成,这将成为后续研究的突破点和创新点。
对从甘氨酸合成甜菜碱的嗜盐菌而言,甜菜碱是否也有明显促进其在高盐环境中生长的效果呢?嗜盐放线多孢菌A. halophila ATCC27976T和盐绿需盐红螺菌E. halochloris ATCC35916T (现为Halorhodospira halochloris AT)是目前研究较为深入的采用甘氨酸甲基化途径的嗜盐细菌,该研究证实了两株嗜盐菌均可以在高盐环境中积累甜菜碱,4.0 mol/L NaCl环境中细胞内甜菜碱比2.0 mol/L NaCl环境中高2倍,并且已经完成相关合成蛋白的功能测定,表明该菌株在高盐刺激下甜菜碱合成蛋白发挥作用,促进甜菜碱的合成[11, 15-16]。厚壁菌门中嗜热盐碱厌氧菌Natranaerobius thermophilus JWNM-WN-LFT是一株厌氧、嗜盐、嗜碱、嗜热细菌,最佳生长条件为3.9 mol/L Na+ (23% NaCl,W/V),pH 9.5,温度53 ℃,其独特的生存条件表明其细胞内必定存在不同于常见中度嗜盐菌的机制特征。据报道[35-38],N. thermophilus中携带甜菜碱从头合成途径相关基因,本研究组对N. thermophilus不同嗜极条件下胞内甜菜碱进行了分析(数据未发表),结果表明N. thermophilus细胞内甜菜碱浓度随着盐浓度的增加而升高,且浓度增长幅度较大,如Na+浓度为4.5 mol/L (相当于约27% NaCl)时,甜菜碱浓度是Na+浓度为3.3 mol/L时(约19% NaCl)的3倍。对于同属于硬壁菌门的革兰氏阳性中度嗜盐菌盐反硝化枝芽孢杆菌Virgibacillus halodenitrificans PDB-F2来说,甜菜碱显著促进了菌株在6%–15% NaCl中的生长速度,15% NaCl中尤为明显,增长速度是对照组的1.28倍。这些结果表明甜菜碱对嗜盐菌适应盐环境及响应高盐胁迫冲击来说至关重要[39]。对于生存时间较久的嗜盐古菌来说,其“相容性溶质机制”还未得到明确的认识和较为深入的研究。葡萄牙甲烷嗜盐菌Methanohalophilus portucalensis PDF1T是目前研究最为彻底的采用甘氨酸途径合成甜菜碱的嗜盐古菌,该研究除了证明M. portucalensis在高盐环境中能合成甜菜碱外,若将相关蛋白基因转入到缺陷型菌株E.coli MKH13中,在盐环境下携带重组蛋白的菌株比缺陷型菌株在生长时间上明显加快(0.8 mol/L NaCl下提前约10 h)[10]。
甜菜碱的重要作用不仅仅局限于盐环境,嗜热盐碱厌氧菌中甜菜碱相关合成基因的发现,我们推测该基因表达的蛋白可能存在耐受多种极端环境的特性。甜菜碱除了有作为渗透保护剂的作用外,还可作为酶功能的有效稳定剂,提供对盐度、高温、冻融处理甚至干燥等胁迫的抵抗作用[2, 29, 40]。在低渗条件下,甜菜碱可以降解为二甲基甘氨酸、肌氨酸和甘氨酸等[6],降低细胞内渗透压并作为碳源或氮源等能源物质供应细胞生长需求。
4 甜菜碱合成基因在生物技术中的应用虽然甜菜碱合成基因的功能鉴定方面的科学报道较多,但是其在现代生物技术应用领域的报道还较少。以下就其在基因工程菌的构建和转基因农作物中的应用作一些介绍。
(1) 胆碱途径合成基因的应用。Cánovas等(2000)将伸长盐单胞菌H. elongate DSM3043的betIBA基因导入缺陷型大肠杆菌MKH13菌株中,该菌株在0.5 mol/L盐度下,能够合成甜菜碱。Gu等(2008)将达坂喜盐芽孢杆菌H.dabanensis D-8T中的gbsAB基因在大肠杆菌PD141中进行表达,在含有1 mmol/L胆碱的0.7 mol/L盐度下培养,能够检测到甜菜碱。Prasad等将嗜盐节杆菌Arthrobacter pascens中的胆碱氧化酶基因(codeA)转入荠菜中,发现可以促进种子萌发和植株生长。在0.2 mol/L Na+环境中,野生型植物无种子萌发,而携带codeA基因的植株种子萌发率可达40%[41]。Mohanty等将codeA基因转入水稻中,50%的转基因植株在盐胁迫条件下存活并有种子生成[42];Su等将带有胁迫诱导启动子的cox基因导入水稻中,发现其甜菜碱的积累是组成型启动子植株2倍[43]。此外,在拟南芥、玉米和番茄中,发现甜菜碱基因的异源表达对植物的生殖器官也有保护作用[44]。这可能是由于甜菜碱从植物其他组织(如叶子)中转移流入到生殖器官中,从而在生殖器官内积聚了相对高浓度的甜菜碱,最终对其起到了保护作用。这说明甘氨酸甜菜碱合成基因的异源表达可以保护或增强植物多个组织器官的功能。
(2) 甘氨酸途径合成基因的应用。迄今为止,国内外已有多篇报道涉及将甘氨酸甜菜碱途径合成基因导入植物中[45-48]。目前,盐生隐杆藻A. halophytica中的甘氨酸甜菜碱从头合成基因的异源表达是转基因领域研究的重点。将盐隐生杆藻中甜菜碱合成基因ApGSMT及ApDMT转入鱼腥藻Anabaena sp. PCC7120中,在0.12 mol/L NaCl条件下,转基因鱼腥藻的生长速率是野生型的2倍[48-49]。将盐隐生杆藻的甜菜碱合成基因ApGSMT和ApDMT转入水稻中,转基因植株对盐胁迫的抗性高于野生型植株,在0.15 mol/L NaCl下,转基因植株表达的甜菜碱浓度为普通水稻的4–6倍。通过检测叶片中叶绿素的含量,从而确定盐胁迫对于植株的影响,在0.2 mol/L NaCl环境中,转基因植株叶片中叶绿素含量是野生型的2.6倍,在0.4 mol/L NaCl下,高达6倍[46]。最近,Song等将盐隐生杆菌中ApGSMT2g和ApDMT2g(编码甜菜碱合成酶,此报道采用此名称)转入棉花中,结果表明转基因植株在0.15–0.20 mol/L NaCl条件下甜菜碱获得率是野生型的2倍[47]。此外,将M. portucalensis FDF1T中的甜菜碱合成基因Mpgsmt-sdmt导入大肠杆菌MKH13中,在0.7–0.8 mol/L盐条件下,携带合成基因的大肠杆菌共表达MpGSMT-SDMT的滞后时间缩短,对外界环境的盐适应能力增强[10]。若将MpGSMT和MpDMT基因转入到拟南芥中,则转基因植株在0.15–0.30 mol/L NaCl环境中相较于对照组长势较好,叶片数量及根长度均约为对照组2倍[45]。以上甘氨酸甜菜碱合成基因的拓展研究有利于耐盐基因工程菌的构建,将甜菜碱合成基因导入某些不耐盐工程菌株,可使菌株在高渗环境中发挥其生理功能,更好地应用于工农业生产实践。细胞内生物合成的甜菜碱在植物抗逆性中可发挥重要的作用,对于在盐碱环境中难以生存的经济作物,可以将甜菜碱合成基因导入植株内,从而合成甜菜碱,以增强植株抵抗盐胁迫的能力,有益于植株的生长及适应。
5 展望甜菜碱作为一种很重要的相容性溶质,在嗜盐菌长期盐适应及短期盐胁迫过程中发挥关键性的作用。甜菜碱的生物合成有胆碱氧化途径和甘氨酸甲基化途径。目前较多的研究集中于单一合成途径,但我们发现还有一部分嗜盐细菌拥有2种合成途径,这一有趣的发现引发了我们的思考:这两种合成途径在高盐环境下是如何调节的,是否存在某一盐浓度阈值来触发不同的生物合成机制,又或是两种途径在盐环境中相互影响?这些问题有待解决。现在对于能生物合成甜菜碱的嗜盐菌(不同类别)之间的亲缘关系还缺乏系统的认识和比较,甜菜碱合成基因在各类细胞中的功能还有待深入研究。甜菜碱和其他相容性溶质相比有什么优势,嗜盐菌更偏向于利用哪种相容性溶质作为渗透保护剂也值得大家的深入思考。
由于嗜盐菌的盐适应机制是非常复杂的过程,有时包括几种相容性溶质同时发挥作用,这几种相容性溶质如何发挥作用以及它们之间存在的相互影响及作用,也是以后需要深入研究的重点。对于采用其他相容性溶质作为渗透调节剂的嗜盐菌,甜菜碱是否会对其产生渗透保护作用,又会对其本身合成的相容性溶质产生什么影响,目前此方面鲜有报道。在构建基因工程菌株方面,应用甜菜碱合成基因在现代食品工业、发酵工业、污水处理和石油污染生物修复中发挥作用,开发转基因植物以抵抗盐碱地环境是今后研究的重点内容。我国盐碱地的改良和耐盐碱农作物的开发也是改善盐碱地问题、提高农业水平的关键。综上,希望本综述提供的一些研究结果、观点以及思路能为该领域的深入研究尽一点绵薄之力,以期甜菜碱生物合成基因功能等方面的科学研究成果更加丰硕。
| [1] | Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiology and Molecular Biology Reviews, 1998, 62(2): 504-544. |
| [2] | Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiology Reviews, 2018, 42(3): 353-375. DOI:10.1093/femsre/fuy009 |
| [3] | Zhao BS, Yan YC, Chen SL. How could haloalkaliphilic microorganisms contribute to biotechnology?. Canadian Journal of Microbiology, 2014, 60(11): 717-727. DOI:10.1139/cjm-2014-0233 |
| [4] |
Zhao BS, Yang LF, Wang L, Lu WD, Yang SS. Study progress on compatible solutes in moderately halophilic bacteria. Acta Microbiologica Sinica, 2007, 47(5): 937-941.
(in Chinese) 赵百锁, 杨礼富, 王磊, 卢伟东, 杨苏声. 中度嗜盐菌相容性溶质机制的研究进展. 微生物学报, 2007, 47(5): 937-941. DOI:10.3321/j.issn:0001-6209.2007.05.037 |
| [5] | Roberts MF. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Systems, 2005, 1: 5. DOI:10.1186/1746-1448-1-5 |
| [6] | Shao YH, Guo LZ, Zhang YQ, Yu H, Zhao BS, Pang HQ, Lu WD. Glycine betaine monooxygenase, an unusual Rieske-type oxygenase system, catalyzes the oxidative N-demethylation of glycine betaine in Chromohalobacter salexigens DSM 3043. Applied and Environmental Microbiology, 2018, 84(13): e00377-18. DOI:10.1128/AEM.00377-18 |
| [7] | Mansour MMF, Ali EF. Glycinebetaine in saline conditions:an assessment of the current state of knowledge. Acta Physiologiae Plantarum, 2017, 39(2): 56. DOI:10.1007/s11738-017-2357-1 |
| [8] |
Zhao BS, Yang LF, Song L, Wang H. Biotechnology applications in moderately halophilic eubacteria. Microbiology, 2007, 34(2): 359-362.
(in Chinese) 赵百锁, 杨礼富, 宋蕾, 王慧. 中度嗜盐菌在生物技术中的应用. 微生物学通报, 2007, 34(2): 359-362. DOI:10.3969/j.issn.0253-2654.2007.02.038 |
| [9] |
Zhao BS, Li J. Biodiversity of culture-dependent haloalkaliphilic microorganisms. Acta Microbiologica Sinica, 2017, 57(9): 1409-1420.
(in Chinese) 赵百锁, 李俊. 可培养盐碱菌多样性的研究进展. 微生物学报, 2017, 57(9): 1409-1420. |
| [10] | Lai SJ, Lai MC. Characterization and regulation of the osmolyte betaine synthesizing enzymes GSMT and SDMT from halophilic methanogen Methanohalophilus portucalensis. PLoS One, 2011, 6(9): e25090. DOI:10.1371/journal.pone.0025090 |
| [11] | Nyyssölä A, Kerovuo J, Kaukinen P, von Weymarn N, Reinikainen T. Extreme halophiles synthesize betaine from glycine by methylation. Journal of Biological Chemistry, 2000, 275(29): 22196-22201. DOI:10.1074/jbc.M910111199 |
| [12] | Cánovas D, Vargas C, Kneip S, Morón MJ, Ventosa A, Bremer E, Nieto JJ. Genes for the synthesis of the osmoprotectant glycine betaine from choline in the moderately halophilic bacterium Halomonas elongata DSM 3043. Microbiology, 2000, 146(2): 455-463. DOI:10.1099/00221287-146-2-455 |
| [13] | Gadda G, McAllister-Wilkins EE. Cloning, expression, and purification of choline dehydrogenase from the moderate halophile Halomonas elongata. Applied and Environmental Microbiology, 2003, 69(4): 2126-2132. |
| [14] | Gu ZJ, Wang L, Le Rudulier D, Zhang B, Yang SS. Characterization of the glycine betaine biosynthetic genes in the moderately halophilic bacterium Halobacillus dabanensis D-8T. Current Microbiology, 2008, 57(4): 306-311. DOI:10.1007/s00284-008-9194-9 |
| [15] | Nyyssölä A, Leisola M. Actinopolyspora halophila has two separate pathways for betaine synthesis. Archives of Microbiology, 2001, 176(4): 294-300. DOI:10.1007/s002030100325 |
| [16] | Nyyssölä A, Reinikainen T, Leisola M. Characterization of glycine sarcosine N-methyltransferase and sarcosine dimethylglycine N-methyltransferase. Applied and Environmental Microbiology, 2001, 67(5): 2044-2050. DOI:10.1128/AEM.67.5.2044-2050.2001 |
| [17] | Waditee R, Tanaka Y, Aoki K, Hibino T, Jikuya H, Takano J, Takabe T, Takabe T. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. Journal of Biological Chemistry, 2003, 278(7): 4932-4942. DOI:10.1074/jbc.M210970200 |
| [18] | Lechtenfeld M, Heine J, Sameith J, Kremp F, Müller V. Glycine betaine metabolism in the acetogenic bacterium Acetobacterium woodii. Environmental Microbiology, 2018, 20(12): 4512-4525. DOI:10.1111/1462-2920.14389 |
| [19] | Munoz R, Rosselló-Móra R, Amann R. Revised phylogeny of Bacteroidetes and proposal of sixteen new taxa and two new combinations including Rhodothermaeota phyl. nov. Systematic and Applied Microbiology, 2016, 39(5): 281-296. DOI:10.1016/j.syapm.2016.04.004 |
| [20] | Zhang SS, Pan J, Lu WD, Yan YC, Wang HS, Wiegel J, Zhao BS. Halomonas urumqiensis sp. nov., a moderately halophilic bacterium isolated from a saline-alkaline lake. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(5): 1962-1969. DOI:10.1099/ijsem.0.000975 |
| [21] | Zhang SS, Li ZJ, Yan YC, Zhang CL, Li J, Zhao BS. Bacillus urumqiensis sp. nov., a moderately haloalkaliphilic bacterium isolated from a salt lake. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(6): 2305-2312. DOI:10.1099/ijsem.0.001028 |
| [22] | Qiu DC, Liao ZY, Lu WD, Wang HS, Li J, Zhao BS. Draft genome sequence of Alkalicoccus saliphilus DSM 15402T, a haloalkaliphilic bacterium isolated from a mineral pool. Microbiology Resource Announcements, 2019, 8(24): e00266-19. DOI:10.1128/MRA.00266-19 |
| [23] | Zhao B, Holtzapple M, Lu W. Genus Alkalicoccus//Bergey's Manual of Systematics of Archaea and Bacteria. John Wilkey & Sons, Inc., in association with Bergey's Manual Trust. (In press). |
| [24] | Fendrihan S, Legat A, Pfaffenhuemer M, Gruber C, Weidler G, Gerbl F, Stan-Lotter H. Extremely halophilic archaea and the issue of long-term microbial survival. Reviews in Environmental Science and Bio/Technology, 2006, 5(2/3): 203-218. |
| [25] | Müller V, Spanheimer R, Santos H. Stress response by solute accumulation in archaea. Current Opinion in Microbiology, 2005, 8(6): 729-736. DOI:10.1016/j.mib.2005.10.011 |
| [26] | Lu WD, Chi ZM, Su CD. Identification of glycine betaine as compatible solute in Synechococcus sp. WH8102 and characterization of its N-methyltransferase genes involved in betaine synthesis. Archives of Microbiology, 2006, 186(6): 495-506. DOI:10.1007/s00203-006-0167-8 |
| [27] | Kimura Y, Kawasaki S, Yoshimoto H, Takegawa K. Glycine betaine biosynthesized from glycine provides an osmolyte for cell growth and spore germination during osmotic stress in Myxococcus xanthus. Journal of Bacteriology, 2010, 192(5): 1467-1470. DOI:10.1128/JB.01118-09 |
| [28] | Goh F, Jeon YJ, Barrow K, Neilan BA, Burns BP. Osmoadaptive strategies of the archaeon Halococcus hamelinensis isolated from a hypersaline stromatolite environment. Astrobiology, 2011, 11(6): 529-536. DOI:10.1089/ast.2010.0591 |
| [29] | Zou HB, Chen NN, Shi MX, Xian M, Song YM, Liu JH. The metabolism and biotechnological application of betaine in microorganism. Applied Microbiology and Biotechnology, 2016, 100(9): 3865-3876. DOI:10.1007/s00253-016-7462-3 |
| [30] | Cánovas D, Vargas C, Csonka LN, Ventosa A, Nieto JJ. Osmoprotectants in Halomonas elongata:high-affinity betaine transport system and choline-betaine pathway. Journal of Bacteriology, 1996, 178(24): 7221-7226. DOI:10.1128/JB.178.24.7221-7226.1996 |
| [31] | Cánovas D, Vargas C, Csonka LN, Ventosa A, Nieto JJ. Synthesis of glycine betaine from exogenous choline in the moderately halophilic bacterium Halomonas elongata. Applied and Environmental Microbiology, 1998, 64(10): 4095-4097. DOI:10.1128/AEM.64.10.4095-4097.1998 |
| [32] | Ginzburg M, Sachs L, Ginzburg BZ. Ion metabolism in a halobacterium. I. Influence of age of culture on intracellular concentrations. The Journal of General Physiology, 1970, 55(2): 187-207. DOI:10.1085/jgp.55.2.187 |
| [33] | Matheson AT, Sprott GD, McDonald IJ, Tessier H. Some properties of an unidentified halophile:growth characteristics, internal salt concentration, and morphology. Canadian Journal of Microbiology, 1976, 22(6): 780-786. DOI:10.1139/m76-114 |
| [34] | Waditee-Sirisattha R, Kageyama H, Takabe T. Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiology, 2016, 2(1): 42-54. DOI:10.3934/microbiol.2016.1.42 |
| [35] | Mesbah NM, Hedrick DB, Peacock AD, Rohde M, Wiegel J. Natranaerobius thermophilus gen. nov., sp. nov., a halophilic, alkalithermophilic bacterium from soda lakes of the Wadi An Natrun, Egypt, and proposal of Natranaerobiaceae fam. nov. and Natranaerobiales ord. nov. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(11): 2507-2512. DOI:10.1099/ijs.0.65068-0 |
| [36] | Mesbah NM, Cook GM, Wiegel J. The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+ (K+)/H+ antiporters. Molecular Microbiology, 2009, 74(2): 270-281. DOI:10.1111/j.1365-2958.2009.06845.x |
| [37] | Mesbah NM, Hänelt I, Zhao BS, Müller V. Microbial Adaptation to Saline Environments:Lessons from the Genomes of Natranaerobius thermophilus and Halobacillus halophilus//Halophiles:Genetics and Genomes. UK: Caister Academic Press, 2014. |
| [38] | Zhao BS, Mesbah NM, Dalin E, Goodwin L, Nolan M, Pitluck S, Chertkov O, Brettin TS, Han J, Larimer FW, Land ML, Hauser L, Kyrpides N, Wiegel J. Complete genome sequence of the anaerobic, halophilic alkalithermophile Natranaerobius thermophilus JW/NM-WN-LF. Journal of Bacteriology, 2011, 193(15): 4023-4024. DOI:10.1128/JB.05157-11 |
| [39] | Qian CJ, Li W, Li H, Ou D, Zhuge YY, Liu YD. Responses of genes for the uptake of glycine betaine in Virgibacillus halodenitrificans PDB-F2 under NaCl stress. International Biodeterioration & Biodegradation, 2018, 132: 192-199. |
| [40] | Stadmiller SS, Gorensek-Benitez AH, Guseman AJ, Pielak GJ. Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. Journal of Molecular Biology, 2017, 429(8): 1155-1161. DOI:10.1016/j.jmb.2017.03.001 |
| [41] | Prasad KVSK, Sharmila P, Kumar PA, Pardha-Saradhi P. Transformation of Brassica juncea (L.) Czern with bacterial codA gene enhances its tolerance to salt stress. Molecular Breeding, 2000, 6(5): 489-499. DOI:10.1023/A:1026542109965 |
| [42] | Mohanty A, Kathuria H, Ferjani A, Sakamoto A, Mohanty P, Murata N, Tyagi A. Transgenics of an elite indica rice variety Pusa Basmati 1 harbouring the codA gene are highly tolerant to salt stress. Theoretical and Applied Genetics, 2002, 106(1): 51-57. DOI:10.1007/s00122-002-1063-5 |
| [43] | Su J, Hirji R, Zhang L, He CK, Selvaraj G, Wu R. Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. Journal of Experimental Botany, 2006, 57(5): 1129-1135. DOI:10.1093/jxb/erj133 |
| [44] | Goel D, Singh AK, Yadav V, Babbar SB, Murata N. Transformation of tomato with a bacterial codA gene enhances tolerance to salt and water stresses. Journal of Plant Physiology, 2011, 168(11): 1286-1294. DOI:10.1016/j.jplph.2011.01.010 |
| [45] | Lai SJ, Lai MC, Lee RJ, Chen YH, Yen HE. Transgenic Arabidopsis expressing osmolyte glycine betaine synthesizing enzymes from halophilic methanogen promote tolerance to drought and salt stress. Plant molecular Biology, 2014, 85(4/5): 429-441. |
| [46] | Niu XL, Xiong FJ, Liu J, Sui Y, Zeng ZM, Lu BR, Liu YS. Co-expression of ApGSMT and ApDMT promotes biosynthesis of glycine betaine in rice (Oryza sativa L.) and enhances salt and cold tolerance. Environmental and Experimental Botany, 2014, 104: 16-25. DOI:10.1016/j.envexpbot.2014.03.003 |
| [47] | Song JL, Zhang R, Yue D, Chen XG, Guo ZQ, Cheng C, Hu MH, Zhang JR, Zhang KW. Co-expression of ApGSMT2g and ApDMT2g in cotton enhances salt tolerance and increases seed cotton yield in saline fields. Plant Science, 2018, 274: 369-382. DOI:10.1016/j.plantsci.2018.06.007 |
| [48] | Waditee-Sirisattha R, Singh M, Kageyama H, Sittipol D, Rai AK, Takabe T. Anabaena sp. PCC7120 transformed with glycine methylation genes from Aphanothece halophytica synthesized glycine betaine showing increased tolerance to salt. Archives of Microbiology, 2012, 194(11): 909-914. DOI:10.1007/s00203-012-0824-z |
| [49] | Singh M, Sharma NK, Prasad SB, Yadav SS, Narayan G, Rai AK. The freshwater cyanobacterium Anabaena doliolum transformed with ApGSMT-DMT exhibited enhanced salt tolerance and protection to nitrogenase activity, but became halophilic. Microbiology, 2013, 159(3): 641-648. |
 2020, Vol. 60
2020, Vol. 60