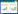中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 方安琪, 贺志理, 王成, 杨超, 颜庆云. 2020
- Anqi Fang, Zhili He, Cheng Wang, Chao Yang, Qingyun Yan. 2020
- 红树林沉积物中微生物驱动硫循环研究进展
- Progress in studying microbially-driven sulfur cycling in mangrove sediments
- 微生物学报, 60(1): 13-25
- Acta Microbiologica Sinica, 60(1): 13-25
-
文章历史
- 收稿日期:2019-03-20
- 修回日期:2019-05-27
- 网络出版日期:2019-10-24
2. 南方海洋科学与工程广东省实验室(珠海), 广东 珠海 519000;
3. 湖南农业大学农学院, 湖南 长沙 410128;
4. 中山大学南海研究院, 广东 广州 510275;
5. 加拿大农业部斯威夫特卡伦特科学技术研究所, Saskatchewan Swift Current S9H 3X2
2. Southern Marine Science and Engineering Guangdong Laboratory(Zhuhai), Zhuhai 519000, Guangdong Province, China;
3. College of Agronomy, Hunan Agricultural University, Changsha 410128, Hunan Province, China;
4. South China Sea Institution, Sun Yat-sen University, Guangzhou 510275, Guangdong Province, China;
5. Agriculture and Agri-Food Canada, Swift Current Research and Development Center, Swift Current S9H 3X2 SK, Canada
红树林湿地分布在热带和亚热带的潮间带,主要由红树植物和半红树植物为主的常绿灌木和小乔木组成,是一种连接陆地和海洋的特殊生态系统[1]。在目前已知的各类生态系统中,红树林湿地是最具生产力的生态系统之一[2],其特点是有机物质和养分周转率高。微生物作为红树林物质循环的重要驱动者[3],在创造和维护这个生物圈的过程中扮演着非常重要的角色[4]。在过去的几十年里,红树林生态系统在各种人类活动的影响下(如森林砍伐、煤矿开采、城市和工业废物肆意排放和原油泄漏等[5-7]),受到了很大的挑战[8-9]。为适应这些影响带来的环境改变,红树林沉积物中的微生物已经演变成适应多种有机和无机污染物的复合功能型微生物[10-11]。有研究表明,微生物结构变量的差异与环境功能变量的差异显著相关[12-13]。因此,微生物群落结构的组成在一定程度上能反映红树林生态系统的生态学功能[14-15]。
红树林作为海陆过渡地带的典型湿地生态系统,其沉积物环境通常具有贫氧、高硫和富营养等特征[16]。红树植物的含硫量通常高于同地带非红树林生态系统中的其他灌木[17],这对红树林沉积物的结构和性质都具有重要影响[18]。微生物在硫循环过程中驱动氧化还原反应,其中氧、碳和铁是主要的反应物,这导致了这些元素循环相互耦合[19]。微生物驱动的硫酸盐还原及其相关化学反应被认为是控制红树林沉积物化学环境的关键过程[20]。本文主要综述红树林沉积物中微生物驱动的无机硫化合物之间的迁移转化,为更好地认识红树林中微生物驱动的硫循环机制提供理论参考。
1 红树林沉积物中微生物驱动的硫循环过程红树林沉积物中普遍富含硫,主要以黄铁矿(80%的Fe以FeS2赋存)和硫酸盐(SO42–)形式存在[16]。SO42–是红树林沉积物内有机物氧化反应中仅次于O2的重要氧化剂。由于沉积物中共生的大量底栖生物和红树植物发达的呼吸根消耗了O2,随着沉积物深度的增加,生物可利用的O2浓度逐渐降低[21]。在有氧层,O2是最重要的氧化剂,有机物的降解主要通过微生物的有氧呼吸来实现;而在缺氧层,SO42–是最重要的氧化剂,有机物主要通过微生物驱动的硫酸盐还原来分解[22]。微生物能够将包括还原反应产生的硫化氢(H2S)在内的硫化物(HS–/S2–)氧化为单质硫(S0),然后S0歧化为SO42–和HS–/S2–[23],从而形成了硫元素的无机代谢循环(图 1)。此外,微生物也会驱动有机硫化合物之间的转化,例如二甲基亚砜(DMSO)可以通过微生物转化为二甲基硫醚(DMS),反之亦然[24]。

|
| 图 1 红树林沉积物中硫元素的生物地球化学循环 Figure 1 Biogeochemical cycle of sulfur in mangrove sediments. |
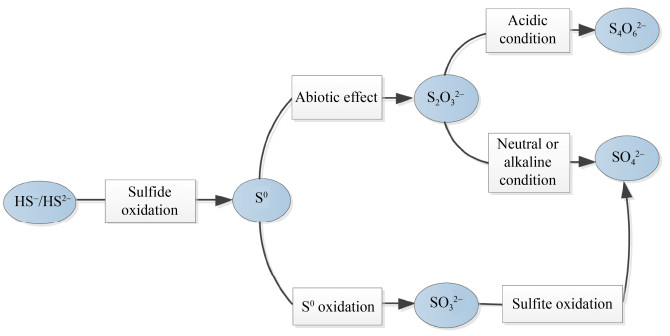
1.1 微生物驱动的硫氧化过程及其影响因素 1.1.1 微生物驱动的硫氧化过程: 硫氧化过程是指将低价态的单质硫或还原性硫化物完全氧化为硫酸盐或部分氧化成更高价态的硫化物[25],主要包括硫化物氧化、单质硫氧化、硫代硫酸盐氧化以及亚硫酸盐氧化[26](图 2)。其中硫化物氧化是指硫氧化菌(sulfur-oxidizing bacteria,SOB)中硫化物氧化酶将HS–/S2–氧化成S0的过程。由于H2S等硫化物影响红树林沉积物中动植物的正常生命活动[27],因此SOB在沉积物的硫化物解毒中起着重要作用。单质硫氧化是指S0在SOB中反向异化硫酸盐还原酶作用下被氧化成亚硫酸盐(SO32–)的过程。硫代硫酸盐氧化是指硫代硫酸盐(S2O32–)在碱性或者中性条件下被SOB氧化成SO42–;而在酸性条件下S2O32–被氧化成连四硫酸盐(S4O62–)[28]。亚硫酸盐氧化是指SO32–在亚硫酸盐氧化酶的作用下被氧化成SO42–。在红树林沉积物的有氧区,硫氧化过程由耗氧SOB进行有氧氧化完成[29];在缺氧区硫氧化过程主要由光养或化能SOB利用光能营养或化学能营养进行厌氧氧化完成[30]。但是在缺氧区由于底栖动物的扰动致使微生境局部含氧,因此在缺氧区可能也存在部分耗氧微生物参与硫氧化。
|
| 图 2 微生物驱动的硫化物代谢过程 Figure 2 Schematic illustration of the metabolism of inorganic sulfur compounds by sulfur oxidizing microbes. |
已报道的SOB主要分布在绿菌纲(Chlorobia)、绿弯菌纲(Chloroflexi)、α-变形菌纲(Alphaproteobacteria)、β-变形菌纲(Betaproteobacteria)、γ-变形菌纲(Gammaproteobacteria)等[31-32]。红树林生态系统中孕育着丰富的SOB (表 1)。如Liang等[33]在中国深圳福田红树林沉积物中发现了一些自由生活和共生的SOB:杆状色菌属(Rhabdochromatium)和Thioalkalivibrio denitrificans SOB自由生活,γ-变形菌纲SOB与双壳类满月蛤总科(Lucinacea)共生;Zhao等[34]在中国福建泉州红树林中发现1种能广泛利用多种碳、氮和硫物质的海洋紫色硫细菌(Marichromatium gracile)新菌株YL28。在巴西原始红树林被原油泄漏污染的沉积物中,SOB有较高的多样性,污染样品中检测到的OTUs的数量显著高于未污染样品,其中报道了着色菌目(Chromatiales)中的着色菌科(Chromatiaceae)和外硫红螺旋菌科(Ectothiorhodospiraceae) 2种SOB[35];Maryeimy等[36]在巴西被石油和城市垃圾污染的红树林中发现存在丰富的β-变形菌纲和γ-变形菌纲的SOB。综上所述,在红树林沉积物中,硫氧化功能菌以变形菌为优势类群,环境的复杂性会增加SOB多样性。
| Study sites | SOB groups | References |
| Futian mangrove in Shenzhen, China | Rhabdochromatium, Thioalkalivibrio | [33] |
| The mangrove in Fujian, China | Marichromatium | [34] |
| The mangrove in Rio de Janeiro, Brazil | Chromatiaceae, Ectothiorhodospiraceae | [35] |
| Mangroves in Sao Paulo, Brazil | Betaproteobacteria, Gammaproteobacteria | [36] |
用于跟踪硫循环相关的生物分子标记主要是基于编码关键酶的功能基因。目前,硫氧化途径的关键功能基因包括sqr (编码硫醌氧化还原酶)、soxB (编码硫氧化酶)[37]和dsrA (编码反向异化硫酸盐还原酶)。在珠江流域的研究中,定量分析显示具有soxB功能基因的SOB比具有sqr和dsrA功能基因的SOB更丰富,是硫氧化的主要贡献者[31]。在具有sqr功能基因的SOB群落中,披毛菌属(Gallionella)、噬氢菌属(Hydrogenophaga)、Limnohabitans、甲基单胞菌属(Methylomonas)、硝化螺菌属(Nitrospira)、红育菌属(Rhodoferax)和硫针菌属(Sulfuritalea)为优势属;对于具有soxB功能基因的SOB群落中,β-变形菌纲脱氯菌属(Dechloromonas)、Limnohabitans、副球菌属(Paracoccus)、硫针菌属和硫杆菌属(Thiobacillus)丰度较高;而硫针菌属、硫体属(Sulfurisoma)和硫杆菌属在具有dsrA功能基因的SOB群落中数量较多[31]。在红树林生境中,上述3种硫氧化途径关键酶编码基因相对应的菌属尚未对比研究,其结果是否一致有待证实。
1.1.2 影响硫氧化过程的主要环境因素: 在红树林沉积物中O2浓度、pH、季节、潮汐周期等因素均会影响硫氧化过程[38]。(1) O2浓度:SOB对O2的依赖性不强,除了需氧型的SOB外,很多厌氧SOB也能在无氧或少氧的条件下进行厌氧氧化。例如,苍白杆菌属(Ochrobactrum)以亚硝酸盐(NO2–)为电子受体在厌氧的环境下进行硫氧化[39]。(2) pH:SOB对pH的适应性非常广泛,目前已知的SOB中性菌、嗜酸菌和嗜碱菌均有报道,但红树林生态系统中的SOB多为中性菌。这些SOB在不同pH条件下利用同一底物时生成的产物会有所差异。例如S2O32–在碱性或者中性条件下被SOB氧化成SO42–;而在酸性条件下则被氧化成S4O62–[28]。(3)季节:沉积物中微生物群落在不同季节有比较大的变化,这些差异性往往是由土壤中可利用养分来决定的。Mishra等[40]发现,在印度红树林雨季有较高含量的土壤养分,致使雨季比旱季有着更丰富的SOB。(4)潮汐周期:在红树林沉积物中,潮汐周期对微生物群落的构成有很大的影响。Zhang等[41]证实位于红树林潮高滩、潮中滩和海水中的微生物群落有显著差异并且呈现梯度分布。由于潮汐致使红树林的环境条件在时空尺度上发生显著变化,特别是盐度、养分和含氧量,因而影响SOB活性。但潮汐周期对SOB活性影响程度尚未研究报道。红树林复杂多变的环境极大地丰富了SOB多样性,随着测序技术和相关分析手段的飞速发展,新物种有待进一步发掘。 1.2 微生物驱动的硫酸盐还原过程及其影响因素 1.2.1 微生物驱动的硫酸盐还原过程: 硫酸盐还原是指在简单有机物降解过程中以SO42–为末端电子受体,最终生成H2S和CO2的还原反应过程[30]。在红树林沉积物与水交界处,硫酸盐还原产生的CO2几乎占据了CO2总排放量的100%[42]。Kristensen等[43]在泰国、巴基斯坦、牙买加等地区的红树林沉积物中都检测到了很高的硫酸盐还原活性。硫酸盐还原菌(sulfate-reducing bacteria,SRB)通常被认为是严格厌氧菌[44],但研究发现部分SRB如脱硫弧菌属(Desulfovibrio)可以在微量O2的环境中生存[45]。在红树林沉积物中,O2仅存在表层几毫米处[46],以厌氧为主体的环境给SRB的生长代谢提供了有利条件。在厌氧环境中,硫酸盐还原是有机物质降解过程中重要的电子转移过程[47]。红树林中有机物的分解主要由发酵过程和SRB驱动的硫酸盐还原过程完成[42],其中超过50%的有机物分解由SRB承担[48]。其基本过程为复杂的碳水化合物经发酵成简单的有机物,SRB再进一步利用SO42–作为有机物降解的电子受体,最终产生H2S和CO2。SRB在厌氧环境中还能够降解复杂的底物,如长链芳香烃和石油衍生烃[49]。因红树林高硫酸盐的特性及其厌氧环境,丰富的SRB及其很强的代谢能力促进了硫元素的快速循环。据统计,已知的SRB目前有60个属的220多个种[50]。依其反应底物分为4类[51-52]:第一类以氢为硫酸盐还原的反应底物,如脱硫肠状菌属(Desulfotomaculum)等;第二类以乙醇、乙酸盐、乳酸盐等较简单的有机物为底物,如脱硫弧菌属、脱硫杆菌属(Desulfobacter)、脱硫叶菌属(Desulfobulbus)和脱硫单胞菌属(Desulfomonas)等;第三类以高级脂肪酸为反应底物,如脱硫线菌属(Desulfonema)等;第四类以芳香族化合物为反应底物,如脱硫球菌属(Desulfococcus)和脱硫八叠球菌属(Desulfosarcina)等。红树林沉积物中缺氧和低氧化还原电位的环境特征为硫酸盐还原提供了有利的条件[53]。Quillet等[54]在英国Medway河口的盐沼中发现脱硫弧菌科(Desulfovibrionaceae)、脱硫杆菌科(Desulfobacteraceae)、脱硫球茎菌科(Desulfobulbaceae)、互营杆菌科(Syntrophobacteraceae)的存在。在巴西红树林沉积物中存在丰富的脱硫杆菌目(Desulfobacterales)、脱硫弧菌目(Desulfovibrionales)以及拟杆菌门(Bacteroidetes)参与硫还原代谢[36, 55]。Ding等[56]在中国海南红树林中分离培养出芽孢杆菌属(Bacillus)、弧菌属(Vibrio)、梭状芽胞杆菌属(Clostridium)、伯克霍尔德菌属(Burkholderia)、希瓦氏菌属(Shewanella)和海杆菌属(Marinobacterium)的SRB。Lyimo等[57]在坦桑尼亚红树林中分离出1种脱硫八叠球菌属的SRB新菌株SD1,其可以二甲基硫化物、甲硫醇、丙酮酸和丁酸盐为还原反应的底物。Gomes等[58]发现在巴西瓜纳巴拉红树林中,2种常见的红树物种海榄雌属和拉关木选择性地构建脱硫杆菌目和除硫单胞菌目(Desulfuromonadales) 2种SRB为根际群落(表 2)。在佛罗里达州,红树林的根际周围,SRB是细菌群落中丰度最高的一类菌群[59]。综上所述,各种研究表明,因红树林沉积物中蕴含大量的硫酸盐,SRB成为红树林沉积物中的一大优势类群。
| Study sites | SRB groups | References |
| Salt marsh in Medway estuary, UK | Desulfovibrionaceae, Desulfobacteraceae, Desulfobulbaceae, Syntrophobacteraceae | [54] |
| Mangroves in Sao Paulo, Brazil | Desulfobacterales, Desulfovibrionales, Bacteroidetes | [36, 55] |
| Mangroves in Hainan, China | Bacillus, Vibrio, Clostridium, Burkholderia, Shewanella, Marinobacterium | [56] |
| Mtoni mangrove in Dar es Salaam, Tanzania | Desulfosarcina | [57] |
| The mangrove in Guanabara Bay, Brazil | Desulfobacterales, Desulfuromonadales | [58] |
硫酸盐还原途径中的关键基因为aprAB (编码5′-腺苷酰硫酸还原酶)和dsrAB (编码亚硫酸盐还原酶)[60-62],aprAB催化5′-腺苷磷酸硫酸酐转化成腺苷单磷酸盐和SO32–,dsrAB催化SO32–还原成S2–[36]。现有大量研究通过使用功能基因aprAB和dsrAB作为分子标记来鉴定各种环境中SRB,例如硫化生物反应器中SRB的代谢活性[63],石油存储库中SRB对金属管道的腐蚀[64]等。同样,功能基因鉴定也可以用来推断在红树林沉积物中SRB的生态作用。Leloup等[65]基于dsrAB功能基因扩增对法国塞纳河河口沉积物中SRB的动态特性进行了研究。Bai等[66]采用高通量功能基因芯片(GeoChip 4.0)分析了中国漳江红树林沉积物中外来入侵物种互花米草相比较本地种有更丰富硫代谢相关微生物群落。目前,利用功能基因标记等分子手段对红树林沉积物中SRB的研究较少,揭示红树林沉积物中硫代谢功能相关微生物的生态演变是今后可突破的方向。
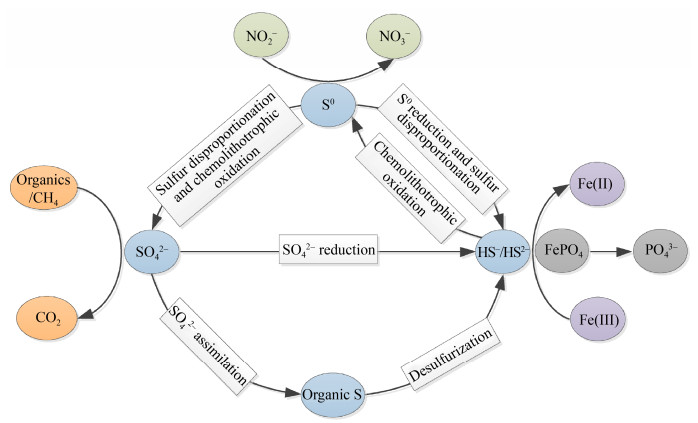
1.2.2 影响硫酸盐还原过程的主要环境因素: 研究表明,影响SRB进行硫酸盐还原过程的几种环境因子分别是有机碳、氧化还原电位、铁离子浓度、O2浓度和底物浓度等[67-69]。(1)有机碳:沉积物中的有机碳是微生物进行硫酸盐还原反应的电子供体,沉积物中SRB可代谢的碳源与硫酸盐还原率呈正相关[53]。(2)氧化还原电位:硫酸盐还原适宜发生于氧化还原电位+100 mv至–250 mv范围内[70]。由于硫酸盐还原剂是电子供体的弱势竞争者,它们在竞争中的相对成功主要取决于需氧生物体由于氧耗尽而剩余的碳[71]。因此,SRB的存在通常指示着氧化还原电位的高低[72]。(3)铁离子浓度:硫酸盐还原产生的H2S先结合Fe2+生成FeS,再结合S0生成FeS2。硫化铁是红树林沉积物中硫元素最主要的赋存形式之一[73]。然而,Attri等[74]实验表明,环境中铁离子浓度也会影响硫酸盐还原活性,过高的铁离子浓度甚至会抑制硫酸盐还原。其中,微生物在铁离子抑制硫酸盐还原反应过程中扮演什么角色有待深入探究。(4) O2浓度:红树林沉积物的氧化还原状态会随潮汐周期而波动。在沉积物表层,大部分SRB的活性因O2的存在受到抑制;在该氧化区域下,硫酸盐还原通常随深度的增加而增强[75],直至有机质含量急剧降低的深层,硫酸盐还原开始减弱。Das等[76]在孟加拉的红树林中发现,SRB的数量随着沉积物深度的增加而增大,在60 cm深度处,SRB的数量达到最大。(5)底物浓度:在红树林沉积物的不同深度上,SRB可利用的各类底物浓度也有差异[77]。深度上不同浓度的底物可以使SRB有效地竞争营养物质[78]。SRB在红树林沉积物中垂直方向上的空间分布是上述各种环境因子综合导致的。 2 微生物驱动硫循环与其他元素循环的耦合硫循环可以耦合碳、氮、磷和金属元素循环[22, 79](图 3),在红树林沉积物生物地球化学循环中起着举足轻重的作用。SRB以有机物为电子供体、以SO42–为末端电子受体进行有机物的矿化作用;其产物H2S被SOB利用进行CO2的固定作用[80]。同时,硫酸盐还原过程耦合甲烷(CH4)厌氧氧化[81-82],促进碳循环的代谢。此外,硫氧化过程也会耦合硝酸盐还原等氮元素的循环[83]。Griffin等[84]第一次报道了无氧光养SOB利用NO2–作为光合作用的电子供体,将NO2–厌氧氧化成硝酸盐(NO3–);Stevens等[85]在海洋中富含NO3–的稀氧区发现了γ-变形菌纲中未培养的硫氧化共生菌。硫循环与金属元素的耦合使HS–/S2–与金属离子(如Zn2+、Cu2+、Hg2+及Pb2+等)形成金属硫化物沉淀[86-87],这一耦合能有效地固定沉积物中的重金属[88]。在硫元素接受和释放电子的过程中,Fe(Ⅲ)被还原成Fe(Ⅱ)形成FeS2,磷溶解释放[18]。在红树林沉积物中,铁、磷和硫元素循环与SRB的活性密切相关[89],铁和磷的可利用性可能取决于SRB的活性[90]。Jian等[91]研究表明,硫酸盐对沉积物中铁和磷元素地球化学循环有显著影响,它显著降低了铁和磷对植物生长的限制,增强了植物对红树林特殊环境的适应力。
|
| 图 3 硫与其他元素循环的耦合 Figure 3 Coupling processes of sulfur and other element cycles. |
在硫酸盐浓度很高的环境中,硫酸盐还原作用会抑制CH4的产生,减少CH4排放,从而对缓解全球变暖具有重要意义。CH4对全球的变暖潜能是CO2的34倍[92-93],湿地是CH4最大的自然源。已有研究表明,SRB与产CH4菌共存于富含硫酸盐的滨海湿地生态系统中并且竞争通用底物[94-96]。由于SRB对乙酸盐[97]、氢气[98-99]和甲酸盐[47]等底物具有更高的亲和力,SRB通常会将这些底物维持在产CH4菌无法利用的较低浓度水平[72],且硫酸盐还原在热力学上比发酵过程及产CH4过程更有利[100],硫酸盐还原在富含硫酸盐的环境中比CH4生成更具优势[99, 101]。因此,SRB驱动的硫酸盐还原过程会影响滨海湿地温室气体的排放,并可能进一步影响区域气候。
3 总结和展望在红树林沉积物中,SRB驱动的硫酸盐还原过程相对于SOB驱动的硫氧化过程更为活跃,其生物多样性更高,在硫代谢循环中占主导地位。现有的针对红树林硫循环的研究主要集中在可培养微生物,对自然环境条件下驱动硫循环的微生物认识还很有限;SOB对硫化物解毒起着重要的作用,但其在红树林O2浓度随潮汐多变的环境中的代谢机理仍需进一步研究。因此,将来微生物驱动硫循环的研究应加强如下几个方面:(1)利用高通量测序技术和基因芯片技术,对具有硫循环功能基因的微生物多样性进行全面分析,探究红树林特殊生态系统中硫代谢特别是硫氧化过程及其关键功能微生物;(2)通过了解不同深度电子受体及环境因子的分布规律,阐明红树林沉积物中微生物驱动的硫循环与其他生物地球化学元素循环的耦合机制;(3)结合红树林沉积物中碳、氮、金属元素循环,利用各种组学方法详细探究微生物驱动的硫循环对地球化学大循环的贡献。通过16S rRNA基因扩增子测序和鸟枪法宏基因组测序发现,互营杆菌属(Syntrophobacter)、硫卵菌属(Sulfurovum)、硝化螺菌属(Nitrospira)、厌氧绳菌属(Anaerolinea)为驱动红树林沉积物中碳、氮、硫循环的主导微生物[102]。这些主导微生物属中都包含硫功能微生物种,对于硫功能微生物耦合各元素循环的具体机制仍需进一步研究。
| [1] | Kathiresan K, Bingham BL. Biology of mangroves and mangrove ecosystems. Advances in Marine Biology, 2001, 40: 81-251. DOI:10.1016/S0065-2881(01)40003-4 |
| [2] | Weiss C, Weiss J, Boy J, Iskandar I, Mikutta R, Guggenberger G. Soil organic carbon stocks in estuarine and marine mangrove ecosystems are driven by nutrient colimitation of P and N. Ecology and Evolution, 2016, 6(14): 5043-5056. DOI:10.1002/ece3.2258 |
| [3] | Thatoi H, Behera BC, Mishra RR, Dutta SK. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: a review. Annals of Microbiology, 2013, 63(1): 1-19. |
| [4] | Basak P, Pramanik A, Sengupta S, Nag S, Bhattacharyya A, Roy D, Pattanayak R, Ghosh A, Chattopadhyay D, Bhattacharyya M. Bacterial diversity assessment of pristine mangrove microbial community from dhulibhashani, sundarbans using 16S rRNA gene tag sequencing. Genomics Data, 2016, 7: 76-78. DOI:10.1016/j.gdata.2015.11.030 |
| [5] | Zhou YW, Zhao B, Peng YS, Chen GZ. Influence of mangrove reforestation on heavy metal accumulation and speciation in intertidal sediments. Marine Pollution Bulletin, 2010, 60(8): 1319-1324. DOI:10.1016/j.marpolbul.2010.03.010 |
| [6] | Peng YS, Diao JM, Zheng MX, Guan DS, Zhang RD, Chen GZ, Lee SY. Early growth adaptability of four mangrove species under the canopy of an introduced mangrove plantation: implications for restoration. Forest Ecology and Management, 2016, 373: 179-188. DOI:10.1016/j.foreco.2016.04.044 |
| [7] | Burns KA, Garrity SD, Levings SC. How many years until mangrove ecosystems recover from catastrophic oil spills?. Marine Pollution Bulletin, 1993, 26(5): 239-248. DOI:10.1016/0025-326X(93)90062-O |
| [8] | Cabral L, Lacerda Júnior GV, Pereira de Sousa ST, Franco Dias AC, Cadete LL, Andreote FD, Hess M, de Oliveira VM. Anthropogenic impact on mangrove sediments triggers differential responses in the heavy metals and antibiotic resistomes of microbial communities. Environmental Pollution, 2016, 216: 460-469. DOI:10.1016/j.envpol.2016.05.078 |
| [9] | Duke NC, Meynecke JO, Dittmann S, Ellison AM, Anger K, Berger U, Cannicci S, Diele K, Ewel KC, Field CD, Koedam N, Lee SY, Marchand C, Nordhaus I, Dahdouh-Guebas F. A world without mangroves?. Science, 2007, 317(5834): 41-42. |
| [10] | Yun JL, Deng YC, Zhang HX. Anthropogenic protection alters the microbiome in intertidal mangrove wetlands in Hainan Island. Applied Microbiology and Biotechnology, 2017, 101(15): 6241-6252. DOI:10.1007/s00253-017-8342-1 |
| [11] | Ottoni JR, Cabral L, Pereira de Sousa ST, Lacerda Junior GV, Domingos DF, Soares Junior FL, Pinheiro da Silva MC, Marcon J, Franco Dias AC, de Melo IS, de Souza AP, Andreote FD, de Oliveira VM. Functional metagenomics of oil-impacted mangrove sediments reveals high abundance of hydrolases of biotechnological interest. World Journal of Microbiology and Biotechnology, 2017, 33(7): 141. DOI:10.1007/s11274-017-2307-5 |
| [12] | Yang Q, Lei AP, Li FL, Liu LN, Zan QJ, Shin PKS, Cheung SG, Tam NFY. Structure and function of soil microbial community in artificially planted Sonneratia apetala and S. caseolaris forests at different stand ages in Shenzhen Bay, China. Marine Pollution Bulletin, 2014, 85(2): 754-763. DOI:10.1016/j.marpolbul.2014.02.024 |
| [13] | Barreto CR, Morrissey EM, Wykoff DD, Chapman SK. Co-occurring mangroves and salt marshes differ in microbial community composition. Wetlands, 2018, 38(3): 497-508. DOI:10.1007/s13157-018-0994-9 |
| [14] | Chen Q, Zhao Q, Li J, Jian SG, Ren H. Mangrove succession enriches the sediment microbial community in South China. Scientific Reports, 2016, 6: 27468. DOI:10.1038/srep27468 |
| [15] | Ghizelini AM, Santana Mendonca-Hagler LC, Macrae A. Microbial diversity in Brazilian mangrove sediments - a mini review. Brazilian Journal of Microbiology, 2012, 43(4): 1242-1254. DOI:10.1590/S1517-83822012000400002 |
| [16] | Ferreira TO, Otero XL, de Souza VS, Vidal-Torrado P, Macías F, Firme LP. Spatial patterns of soil attributes and components in a mangrove system in Southeast Brazil (São Paulo). Journal of Soils and Sediments, 2010, 10(6): 995-1006. DOI:10.1007/s11368-010-0224-4 |
| [17] |
Zhang RG. Study on sulphur accumulation and cycling in mangrove forest in pear river mouth. Tropical and Subtropical Soil Science, 1996, 5(2): 67-73.
(in Chinese) 张汝国. 珠江口红树林硫的累积和循环研究. 热带亚热带土壤科学, 1996, 5(2): 67-73. |
| [18] | Sherman RE, Fahey TJ, Howarth RW. Soil-plant interactions in a neotropical mangrove forest: Iron, phosphorus and sulfur dynamics. Oecologia, 1998, 115(4): 553-563. DOI:10.1007/s004420050553 |
| [19] | Schoonen MAA. Sulfur Cycle. Dordrecht: Springer Netherlands, 1998. |
| [20] | Holmer M, Storkholm P. Sulphate reduction and sulphur cycling in lake sediments: a review. Freshwater Biology, 2001, 46(4): 431-451. DOI:10.1046/j.1365-2427.2001.00687.x |
| [21] | Wu H, Ding ZH, Liu Y, Liu JL, Yan HY, Pan JY, Li LQ, Lin HN, Lin GH, Lu HL. Methylmercury and sulfate-reducing bacteria in mangrove sediments from Jiulong River Estuary, China. Journal of Environmental Sciences, 2011, 23(1): 14-21. DOI:10.1016/S1001-0742(10)60368-3 |
| [22] | Nedwell DB, Blackburn TH, Wiebe WJ. Dynamic nature of the turnover of organic carbon, nitrogen and sulphur in the sediments of a Jamaican mangrove forest. Marine Ecology Progress Series, 1994, 110(2/3): 223-231. |
| [23] | Pellerin A, Bui TH, Rough M, Mucci A, Canfield DE, Wing BA. Mass-dependent sulfur isotope fractionation during reoxidative sulfur cycling: a case study from Mangrove Lake, Bermuda. Geochimica et Cosmochimica Acta, 2015, 149: 152-164. DOI:10.1016/j.gca.2014.11.007 |
| [24] | Koch T, Dahl C. A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. The ISME Journal, 2018, 12(10): 2479-2491. DOI:10.1038/s41396-018-0209-7 |
| [25] | Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiology Reviews, 2009, 33(6): 999-1043. DOI:10.1111/j.1574-6976.2009.00187.x |
| [26] |
Liu Y, Jiang LJ, Shao ZZ. Advances in sulfur-oxidizing bacterial taxa and their sulfur oxidation pathways. Acta Microbiologica Sinica, 2018, 58(2): 191-201.
(in Chinese) 刘阳, 姜丽晶, 邵宗泽. 硫氧化细菌的种类及硫氧化途径的研究进展. 微生物学报, 2018, 58(2): 191-201. |
| [27] | Krishnani KK, Kathiravan V, Natarajan M, Kailasam M, Pillai SM. Diversity of sulfur-oxidizing bacteria in greenwater system of coastal aquaculture. Applied Biochemistry and Biotechnology, 2010, 162(5): 1225-1237. DOI:10.1007/s12010-009-8886-3 |
| [28] | Dahl C. Sulfur metabolism in phototrophic bacteria//Hallenbeck PC. Modern Topics in the Phototrophic Prokaryotes: Metabolism, Bioenergetics, and Omics. Cham: Springer International Publishing, 2017: 27–66. |
| [29] | Anandham R, Indiragandhi P, Madhaiyan M, Ryu KY, Jee HJ, Sa TM. Chemolithoautotrophic oxidation of thiosulfate and phylogenetic distribution of sulfur oxidation gene (soxB) in rhizobacteria isolated from crop plants. Research in Microbiology, 2008, 159(9/10): 579-589. |
| [30] | Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nature Reviews Microbiology, 2008, 6(6): 441-454. DOI:10.1038/nrmicro1892 |
| [31] | Luo JF, Tan XQ, Liu KX, Lin WT. Survey of sulfur-oxidizing bacterial community in the Pearl River water using soxB, sqr, and dsrA as molecular biomarkers. 3 Biotech, 2018, 8(1): 73. DOI:10.1007/s13205-017-1077-y |
| [32] | Belila A, Snoussi M, Hassan A. Rapid qualitative characterization of bacterial community in eutrophicated wastewater stabilization plant by T-RFLP method based on 16S rRNA genes. World Journal of Microbiology and Biotechnology, 2012, 28(1): 135-143. DOI:10.1007/s11274-011-0802-7 |
| [33] | Liang JB, Chen YQ, Lan CY, Tam NFY, Zan QJ, Huang LN. Recovery of novel bacterial diversity from mangrove sediment. Marine Biology, 2007, 150(5): 739-747. DOI:10.1007/s00227-006-0377-2 |
| [34] |
Zhao JY, Fu YN, Zhao CG, Yang SP, Qu YB, Jiao NZ. Identification and characterization of a purple sulfur bacterium from mangrove with rhodopin as predominant carotenoid. Acta Microbiologica Sinica, 2011, 51(10): 1318-1325.
(in Chinese) 赵江艳, 傅英楠, 赵春贵, 杨素萍, 曲音波, 焦念志. 一株高含玫红品的红树林海洋紫色硫细菌分离鉴定及特性. 微生物学报, 2011, 51(10): 1318-1325. |
| [35] | dos Santos HF, Cury JC, do Carmo FL, dos Santos AL, Tiedje J, van Elsas JD, Rosado AS, Peixoto RS. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One, 2011, 6(3): e16943. DOI:10.1371/journal.pone.0016943 |
| [36] | Varon-Lopez M, Dias ACF, Fasanella CC, Durrer A, Melo IS, Kuramae EE, Andreote FD. Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments. Environmental Microbiology, 2014, 16(3): 845-855. DOI:10.1111/1462-2920.12237 |
| [37] | Tourova TP, Slobodova NV, Bumazhkin BK, Kolganova TV, Muyzer G, Sorokin DY. Analysis of community composition of sulfur-oxidizing bacteria in hypersaline and soda lakes using soxB as a functional molecular marker. FEMS Microbiology Ecology, 2013, 84(2): 280-289. DOI:10.1111/1574-6941.12056 |
| [38] | Deborde J, Marchand C, Molnar N, Patrona LD, Meziane T. Concentrations and fractionation of carbon, iron, sulfur, nitrogen and phosphorus in mangrove sediments along an intertidal gradient (semi-arid climate, New Caledonia). Journal of Marine Science and Engineering, 2015, 3(1): 52-72. DOI:10.3390/jmse3010052 |
| [39] | Mahmood Q, Hu BL, Cai J, Zheng P, Azim MR, Jilani G, Islam E. Isolation of Ochrobactrum sp. QZ2 from sulfide and nitrite treatment system. Journal of Hazardous Materials, 2009, 165(1/3): 558-565. |
| [40] | Mishra RR, Swain MR, Dangar TK, Thatoi H. Diversity and seasonal fluctuation of predominant microbial communities in Bhitarkanika, a tropical mangrove ecosystem in India. Revista De Biologia Tropical, 2012, 60(2): 909-924. |
| [41] | Zhang XY, Hu BX, Ren HJ, Zhang J. Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Science of the Total Environment, 2018, 633: 518-528. DOI:10.1016/j.scitotenv.2018.03.158 |
| [42] | Kristensen E, Holmer M, Bussarawit N. Benthic metabolism and sulfate reduction in a Southeast Asian mangrove swamp. Marine Ecology Progress Series, 1991, 73: 93-103. DOI:10.3354/meps073093 |
| [43] | Kristensen E, Holmer M, Banta GT, Jensen MH, Hansen K. Carbon, nitrogen and sulfur cycling in sediments of the Ao Nam Bor mangrove forest, Phuket, Thailand: a review. Phuket Marine Biological Center Research Bulletin, 1995, 60: 37-64. |
| [44] | Eschemann A, Kühl M, Cypionka H. Aerotaxis in Desulfovibrio. Environmental Microbiology, 1999, 1(6): 489-494. DOI:10.1046/j.1462-2920.1999.00057.x |
| [45] | Cypionka H. Oxygen respiration by Desulfovibrio species. Annual Review of Microbiology, 2000, 54: 827-848. DOI:10.1146/annurev.micro.54.1.827 |
| [46] | Balk M, Keuskamp JA, Laanbroek HJ. Potential for sulfate reduction in mangrove forest soils: comparison between two dominant species of the Americas. Frontiers in Microbiology, 2016, 7: 1855. |
| [47] | Lyimo TJ, Pol A, den Camp HJMO. Sulfate reduction and methanogenesis in sediments of Mtoni mangrove forest, Tanzania. Ambio, 2002, 31(7/8): 614-616. |
| [48] | Jørgensen BB. The sulfur cycle of a coastal marine sediment (Limfjorden, Denmark). Limnology and Oceanography, 1977, 22(5): 814-832. DOI:10.4319/lo.1977.22.5.0814 |
| [49] | Pérez-Jiménez JR, Kerkhof LJ. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Applied and Environmental Microbiology, 2005, 71(2): 1004-1011. DOI:10.1128/AEM.71.2.1004-1011.2005 |
| [50] | Barton LL, Fauque GD. Biochemistry, physiology and biotechnology of sulfate-reducing bacteria. Advances in Applied Microbiology, 2009, 68: 41-98. DOI:10.1016/S0065-2164(09)01202-7 |
| [51] | Kleindienst S, Herbst FA, Stagars M, von Netzer F, von Bergen M, Seifert J, Peplies J, Amann R, Musat F, Lueders T, Knittel K. Diverse sulfate-reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps. The ISME Journal, 2014, 8(10): 2029-2044. DOI:10.1038/ismej.2014.51 |
| [52] | van Houten RT, Yun SY, Lettinga G. Thermophilic sulphate and sulphite reduction in lab-scale gas-lift reactors using H2 and CO2 as energy and carbon source. Biotechnology and Bioengineering, 1997, 55(5): 807-814. DOI:10.1002/(SICI)1097-0290(19970905)55:5<807::AID-BIT11>3.0.CO;2-8 |
| [53] | Taketani RG, Yoshiura CA, Dias ACF, Andreote FD, Tsai SM. Diversity and identification of methanogenic archaea and sulphate-reducing bacteria in sediments from a pristine tropical mangrove. Antonie van Leeuwenhoek, 2010, 97(4): 401-411. DOI:10.1007/s10482-010-9422-8 |
| [54] | Quillet L, Besaury L, Popova M, Paissé S, Deloffre J, Ouddane B. Abundance, diversity and activity of sulfate-reducing prokaryotes in heavy metal-contaminated sediment from a salt marsh in the Medway estuary (UK). Marine Biotechnology, 2012, 14(3): 363-381. DOI:10.1007/s10126-011-9420-5 |
| [55] | Dini Andreote F, Jiménez DJ, Chaves D, Dias ACF, Luvizotto DM, Dini-Andreote F, Fasanella CC, Lopez MV, Baena S, Taketani RG, de Melo IS. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One, 2012, 7(6): e38600. DOI:10.1371/journal.pone.0038600 |
| [56] |
Ding H, Yao SP, Liu GJ, Liu CH. Diversity and vertical distribution of culturable sulfate-reducing bacteria in coastal mangrove swamps from Hainan island, China. Geological Journal of China Universities, 2016, 22(4): 621-630.
(in Chinese) 丁海, 姚素平, 刘桂建, 刘常宏. 海南红树林湿地可培养硫酸盐还原菌的垂直分布特征研究. 高校地质学报, 2016, 22(4): 621-630. |
| [57] | Lyimo TJ, Pol A, Harhangi HR, Jetten MSM, den Camp HJMO. Anaerobic oxidation of dimethylsulfide and methanethiol in mangrove sediments is dominated by sulfate-reducing bacteria. FEMS Microbiology Ecology, 2009, 70(3): 483-492. DOI:10.1111/j.1574-6941.2009.00765.x |
| [58] | Gomes NCM, Cleary DFR, Pires ACC, Almeida A, Cunha A, Mendonça-Hagler LCS, Smalla K. Assessing variation in bacterial composition between the rhizospheres of two mangrove tree species. Estuarine, Coastal and Shelf Science, 2014, 139: 40-45. DOI:10.1016/j.ecss.2013.12.022 |
| [59] | Zuberer DA, Silver WS. Biological dinitrogen fixation (acetylene reduction) associated with Florida mangroves. Applied and Environmental Microbiology, 1978, 35(3): 567-575. DOI:10.1128/AEM.35.3.567-575.1978 |
| [60] | Blazejak A, Schippers A. Real-time PCR quantification and diversity analysis of the functional genes aprA and dsrA of sulfate-reducing prokaryotes in marine sediments of the Peru continental margin and the Black Sea. Frontiers in Microbiology, 2011, 2: 253. |
| [61] | Geets J, Borremans B, Diels L, Springael D, Vangronsveld J, Van der Lelie D, Vanbroekhoven K. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. Journal of Microbiological Methods, 2006, 66(2): 194-205. DOI:10.1016/j.mimet.2005.11.002 |
| [62] | Pelikan C, Herbold CW, Hausmann B, Müller AL, Pester M, Loy A. Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics. Environmental Microbiology, 2016, 18(9): 2994-3009. DOI:10.1111/1462-2920.13139 |
| [63] | Dar SA, Yao L, van Dongen U, Kuenen JG, Muyzer G. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Applied and Environmental Microbiology, 2007, 73(2): 594-604. DOI:10.1128/AEM.01875-06 |
| [64] | Tian HM, Gao PK, Chen ZH, Li YS, Li Y, Wang YS, Zhou JF, Li GQ, Ma T. Compositions and abundances of sulfate-reducing and sulfur-oxidizing microorganisms in water-flooded petroleum reservoirs with different temperatures in China. Frontiers in Microbiology, 2017, 8: 143. |
| [65] | Leloup J, Petit F, Boust PD, Deloffre J, Bally G, Clarisse O, Quillet L. Dynamics of sulfate-reducing microorganisms (dsrAB genes) in two contrasting mudflats of the Seine estuary (France). Microbial Ecology, 2005, 50(3): 307-314. DOI:10.1007/s00248-004-0034-6 |
| [66] | Bai SJ, Li JW, He ZL, van Nostrand JD, Tian Y, Lin GH, Zhou JZ, Zheng TL. GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Applied Microbiology and Biotechnology, 2013, 97(15): 7035-7048. DOI:10.1007/s00253-012-4496-z |
| [67] | Kostka JE, Roychoudhury A, van Cappellen P. Rates and controls of anaerobic microbial respiration across spatial and temporal gradients in saltmarsh sediments. Biogeochemistry, 2002, 60(1): 49-76. DOI:10.1023/A:1016525216426 |
| [68] | Brandt KK, Vester F, Jensen AN, Ingvorsen K. Sulfate reduction dynamics and enumeration of sulfate-reducing bacteria in hypersaline sediments of the great salt lake (Utah, USA). Microbial Ecology, 2001, 41(1): 1-11. |
| [69] | Pallud C, van Cappellen P. Kinetics of microbial sulfate reduction in estuarine sediments. Geochimica et Cosmochimica Acta, 2006, 70(5): 1148-1162. DOI:10.1016/j.gca.2005.11.002 |
| [70] | Connell WE, Patrick Jr WH. Sulfate reduction in soil: effects of redox potential and pH. Science, 1968, 159(3810): 86-87. DOI:10.1126/science.159.3810.86 |
| [71] | Balk M, Keuskamp JA, Laanbroek HJ. Potential activity, size, and structure of sulfate-reducing microbial communities in an exposed, grazed and a sheltered, non-grazed mangrove stand at the Red Sea coast. Frontiers in Microbiology, 2015, 6: 1478. |
| [72] | Dar SA, Kleerebezem R, Stams AJM, Kuenen JG, Muyzer G. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Applied Microbiology and Biotechnology, 2008, 78(6): 1045-1055. DOI:10.1007/s00253-008-1391-8 |
| [73] |
Liao JL, Yao SP, Ding H. The characteristics and controlling factors of sulfur in the sediments of coastal mangrove peat. Geological Journal of China Universities, 2008, 14(4): 620-630.
(in Chinese) 廖家隆, 姚素平, 丁海. 滨海红树林泥炭沉积物中硫的赋存特点及其控制因素. 高校地质学报, 2008, 14(4): 620-630. DOI:10.3969/j.issn.1006-7493.2008.04.016 |
| [74] | Attri K, Kerkar S, LokaBharathi PA. Ambient iron concentration regulates the sulfate reducing activity in the mangrove swamps of Diwar, Goa, India. Estuarine, Coastal and Shelf Science, 2011, 95(1): 156-164. DOI:10.1016/j.ecss.2011.08.030 |
| [75] | Kristensen E, Mangion P, Tang M, Flindt MR, Holmer M, Ulomi S. Microbial carbon oxidation rates and pathways in sediments of two Tanzanian mangrove forests. Biogeochemistry, 2011, 103(1/3): 143-158. |
| [76] | Das S, De M, Ganguly D, Maiti TK, Mukherjee A, Jana TK, De TK. Depth integrated microbial community and physico-chemical properties in mangrove soil of Sundarban, India. Advances in Microbiology, 2012, 2(3): 234-240. DOI:10.4236/aim.2012.23028 |
| [77] | Saxena D, Lokabharathi PA, Chandramohan D. Sulfate reducing bacteria from mangrove swamps of goa, central west coast of India. Indian Journal of Marine Sciences, 1988, 17(2): 153-157. |
| [78] | Loka Bharathi PA, Oak S, Chandramohan D. Sulfate-reducing bacteria from mangrove swamps Ⅱ: Their ecology and physiology. Oceanologica Acta, 1991, 14(2): 163-171. |
| [79] | Burgin AJ, Yang WH, Hamilton SK, Silver WL. Beyond carbon and nitrogen: how the microbial energy economy couples elemental cycles in diverse ecosystems. Frontiers in Ecology and the Environment, 2011, 9(1): 44-52. |
| [80] | Zhou JZ, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou AF, He ZL, Van Nostrand JD, Hazen TC, Stahl DA, Wall JD, Arkin AP. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nature Reviews Microbiology, 2011, 9(6): 452-466. DOI:10.1038/nrmicro2575 |
| [81] | Antler G, Pellerin A. A critical look at the combined use of sulfur and oxygen isotopes to study microbial metabolisms in methane-rich environments. Frontiers in Microbiology, 2018, 9: 519. DOI:10.3389/fmicb.2018.00519 |
| [82] | Lloyd KG, Lapham L, Teske A. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Applied and Environmental Microbiology, 2006, 72(11): 7218-7230. DOI:10.1128/AEM.00886-06 |
| [83] | Canfield DE, Stewart FJ, Thamdrup B, de Brabandere L, Dalsgaard T, Delong EF, Revsbech NP, Ulloa O. A cryptic sulfur cycle in oxygen-minimum-zone waters off the chilean coast. Science, 2010, 330(6009): 1375-1378. DOI:10.1126/science.1196889 |
| [84] | Griffin BM, Schott J, Schink B. Nitrite, an electron donor for anoxygenic photosynthesis. Science, 2007, 316(5833): 1870. DOI:10.1126/science.1139478 |
| [85] | Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environmental Microbiology, 2008, 10(5): 1244-1259. DOI:10.1111/j.1462-2920.2007.01539.x |
| [86] | Chai MW, Shen XX, Li RL, Qiu GY. The risk assessment of heavy metals in Futian mangrove forest sediment in Shenzhen Bay (South China) based on SEM-AVS analysis. Marine Pollution Bulletin, 2015, 97(1/2): 431-439. |
| [87] | Holmer M, Kristensen E, Banta G, Hansen K, Jensen MH, Bussawarit N. Biogeochemical cycling of sulfur and iron in sediments of a south-east Asian mangrove, Phuket Island, Thailand. Biogeochemistry, 1994, 26(3): 145. |
| [88] | Zhou YW, Peng YS, Li XL, Chen GZ. Accumulation and partitioning of heavy metals in mangrove rhizosphere sediments. Environmental Earth Sciences, 2011, 64(3): 799-807. DOI:10.1007/s12665-011-0904-4 |
| [89] | Queiroz HM, Nóbrega GN, Otero XL, Ferreira TO. Are acid volatile sulfides (AVS) important trace metals sinks in semi-arid mangroves?. Marine Pollution Bulletin, 2018, 126: 318-322. DOI:10.1016/j.marpolbul.2017.11.020 |
| [90] | Ferreira TO, Otero XL, Vidal-Torrado P, Macías F. Effects of bioturbation by root and crab activity on iron and sulfur biogeochemistry in mangrove substrate. Geoderma, 2007, 142(1/2): 36-46. |
| [91] | Li J, Yu JY, Liu JC, Yan CL, Lu HL, Spencer KL. The effects of sulfur amendments on the geochemistry of sulfur, phosphorus and iron in the mangrove plant (Kandelia obovata (S. L.)) rhizosphere. Marine Pollution Bulletin, 2017, 114(2): 733-741. DOI:10.1016/j.marpolbul.2016.10.070 |
| [92] | Bridgham SD, Cadillo-Quiroz H, Keller JK, Zhuang QL. Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Global Change Biology, 2013, 19(5): 1325-1346. DOI:10.1111/gcb.12131 |
| [93] | Bhatia A, Pathak H, Jain N, Singh PK, Singh AK. Global warming potential of manure amended soils under rice-wheat system in the Indo-Gangetic plains. Atmospheric Environment, 2005, 39(37): 6976-6984. DOI:10.1016/j.atmosenv.2005.07.052 |
| [94] | Oremland RS, Marsh LM, Polcin S. Methane production and simultaneous sulphate reduction in anoxic, salt marsh sediments. Nature, 1982, 296(5853): 143-145. DOI:10.1038/296143a0 |
| [95] | Kiene RP, Oremland RS, Catena A, Miller LG, Capone DG. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Applied and Environmental Microbiology, 1986, 52(5): 1037-1045. DOI:10.1128/AEM.52.5.1037-1045.1986 |
| [96] | Winfrey MR, Ward DM. Substrates for sulfate reduction and methane production in intertidal sediments. Applied and Environmental Microbiology, 1983, 45(1): 193-199. DOI:10.1128/AEM.45.1.193-199.1983 |
| [97] | Schönheit P, Kristjansson JK, Thauer RK. Kinetic mechanism for the ability of sulfate reducers to out-compete methanogens for acetate. Archives of Microbiology, 1982, 132(3): 285-288. DOI:10.1007/BF00407967 |
| [98] | Nedwell DB, Banat IM. Hydrogen as an electron donor for sulfate-reducing bacteria in slurries of salt marsh sediment. Microbial Ecology, 1981, 7(4): 305-313. DOI:10.1007/BF02341425 |
| [99] | Abram JW, Nedwell DB. Inhibition of methanogenesis by sulphate reducing bacteria competing for transferred hydrogen. Archives of Microbiology, 1978, 117(1): 89-92. DOI:10.1007/BF00689356 |
| [100] | Pester M, Knorr KH, Friedrich MW, Wagner M, Loy A. Sulfate-reducing microorganisms in wetlands-fameless actors in carbon cycling and climate change. Frontiers in Microbiology, 2012, 3: 72. |
| [101] | Lovley DR, Dwyer DF, Klug MJ. Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Applied and Environmental Microbiology, 1982, 43(6): 1373-1379. DOI:10.1128/AEM.43.6.1373-1379.1982 |
| [102] | Lin XL, Hetharua B, Lin L, Xu H, Zheng TL, He ZL, Tian Y. Mangrove sediment microbiome: adaptive microbial assemblages and their routed biogeochemical processes in Yunxiao mangrove national nature reserve, China. Microbial Ecology, 2019, 78(1): 57-69. DOI:10.1007/s00248-018-1261-6 |
 2020, Vol. 60
2020, Vol. 60