中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王也民, 张家杰, 喻玮. 2020
- Yemin Wang, Jiajie Zhang, Wei Yu. 2020
- 细菌在胃肠道肿瘤发生中的作用及其机制
- Role of bacteria in the development and progression of gastrointestinal tumors
- 微生物学报, 60(3): 441-451
- Acta Microbiologica Sinica, 60(3): 441-451
-
文章历史
- 收稿日期:2019-06-12
- 修回日期:2019-07-02
- 网络出版日期:2019-12-11
2. 浙江省人民医院, 杭州医学院附属人民医院, 感染病科, 浙江 杭州 310014;
3. 浙江大学医学院附属第一医院, 传染病诊治国家重点实验室, 国家感染性疾病临床研究中心, 感染性疾病诊治协同创新中心, 浙江 杭州 310003
2. Department of Infectious Diseases, Zhejiang Provincial People's Hospital, People's Hospital of Hangzhou Medical College, Hangzhou 310014, Zhejiang Province, China;
3. State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, Zhejiang Province, China
胃肠道肿瘤包括食道、胆囊、肝脏、胰腺、胃、小肠、大肠、直肠和肛门等消化系统器官的肿瘤,其中结直肠癌(Colorectal cancer,CRC)最常见。2018年,美国诊断为CRC的患者约140250例,预估死亡人数约50630例[1-2]。且随着年龄的增长,西方和亚洲人群患CRC的风险也随之增加[3]。现有研究表明胃肠道肿瘤的发生是遗传和表观遗传变化逐渐积累和共同作用的结果,这些变化受宿主免疫、饮食、环境和微生物的影响[4]。
Sender等报道70 kg标准体重参考人体内约含有3.8×1013个细菌,其中大多数细菌都存在于人体消化道内[5]。通常,饮食、生活方式和药物都会影响肠道微生态的组成。最新研究数据表明,肠道微生态在胃肠道肿瘤的发生和发展中发挥至关重要的作用[6-7]。其中,已有多项研究证实幽门螺杆菌(Helicobacter pylori,Hp)与胃癌的发生密切相关,而肠致病性大肠杆菌(Enteropathogenic Escherichia coli,EPEC)、产肠毒素的脆弱拟杆菌(Enterotoxigenic Bacteroides fragilis,ETBF)、具核梭杆菌(Fusobacterium nucleatum,Fn)等肠道菌群则与CRC的发生发展密切相关[8-10]。Yachida等[11]通过多组学研究发现肠道微生物种类、数量以及代谢标志物的变化发生在CRC的早期阶段,且在CRC的发展过程中动态改变。然而,究竟这些细菌在CRC发生中是起辅助作用还是起主导作用尚不明确。因此,全面了解细菌在胃肠道肿瘤发生发展中的作用机制,可以为胃肠道肿瘤诊治提供新的预防和治疗策略。
1 毒力因子目前多种研究表明持续携带Hp会增加胃腺癌的发生风险[9]。Hp典型的细菌毒力因子包括空泡性细胞毒素A (Vacuolating cytotoxin A,vacA)和细胞毒素相关基因A (Cytotoxin-associated gene A,cagA)。VacA可引起细胞内空泡形成、细胞膜电位去极化、上皮细胞通透性增加和凋亡、以及上皮细胞从基底膜分离[12]。Sundrud等[13]发现VacA可通过抑制T细胞核内因子(Nuclear factor of activated T cells,NFAT)活化来阻断活化T细胞分泌IL-2,并可诱导调节性T细胞(Treg),从而使Hp逃避适应性免疫反应,导致Hp定殖及慢性感染。此外,VacA还可降低细胞内谷胱甘肽水平而引起自噬,导致活性氧(Reactive oxygen species,ROS)积累和AKT活化,参与胃癌的发生[14]。cagA基因位于一个40 kb的DNA片段内,该片段被称为cag致病性岛,编码一个Ⅳ型分泌系统(Type Ⅳ secretion system,T4SS),并通过该系统分泌到胃上皮细胞,在Hp相关胃病中起关键作用[15]。磷酸化CagA与蛋白酪氨酸磷酸酶(Protein tyrosine phosphatase-2,SHP-2),生长因子受体结合蛋白2 (Growth factor receptor-bound protein 2,Grb2),CT10激酶调节子样蛋白(CT10 regulator of kinase like protein,CRKL),C-Src酪氨酸激酶(C-Terminal Src kinase,Csk),受体酪氨酸激酶Met (Receptor tyrosine kinase Met,c-Met),紧密连接蛋白-1 (Zonula occludens-1,ZO-1)相互作用,激活ERK/MAPK导致上皮结构和完整性失调,并可通过PI3K-AKT、Wnt和NF-κB信号通路来促进细胞增殖,同时抑制p53蛋白降低上皮细胞凋亡[16-17]。此外,CagA还可诱导胃上皮细胞的上皮细胞-间充质细胞转换(Epithelial-mesenchymal transition,EMT),参与肿瘤干细胞的产生[18-19]。
目前,在多种革兰氏阴性致病菌中(包括放线菌、弯曲杆菌、螺旋杆菌及大肠杆菌)分离出能使细胞质和细胞核增大的不耐热毒素,即细胞致死性扩张毒素(Cytolethal distending toxin,CDT),可导致DNA双链断裂,介导不可逆的细胞周期停滞和凋亡,从而发挥遗传毒性损害[20-21]。Graillot等[22]通过进一步研究发现,在CRC发病机制中,CDT本身不直接导致CRC的发生,但可能通过不同机制对癌前病变发挥促进作用。
大肠杆菌是人类肠道微生态的重要成员之一,常在宿主出生后几天内开始定居肠道,并在宿主的整个生命周期中持续存在[23]。大肠杆菌可分成A、B1、B2、D4个亚型,其中B2亚型在结肠中最常见。现有研究发现34%的B2亚型大肠杆菌携带一种保守的基因岛—“聚酮合成酶岛(Polyketide synthase island,pks island)”,编码非核糖体多肽合成酶(Nonribosomal peptide synthetases,NRPS)和聚酮合成酶(Polyketide synthetases,PKS),可产生一种多聚乙酰一肽的基因毒性物质(Colibactin),进而导致机体DNA损伤,参与肿瘤的发生及发展[24]。体外实验证实,真核细胞感染pks+大肠杆菌会导致DNA双链断裂,影响基因组不稳定性,同时激活DNA损伤修复通路(包括ATM–CHK–CDC25– CDK1通路和H2AX Ser139位点磷酸化),导致细胞周期停滞,最终导致细胞死亡[24]。Arthur等[25]在动物实验中发现,pks+大肠杆菌促进Interleukin(IL)-10-/-小鼠的结肠肿瘤发生。现有研究认为PSK的基因毒性效应需要细菌与宿主细胞接触,而肠道的炎症可减少保护性粘蛋白和抗菌肽的产生,促进pks+大肠杆菌的入侵[23-24, 26-27]。然而,随着时间的推移,炎症并不会促进肠腔内大肠杆菌丰度的显著增加[27]。因此可见,炎症-微生物活性-肿瘤发生发展之间存在着复杂的相互作用,需要进一步研究阐明随时间变化宿主产生CRC的具体机制。
脆弱拟杆菌约占正常结肠菌群的0.1%,主要定殖于结肠中,其中ETBF可分泌一种依赖锌的金属蛋白,即脆弱拟杆菌肠毒素(Bacteroides fragilis enterotoxin,BFT)引起组织细胞损伤。现有研究发现,BFT和肠上皮细胞特异性受体结合,激活Wnt、NF-κB、STAT3等多个信号通路,导致高水平的IL-17产生,进而促进CXCL1招募大量单核细胞样髓源性免疫抑制细胞(Monocytic myeloid-derived suppressor cells,MO-MDSCs)累积,上调原癌基因表达,水解肠上皮细胞的钙粘蛋白,破坏细胞间紧密连接,介导炎症和肿瘤的发生[28-30]。
2012年,Castellarin等[10]发现CRC组织中存在Fn。Yachida等[11]通过粪便元基因组研究发现Fn的相对丰度从CRC粘膜内癌至晚期病变持续增加。进一步研究发现,Fn表面表达FadA毒力因子,可与E-钙粘蛋白结合,激活β-连环蛋白(β-catenin)、NF-κB和Wnt信号通路,形成促炎性肿瘤微环境,进而促进CRC的发生和发展[31-32]。此外,Gur等[33]发现了细菌依赖的肿瘤免疫逃避机制,即Fn可产生Fap2蛋白作用于T细胞免疫球蛋白和免疫受体酪氨酸抑制基序(Immunoreceptor tyrosine-based inhibitory motif,ITIM)结构域蛋白(T cell immunoreceptor with Ig and ITIM domains,TIGIT)抑制免疫细胞活性[32]。
DNA错配修复(Mismatch repair,MMR)系统可以纠正DNA复制错误,是DNA损伤修复的多种途径之一。Maddocks等[8]通过实验发现,在CRC患者肠道内的EPEC可分泌线粒体靶向的效应蛋白EspF,导致结肠上皮细胞内的MMR缺失,使DNA无法完成自身修复,著提高宿主细胞的自发突变率,此外还可增加氧自由基的水平,从而诱发CRC的发生。
染色体不稳定性(Chromosomal instability,CIN)是肿瘤演进的主要驱动因素[34]。而人体肠道中的共生粪肠球菌能够通过下调DNA损伤修复功能,并通过膜相关的自氧化作用产生大量ROS,诱导上皮细胞的CIN和线粒体功能障碍,与多发性腺瘤性息肉和CRC的发生密切相关[35-36]。通过动物实验发现,粪肠球菌感染结肠巨噬细胞后,通过旁观者效应(Bystander effects,BSE),诱导环氧合酶-2 (Cyclooxygenases-2,COX-2)表达,增加4-羟基-2-壬醛产生,促进IL-10敲除小鼠CRC的发生[37]。
2 生物膜肠道通常被保护性粘液层覆盖,可阻止大多数细菌与宿主结肠上皮直接接触。然而,Dejea等[38]通过研究发现,CRC患者肠道内存在能够侵入肠道粘液层的特定细菌,导致粘膜微生物群和结肠上皮细胞之间的接触增加,进而形成侵入性生物膜。与没有生物膜的患者相比,有生物膜的患者患CRC的风险要高出5倍以上。该研究还提出生物膜主要可增强上皮细胞通透性,促进原癌组织炎症改变,最终刺激上皮细胞发生瘤变。进一步通过16S rRNA测序发现,在CRC患者的肠道粘液层中细菌生物膜多在右侧结肠近端形成,而不是左侧远端[39-40]。
家族性腺瘤性息肉病(Familial adenomatous polyposis,FAP)早期多为良性病变。Dejea等[41]通过对6名FAP患者结肠粘膜的研究发现大肠杆菌和脆弱拟杆菌可组成斑块状生物膜。进一步研究发现pks+大肠杆菌和ETBF是形成生物膜的主要菌株,且与正常人相比,FAP结肠粘膜中大肠杆菌colibactin毒素基因(clbB)和脆弱拟杆菌毒素基因(bft)高度富集。在成瘤小鼠模型中,表达colibactin毒素的大肠杆菌和产肠毒素的脆弱拟杆菌共同定殖可导致结肠中的IL-17和结肠上皮中的DNA损伤增加,加快肿瘤的发生[41]。值得注意的是,该研究强调只有这两类细菌同时存在,才会增加肿瘤发生的风险。其中ETBF消化粘液层,促使pks+大肠杆菌大量入侵肠道粘膜,介导炎症反应和上皮细胞DNA突变,从而增加结肠肿瘤的发生风险。
3 代谢产物肠道菌群在参与机体对食物或外源物质共代谢中产生大量小分子物质,对宿主细胞和肠道菌群间的信息传递起着关键作用。不同菌种可分别产生短链脂肪酸(Short-chain fatty acids,SCFAs)、胆汁酸、苯甲酰和苯基衍生物、吲哚衍生物、胆碱、多酚类、维生素、氨基酸、脂质、激素类及多胺类等多种代谢产物,直接或间接的影响基因调控、代谢网络调节及微生物细胞的生理功能[42]。最新研究证实在多发性息肉样腺瘤及粘膜内癌中,支链氨基酸、苯丙氨酸及胆汁酸显著增加[11]。且在CRC发生的过程中,多种代谢分子和代谢通路发生显著变化,主要包括胆碱代谢途径、氨基酸降解途径、糖异生途径、糖蛋白和有机酸代谢途径[11, 43-44]。
SCFAs主要包括乙酸、丁酸和丙酸,这些代谢物不仅是肠道微生物的重要能量来源,也是肠道上皮细胞(Intestinal epithelial cells,IECs)的重要能量来源,此外,其还具有不同的免疫调节功能[45]。现有研究证明,SCFAs通过阻断NF-κB信号通路的激活,诱导调节性T细胞分化,发挥致肿瘤作用[46]。然而关于丁酸盐的研究,目前存在争论,有研究认为丁酸盐不仅可通过β-氧化促进能量代谢,维持肠腔低氧环境,还可以激活肠道细胞内的过氧化物酶体增殖物激活受体γ (Peroxisome proliferator-activated receptor γ,PPAR-γ),抑制nos2基因的表达及诱导型一氧化氮合成酶(inducible nitric oxide synthase,iNOS)合成,减少硝酸盐的产生,限制兼性厌氧菌增殖,甚至可抑制结肠的炎症和癌变[47-48]。但另一些研究却提出相反结论,通过动物实验证实丁酸盐可以介导结肠上皮细胞过度增殖以及DNA错配修复缺陷[49]。这可能与丁酸盐局部浓度及其与其他代谢物的相互作用有关。
胆汁酸主要包括胆酸和鹅去氧胆酸,在肝脏合成后与甘氨酸或牛磺酸结合并排泄到十二指肠以促进脂肪消化。结合胆汁酸在小肠和结肠远端肠道微生物的作用下产生次级胆汁酸,即牛磺脱氧胆酸(Taurodeoxycholic acid,TDCA)和脱氧胆酸(Deoxycholic acid,DCA)。现有研究发现次级胆汁酸可以诱导结肠上皮细胞增殖和凋亡,其中TDCA主要通过核转录因子RelA磷酸化诱导IL-8基因表达[50],DCA则是通过阻断NF-κB信号通路的激活和RelA核易位发挥作用。
Dodd等[51]描述了肠道共生菌生孢梭菌(Clostridium sporogenes)产生吲哚丙酸(Indolepropionic acid,IPA)的代谢途径,并通过动物实验证实芳香族氨基酸代谢所产生的次级代谢产物影响肠道的通透性和全身免疫状态。此外,多胺也被证实能够促进肿瘤增殖,抑制抗肿瘤免疫,参与肿瘤细胞侵袭和转移[52]。
4 机体免疫调节天然免疫系统非特异地识别微生物的保守结构,能对病原微生物做出快速反应。当机体遭受病原微生物或疾病相关宿主分子侵害时,能通过宿主模式识别受体(Pattern recognition receptor,PRR)感应病原相关分子模式(Pathogen-associated molecular patterns,PAMPs),发挥免疫调节作用[53]。
细胞表面PRR包括甘露糖受体(Mannose receptor,MR)、清道夫受体(Scavenger receptor,SR)和Toll样受体(Toll like receptor,TLR)。细胞内PRR包括TLR、核苷酸结合寡聚化结构域(Nucleotide binding oligomerization domain,NOD)样受体(NLRs)和视黄酸诱导基因蛋白1 (Retinoic acid-inducible gene-1,RIG-1)样受体(RLR)。
不同的PAMPs可与不同的TLR结合,通常革兰阴性菌的脂多糖(Lipopolysaccharide,LPS)与TLR4相结合;细菌脂蛋白、脂磷壁酸和真菌的酵母多糖与TLR1、TLR2和TLR6结合;细菌鞭毛蛋白激活TLR5[54]。TLR可激活NF-κB、STAT3、MAPK、JUN N-末端激酶(JNK)、p38、ERKS和干扰素调节因子信号通路,在免疫调节中发挥重要作用[55]。
目前,多项研究已证实TLR有助于多种器官肿瘤的发生发展。Luo等[56]通过小鼠结肠腺癌转移模型证实LPS注射后促进转移性肿瘤形成,主要机制是导致肿瘤细胞中NF-κB调节的抗凋亡因子(BCL-XL、cIAP1和cIAP2)上调。髓样分化因子(Myeloid differentiation primary response gene 88,MyD88)也是TLR信号通路中的一个关键分子。Rakoff-Nahoum等[57]发现MyD88依赖的信号传导控制肠道肿瘤发生中的多个正向调节因子的表达,包括COX-2、基质金属蛋白酶(Matrix metalloprotease,MMP)、细胞溶质磷脂酶(Cytosolic phospholipase A2,cPLA2),在自发和致癌诱导的肠道肿瘤发展中起到关键作用。
NLR主要识别细菌肽聚糖的细胞内片段,其中NOD-1和NOD-2在细菌感染和免疫平衡中发挥重要作用,主要通过激活NF-κB和MAPK依赖的基因转录[58]。在肠道细菌感染时,NOD2可诱导树突状细胞产生IL-23,进一步激活Th17细胞介导免疫反应[58]。在小鼠模型中,NOD1缺陷可导致上皮细胞凋亡和肠通透性增加,导致CRC发生[59]。而NOD2则通过上调结肠上皮细胞中IL-6的表达,诱导微生物失调,并促进炎症性CRC的发展[60]。前瞻性胃癌队列研究也显示NOD2基因突变与胃癌的发生风险显著相关[61]。具体机制尚不明确,可能是由于NOD2突变引起胃上皮屏障的稳态破坏,导致Hp感染风险增加,进而导致局部炎症加重,炎性微环境进一步恶化诱发肿瘤形成。此外,结肠上皮细胞缺乏Nod样受体家族含pyrin结构域蛋白(NOD-like receptor family pyrin domain containing 6,NLRP6)可通过趋化因子CCL5的上调诱导炎症,调节肿瘤微环境中IL-6的表达,促进上皮细胞增殖,导致CRC形成[62]。最近的研究表明,在人类CRC组织标本中,组织梭杆菌DNA的数量级与CD3+T细胞的密度呈负相关性[63]。
5 小结综上所述,细菌通过多种机制参与胃肠道肿瘤的发生及发展(图 1)。目前,胃肠道肿瘤主要通过消化道内镜进行筛查,随着对细菌在胃肠道肿瘤发生发展的作用机制研究,有望通过对特定细菌或细菌致瘤的关键分子进行分析,进而发现胃肠道肿瘤早期筛查的有效手段,为预防提供新策略。然而,饮食、生活方式、药物等多种因素均可影响胃肠道微生物种群的组成,因此有必要通过进一步研究确定这些可变因素对微生物群和胃肠道肿瘤发生的影响。此外,细菌对化疗药物疗效也有影响,因此,药学、微生物学及肿瘤学等多学科跨专业合作研究将有助于为胃肠道肿瘤治疗提供新思路及有价值的数据。
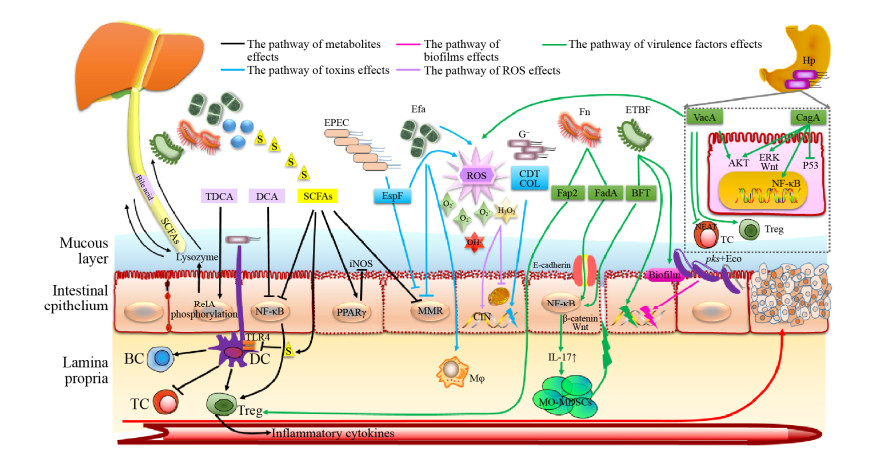
|
| 图 1 细菌参与胃肠道肿瘤的发生及发展 Figure 1 Bacteria participate in the occurrence and development of gastrointestinal tumors. TDCA : Taurodeoxycholic acid; DCA: Deoxycholic acid; SCFAs: Short-chain fatty acids; EPEC: Enteropathogenic Escherichia coli; Efa: Enterococcus faecalis; Fn: Fusobacterium nucleatum; G–: Gram-negative bacteria; ETBF: Enterotoxigenic Bacteroides fragilis; Hp: Helicobacter pylori; Eco: Escherichia coli; Tc: T lymphocyte; Bc: B lymphocyte; Dc: Dendritic cell; Mφ: Macrophage; MO-MDSCs: Monocytic myeloid-derived suppressor cells; PPARγ: Peroxisome proliferator-activated receptor γ; MMR: Mismatch repair; CIN: Chromosomal instability. |
| [1] | Carroll R, Zhao SS. Trends in colorectal cancer incidence and survival in Iowa SEER data: the timing of it all. Clinical Colorectal Cancer, 2018, 18(2): e261-e274. DOI:10.1016/j.clcc.2018.12.001 |
| [2] | Lopes G, Stern MC, Temin S, Sharara AI, Cervantes A, Costas-Chavarri A, Engineer R, Hamashima C, Ho GF, Huitzil FD, Moghani MM, Nandakumar G, Shah MA, Teh C, Manjarrez SEV, Verjee A, Yantiss R, Correa MC. Early detection for colorectal cancer: ASCO resource-stratified guideline. Journal of Global Oncology, 2019, 5: 1-22. DOI:10.1200/JOP.19.00010 |
| [3] | Chung RYN, Tsoi KKF, Kyaw MH, Lui AR, Lai FTT, Sung JY. A population-based age-period-cohort study of colorectal cancer incidence comparing Asia against the West. Cancer Epidemiology, 2019, 59: 29-36. DOI:10.1016/j.canep.2019.01.007 |
| [4] | Song MY, Garrett WS, Chan AT. Nutrients, foods, and colorectal cancer prevention. Gastroenterology, 2015, 148(6): 1244-1260. DOI:10.1053/j.gastro.2014.12.035 |
| [5] | Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biology, 2016, 14(8): e1002533. DOI:10.1371/journal.pbio.1002533 |
| [6] | Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature, 2016, 529(7585): 212-215. DOI:10.1038/nature16504 |
| [7] | Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, LifeLines cohort study, Weersma RK, Feskens EJ, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu JY. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science, 2016, 352(6285): 56-569. DOI:10.1126/science.aad3369 |
| [8] | Maddocks ODK, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio, 2013, 4(3): e00152-13. DOI:10.1128/mBio.00152-13 |
| [9] | Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Current Opinion in Microbiology, 2007, 10(1): 76-81. DOI:10.1016/j.mib.2006.12.004 |
| [10] | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research, 2012, 22(2): 299-306. DOI:10.1101/gr.126516.111 |
| [11] | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature Medicine, 2019, 25(6): 968-976. DOI:10.1038/s41591-019-0458-7 |
| [12] | Montecucco C, de Bernard M. Immunosuppressive and proinflammatory activities of the VacA toxin of Helicobacter pylori. The Journal of Experimental Medicine, 2003, 198(12): 1767-1771. DOI:10.1084/jem.20031839 |
| [13] | Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(20): 7727-7732. DOI:10.1073/pnas.0401528101 |
| [14] | Tsugawa H, Suzuki H, Saya H, Hatakeyama M, Hirayama T, Hirata K, Nagano O, Matsuzaki J, Hibi T. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host & Microbe, 2012, 12(6): 764-777. DOI:10.1016/j.chom.2012.10.014 |
| [15] | Backert S, Meyer TF. Type Ⅳ secretion systems and their effectors in bacterial pathogenesis. Current Opinion in Microbiology, 2006, 9(2): 207-217. DOI:10.1016/j.mib.2006.02.008 |
| [16] | Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D, Berg DE, Sasakawa C. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host & Microbe, 2009, 5(1): 23-34. DOI:10.1016/j.chom.2008.11.010 |
| [17] | Wei JX, Noto JM, Zaika E, Romero-Gallo J, Piazuelo MB, Schneider B, El-Rifai W, Correa P, Peek RM, Zaika AI. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut, 2015, 64(7): 1040-1048. DOI:10.1136/gutjnl-2014-307295 |
| [18] | Bessède E, Staedel C, Acuña Amador LA, Nguyen PH, Chambonnier L, Hatakeyama M, Belleannée G, Mégraud F, Varon C. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene, 2014, 33(32): 4123-4131. DOI:10.1038/onc.2013.380 |
| [19] | Kountouras J, Kapetanakis N, Zavos C, Polyzos SA, Romiopoulos I, Tsiaousi E, Anastasiadou K, Giorgakis N, Vardaka E, Nikolaidou C, Venizelos I, Katsinelos P. Helicobacter pylori might contribute to cancer and/or bone marrow-derived stem cell-related gastrointestinal oncogenesis. Oncogene, 2015, 34(5): 670. DOI:10.1038/onc.2013.602 |
| [20] | Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology, 2011, 157(7): 1851-1875. DOI:10.1099/mic.0.049536-0 |
| [21] | He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, Pons B, Mirey G, Vignard J, Hendrixson DR, Jobin C. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut, 2019, 68(2): 289-300. DOI:10.1136/gutjnl-2018-317200 |
| [22] | Graillot V, Dormoy I, Dupuy J, Shay JW, Huc L, Mirey G, Vignard J. Genotoxicity of Cytolethal Distending Toxin (CDT) on isogenic human colorectal cell lines: potential promoting effects for colorectal carcinogenesis. Frontiers in Cellular and Infection Microbiology, 2016, 6: 34. DOI:10.3389/fcimb.2016.00034 |
| [23] | Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(25): 11537-11542. DOI:10.1073/pnas.1001261107 |
| [24] | Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science, 2006, 313(5788): 848-851. DOI:10.1126/science.1127059 |
| [25] | Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science, 2012, 338(6103): 120-123. DOI:10.1126/science.1224820 |
| [26] | Inaba Y, Ashida T, Ito T, Ishikawa C, Tanabe H, Maemoto A, Watari J, Ayabe T, Mizukami Y, Fujiya M, Kohgo Y. Expression of the antimicrobial peptide α-defensin/cryptdins in intestinal crypts decreases at the initial phase of intestinal inflammation in a model of inflammatory bowel disease, IL-10-deficient mice. Inflammatory Bowel Diseases, 2010, 16(9): 1488-1495. DOI:10.1002/ibd.21253 |
| [27] | Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nature Communications, 2014, 5(1): 4724. DOI:10.1038/ncomms5724 |
| [28] | Riegler M, Lotz M, Sears C, Pothoulakis C, Castagliuolo I, Wang CC, Sedivy R, Sogukoglu T, Cosentini E, Bischof G, Feil W, Teleky B, Hamilton G, LaMont JT, Wenzl E. Bacteroides fragilis toxin 2 damages human colonic mucosa in vitro. Gut, 1999, 44(4): 504-510. DOI:10.1136/gut.44.4.504 |
| [29] | Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P, Ganguly S, Fu J, Pardoll DM, Sears CL, Housseau F. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunology, 2017, 10(2): 421-433. DOI:10.1038/mi.2016.53 |
| [30] | Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan HN, Wu XQ, Ganguly S, Lebid A, Metz P, van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan FY, Wu SG, Sears CL, Housseau F. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host & Microbe, 2018, 23(2): 203-214.e5. |
| [31] | Rubinstein MR, Wang XW, Liu W, Hao YJ, Cai GF, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host & Microbe, 2013, 14(2): 195-206. DOI:10.1016/j.chom.2013.07.012 |
| [32] | Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nature Reviews Microbiology, 2019, 17(3): 156-166. DOI:10.1038/s41579-018-0129-6 |
| [33] | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity, 2015, 42(2): 344-355. DOI:10.1016/j.immuni.2015.01.010 |
| [34] | Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, Duran M, Pauli C, Shaw C, Chadalavada K, Rajasekhar VK, Genovese G, Venkatesan S, Birkbak NJ, McGranahan N, Lundquist M, LaPlant Q, Healey JH, Elemento O, Chung CH, Lee NY, Imielenski M, Nanjangud G, Pe'er D, Cleveland DW, Powell SN, Lammerding J, Swanton C, Cantley LC. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature, 2018, 553(7689): 467-472. DOI:10.1038/nature25432 |
| [35] | Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis, 2002, 23(3): 529-536. DOI:10.1093/carcin/23.3.529 |
| [36] | Strickertsson JAB, Rasmussen LJ, Friis-Hansen L. Enterococcus faecalis infection and reactive oxygen species down-regulates the miR-17-92 cluster in gastric adenocarcinoma cell culture. Genes, 2014, 5(3): 726-738. DOI:10.3390/genes5030726 |
| [37] | Wang XM, Allen TD, Yang YH, Moore DR, Huycke MM. Cyclooxygenase-2 generates the endogenous mutagen trans-4-hydroxy-2-nonenal in Enterococcus faecalis-infected macrophages. Cancer Prevention Research, 2013, 6(3): 206-216. DOI:10.1158/1940-6207.CAPR-12-0350 |
| [38] | Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, Peterson SN, Snesrud EC, Borisy GG, Lazarev M, Stein E, Vadivelu J, Roslani AC, Malik AA, Wanyiri JW, Goh KL, Thevambiga I, Fu K, Wan FY, Llosa N, Housseau F, Romans K, Wu XQ, McAllister FM, Wu SG, Vogelstein B, Kinzler KW, Pardoll DM, Sears CL. Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(51): 18321-18326. DOI:10.1073/pnas.1406199111 |
| [39] | Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, Roslani AC, Wick EC, Mongodin EF, Loke MF, Thulasi K, Gan HM, Goh KL, Chong HY, Kumar S, Wanyiri JW, Sears CL. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. npj Biofilms and Microbiomes, 2017, 3(1): 34. DOI:10.1038/s41522-017-0040-3 |
| [40] | Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Medicine, 2019, 11(1): 11. DOI:10.1186/s13073-019-0621-2 |
| [41] | Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu XQ, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu SG, Pardoll DM, Housseau F, Sears CL. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science, 2018, 359(6375): 592-597. DOI:10.1126/science.aah3648 |
| [42] | Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science, 2012, 336(6086): 1262-1267. DOI:10.1126/science.1223813 |
| [43] | Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, Sunagawa S, Coelho LP, Schrotz-King P, Vogtmann E, Habermann N, Niméus E, Thomas AM, Manghi P, Gandini S, Serrano D, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Waldron L, Naccarati A, Segata N, Sinha R, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature Medicine, 2019, 25(4): 679-689. DOI:10.1038/s41591-019-0406-6 |
| [44] | Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nature Medicine, 2019, 25(4): 667-678. DOI:10.1038/s41591-019-0405-7 |
| [45] | Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nature Reviews Immunology, 2016, 16(6): 341-352. DOI:10.1038/nri.2016.42 |
| [46] | Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science, 2013, 341(6145): 569-573. DOI:10.1126/science.1241165 |
| [47] | Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao YD, Litvak Y, Lopez CA, Xu GG, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science, 2017, 357(6351): 570-575. DOI:10.1126/science.aam9949 |
| [48] | Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell, 2018, 33(6): 954-964. DOI:10.1016/j.ccell.2018.03.004 |
| [49] | Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S, Green B, Geddes K, Pezo RC, Navarre WW, Milosevic M, Wilson BC, Girardin SE, Wolever TMS, Edelmann W, Guttman DS, Philpott DJ, Martin A. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell, 2014, 158(2): 288-299. DOI:10.1016/j.cell.2014.04.051 |
| [50] | Mühlbauer M, Allard B, Bosserhoff AK, Kiessling S, Herfarth H, Rogler G, Schölmerich J, Jobin C, Hellerbrand C. Differential effects of deoxycholic acid and taurodeoxycholic acid on NF-κB signal transduction and IL-8 gene expression in colonic epithelial cells. American Journal of Physiology. Gastrointestinal and Liver Physiology, 2004, 286(6): G1000-G1008. DOI:10.1152/ajpgi.00338.2003 |
| [51] | Dodd D, Spitzer MH, van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature, 2017, 551(7682): 648-652. DOI:10.1038/nature24661 |
| [52] | Soda K. The mechanisms by which polyamines accelerate tumor spread. Journal of Experimental & Clinical Cancer Research, 2011, 30: 95. DOI:10.1186/1756-9966-30-95 |
| [53] | Chu HT, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nature Immunology, 2013, 14(7): 668-675. DOI:10.1038/ni.2635 |
| [54] | Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nature Reviews Immunology, 2010, 10(2): 131-144. DOI:10.1038/nri2707 |
| [55] | Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nature Reviews Cancer, 2009, 9(1): 57-63. DOI:10.1038/nrc2541 |
| [56] | Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-κB in cancer cells converts inflammation- induced tumor growth mediated by TNFα to TRAIL-mediated tumor regression. Cancer Cell, 2004, 6(3): 297-305. DOI:10.1016/j.ccr.2004.08.012 |
| [57] | Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science, 2007, 317(5834): 124-127. DOI:10.1126/science.1140488 |
| [58] | Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nature Reviews Immunology, 2014, 14(1): 9-23. DOI:10.1038/nri3565 |
| [59] | Chen GY, Shaw MH, Redondo G, Núñez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Research, 2008, 68(24): 10060-10067. DOI:10.1158/0008-5472.CAN-08-2061 |
| [60] | Couturier-Maillard A, Secher T, Rehman A, Normand S, de Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Nùñez G, Chen G, Rosenstiel P, Chamaillard M. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. The Journal of Clinical Investigation, 2013, 123(2): 700-711. DOI:10.1172/JCI62236 |
| [61] | Companioni O, Bonet C, Muñoz X, Weiderpass E, Panico S, Tumino R, Palli D, Agnoli C, Vineis P, Boutron-Ruault MC, Racine A, Clavel-Chapelon F, Travis RC, Khaw KT, Riboli E, Murphy N, Vergnaud AC, Trichopoulou A, Benetou V, Trichopoulos D, Lund E, Johansen D, Lindkvist B, Johansson M, Sund M, Ardanaz E, Sánchez-Cantalejo E, Huerta JM, Dorronsoro M, Ramón Quirós J, Tjonneland A, Mortensen LM, Overvad K, Chang-Claude J, Rizzato C, Boeing H, de Mesquita HBB, Siersema P, Peeters PHM, Numans ME, Carneiro F, Licaj I, Freisling H, Sala N, González CA. Polymorphisms of Helicobacter pylori signaling pathway genes and gastric cancer risk in the European Prospective Investigation into Cancer-Eurgast cohort. International Journal of Cancer, 2014, 134(1): 92-101. DOI:10.1002/ijc.28357 |
| [62] | Hu B, Elinav E, Huber S, Strowig T, Hao LM, Hafemann A, Jin CC, Wunderlich C, Wunderlich T, Eisenbarth SC, Flavell RA. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(24): 9862-9867. DOI:10.1073/pnas.1307575110 |
| [63] | Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, Kostic AD, Giannakis M, Watanabe H, Bullman S, Milner DA, Harris CC, Giovannucci E, Garraway LA, Freeman GJ, Dranoff G, Chan AT, Garrett WS, Huttenhower C, Fuchs CS, Ogino S. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncology, 2015, 1(5): 653-61. DOI:10.1001/jamaoncol.2015.1377 |
 2020, Vol. 60
2020, Vol. 60




