中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 周阳, 黎印, 金巍, 成艳芬, 朱伟云. 2020
- Yang Zhou, Yin Li, Wei Jin, Yanfen Cheng, Weiyun Zhu. 2020
- 瘤胃甲烷菌第七目的研究进展
- Progress of Methanomassiliicoccales in the rumen
- 微生物学报, 60(1): 1-12
- Acta Microbiologica Sinica, 60(1): 1-12
-
文章历史
- 收稿日期:2019-03-16
- 修回日期:2019-06-10
- 网络出版日期:2019-07-01
2. 南京农业大学动物科学类国家级实验教学中心, 江苏 南京 210095
2. National Experimental Teaching Center for Animal Science, Nanjing Agricultural University, Nanjing 210095, Jiangsu Province China
近10年来,随着研究的拓展和深入,甲烷菌由6个目扩展到7个目,分别为甲烷火菌目(Methanopyrales)、甲烷球菌目(Methanococcales)、甲烷杆菌目(Methanobacteriales)、甲烷微菌目(Methanomicrobiales)、甲烷八叠球菌目(Methanosarcinales)和甲烷胞菌目(Methanocellales),以及新建立的第七个目Mmc[1]。Mmc在系统进化关系上与古菌热原体目(Thermoplasmatales)相近[2]。这一目甲烷菌分布广泛,已在深海沉积物[3]、稻田[4]、湖泊水体[5]、垃圾填埋场[6]、湿地[7]等自然环境,以及人[8]、猪[9]、牛[10]、羊[11]、袋鼠[12]、白蚁[13]和圣甲虫[14]等动物和昆虫的消化道中发现。在发现和建立这一新的甲烷菌目的过程中,Mmc曾被称为瘤胃古菌C簇(RCC)[15]和Methanoplasmatales[16]。越来越多的研究显示,Mmc是瘤胃中仅次于甲烷杆菌目的第二大甲烷菌菌群[17-18],在一些条件下,甚至可以成为最优势菌群[19]。全面深入了解Mmc菌群,有助于瘤胃甲烷减排新策略的提出。
1 系统进化分类基于16S rRNA基因和mcrA基因构建系统进化树。在进化树中,Mmc主要形成三大簇[20]:“Ca. M. alvus”簇,主要来源于动物消化道(也被称为Gastrointestinal tract clade,肠道簇);“M. luminyensis”簇和“Lake Pavin”簇,主要来自于自然环境(也被称为Environmental clade,环境簇)。然而,菌株Methanomassiliicoccus luminyensis分离于人类粪便,“Candidatus Methanomassiliiciccus intestinalis” Issoire-Mx1富集于人类粪便,Methanomassiliicoccus archaeon RumEn M1(RumEn M1)富集于牛瘤胃,它们均属于“环境簇”;Candidatus Methanomassiliiciccus caenicola富集于厌氧污泥,却属于“肠道簇”(图 1)。Mmc系统进化分支与生境的关系目前还不十分清楚。
由于分离培养较为困难,已分离培养的Mmc菌株极少,缺乏模式菌株,阻碍了进行深入的系统进化分类。Dridi等[21]提出了第一个属水平上的分类“Methanomassiliicoccus”属。目前,该属只有3个种,分别为M. luminyensis、Ca. M. intestinalis和RumEn M1。Seederf等[19]对已发表的Mmc 16S rRNA基因序列进行系统进化分析,并将这些序列在属水平上(95%–97%相似性)进行归类,共形成12个组(临时命名为Group1–Group12)。虽然这一分类体系尚存缺陷,但极大地促进了瘤胃甲烷菌菌群结构的研究[22]。
2 分离培养Mmc严格厌氧,生长缓慢,营养需求复杂,导致对这一目甲烷菌富集和分离都十分困难。目前仅有一株纯菌株M. luminyensis以及一些富集培养物的报道。其中富集培养物“Candidatus Methanomethylophilus alvus” Mx1201[23]和“Candidatus Methanomethylophilus intestinalis” Issoire-Mx1[24]源于人类粪便;“Candidatus Methanoplasma termitum” MpT1[25](Ca. M. termitum)源于白蚁肠道;Methanomassiliicoccus archaeon RumEn M1[7](RumEn M1)、Methanomassiliicoccus archaeon RumEn M2[7](RumEn M2)、“Candidatus Methanomethylophilus” sp. 1R26[26]和Thermoplasmatales archaeon BRNA1(未发表)源于牛瘤胃,Methanogenic archaeon ISO4-G1[27](ISO4-GI)和Methanogenic archaeon ISO4-H5[28](ISO4-H5)源于绵羊瘤胃,Candidatus Methanogranum caenicola[8] (Ca. M.caenicola)源于厌氧污泥。这些富集培养物的获得表明,通过与其他微生物体外共培养的方法能富集得到Mm甲烷菌株。Mmc营养需要复杂,共培养能够提高Mmc甲烷菌抗逆境的能力,从而提高其被富集培养的几率。本实验室研究发现,Mmc菌株仍存活于第62代厌氧真菌体外共培养液中,一些Mmc菌株可能与厌氧真菌存在某种共生机制[29]。辅酶M (2-mercaptoethanesulfonic acid,HS-CoM)是相对分子量最小的辅酶,只存在于大多数甲烷菌中,在甲烷形成过程中充当甲基载体,是产生甲烷的关键酶。但Mmc甲烷菌缺少编码HS-CoM的基因(comABC),在富集分离Mmcs时需额外添加辅酶M才能生长[28, 30-31]。Mmc营养需求还不是很清楚,已有研究结果显示,在培养基中添加酵母膏、钨酸盐以及亚硒酸盐能促进Mmc的生长[8, 16, 21]。体外培养、宏基因组学和基因组学研究发现Mmc甲烷菌严格利用氢气还原甲基化合物(甲醇、一甲胺、二甲胺或二甲基硫)生成甲烷[25, 28],人源菌株M.luminyensis B10、人源富集培养物“Candidatus Methanomethylophilus alvus” Mx1201和“Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1以及瘤胃源富集培养物中Methanomassiliicoccus archaeon RumEn M1、“Candidatus Methanomethylophilus” sp. 1R26、Methangenic archaeon ISO4-G1和Methanogenic archaeon ISO4-H5均能利用氢气还原甲醇、一甲胺、二甲胺、三甲胺生成甲烷。因此,在分离培养甲烷菌的过程中,添加适量的甲基底物有助于这一目甲烷菌的富集。本实验室研究发现,利用甲烷菌与产氢纤维降解菌共分离的方法,在培养基中添加20 mmol/L的三甲胺或60 mmol/L的甲醇,能够在短期内从瘤胃液中富集培养Mmc甲烷菌。
3 生理生化特性对纯菌株和富集培养物的研究结果显示(表 1),Mmc菌株严格厌氧,是不运动、有规则的球型菌,直径大约在0.3–1.0 μm,具有嗜常温性和轻微嗜碱性。在有H2作为电子供体时,Mmc甲烷菌可通过还原C1甲基底物产生CH4。甲酸盐、乙酸盐、CO2、2-丁醇、2-丙醇、环戊醇、2-戊醇、乙醇、正丙醇以及丁二醇不能作为其能量来源产生CH4[8, 21, 25, 28]。但不同菌株可利用的甲基底物存在较大差异。M. luminyensis和ISO4-H5能利用甲醇、一甲胺、二甲胺和三甲胺生成甲烷[21, 28];Ca. M. termitum只能利用甲醇和一甲胺,不能利用二甲胺和三甲胺[25];Thermoplasmatales archaeon BRNA1和Methanomassiliicoccus archaeon RumEn M2的宏基因组测序结果显示,前者可利用一甲胺、二甲胺、三甲胺,但不能利用甲醇[28];后者仅能利用三甲胺[7]。这些底物利用差异可能反映了Mmc菌株对不同生境的适应性,另一方面也反映了不同生境的生理环境差异。
| Characteristic | M. luminyensis[21] | Ca. M. termitum[8] | Ca. M. caenicola[28] | ISO4-H54 |
| Source | Human faeces | Termite guts | Digested sludge | Ovine rumen |
| Oxygen | Strict anaerobe | Strict anaerobe | Strict anaerobe | Strict anaerobe |
| Morphology | Cocci, as single cells | Cocci, as single cells | Cocci, as single cells | Cocci, as single cells |
| Diameter/μm | 0.7–1.0 | 0.5–0.8 | Not reported | 0.3–0.6 |
| Motility | Non-motile | Non-motile | Not reported | Non-motile |
| Optimum TEMP | 37 ℃ | Not reported | Not reported | 38/39 ℃ |
| Optimum pH | 7.6 | Not reported | Not reported | Not reported |
| Salinity | 0.1%–1.0% | Not reported | Not reported | Not reported |
| Substrates | H2+MeOH/MMA/DMA/TMA | H2+MeOH/MMA(not utilize DMA or TMA) | H2+MeOH(MMA, DMA or TMA were not reported) | H2+MeOH/MMA/ DMA/TMA |
| * TEMP: temperature; MeOH: methanol; MMA: monomethylamine; DMA: dimethylamine; TMA: trimethylamine. | ||||
M. luminyensis在420 nm激发光下能产生蓝绿光,但其他Mmc菌株缺少编码辅酶F420的基因,在420 nm激发光下不呈现甲烷菌的特征性自发荧光[21, 28]。甲烷菌的细胞壁主要由假胞壁质组成,UDP-N-乙酰-D-葡萄糖胺是合成假胞壁质的前体物质。肠道簇的Ca. M. termitum、ISO4-H5、Ca. M. alvus缺失了编码UDP-N-乙酰-D-葡萄糖胺的大多数基因,但环境簇的M. luminyensis、Ca. M. intestinalis仍保留合成这种物质的全部基因[25]。M. luminyensis为革兰氏阳性菌,其细胞壁由一层薄的电子浓密层和一层厚的透明层组成[21]。菌株ISO4-H5[28]和Ca. M. termitum[25]没有细胞壁,但具有特殊的双层膜结构,即细胞质由细胞质膜和最外层膜包围。这种不同的生理结构可能与其生存环境存在一定联系。“环境簇”的Mmc菌株可能更需要细胞壁保护细胞适应外界环境,而“肠道簇”的Mmc菌株具有双层膜结构,能够适应肠道渗透压、pH等环境的改变,基因组的分析结果进一步证实了这种观点[25]。
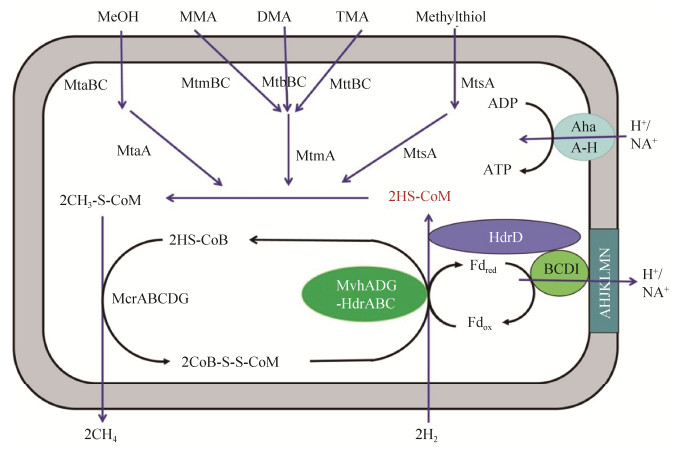
4 甲烷生成途径Mmc缺失了编码甲酰甲烷呋喃脱氢酶、Eha氢化酶、Ehb氢化酶、甲酰四氢甲烷蝶呤环化水解酶、甲酰四氢甲烷蝶呤脱氢酶、甲酰四氢甲烷蝶呤还原酶、F420还原氢化酶、辅酶M甲基转移酶(MtrA-H)基因,这些酶参与将CO2还原成为CH3-S-CoM的代谢路径[25, 28, 32]。这些基因的缺失暗示,Mmc拥有不同的甲烷生成途径。
Mmc属于甲基还原营养型,能以氢作为电子供体,还原C1甲基化合物生成甲烷,Mmc具有特殊的辅酶M甲基转移酶系统(Mt)。每种Mt均由一个具有底物特异性的甲基转移酶MT1、甲基转移酶MT2以及一个具有底物特异性的类咕琳蛋白组成。甲基转移酶MT1可以催化还原态的类咕琳蛋白甲基化,甲基转移酶MT2可将类咕琳蛋白上的甲基基团转移到辅酶M(HS-CoM)上[33]。不同类型的甲基化合物诱导相应Mt的表达。甲醇、一甲胺、二甲胺、三甲胺可以分别诱导类咕琳蛋白mtaC、mtmC、mtbC和mttC基因的表达,以及甲基转移酶MT1 mtaB、mtmB、mMtbB和mttB基因的表达;甲基转移酶MT2的mtaA基因对应的底物为甲醇,mtbA基因对应的底物为一甲胺、二甲胺和三甲胺[33-34]。
在辅酶M甲基转移酶系统(Mt)的作用下,底物的甲基基团转移到HS-CoM的巯基上生成CH3-S-CoM (以甲醇为例),即2CH3OH+2HS- CoM→2CH3-S-CoM[25, 28, 32]。不同类型的甲基化合物诱导相应Mt的表达。随后,辅酶B(HS-CoB)在甲基辅酶M还原酶的催化下还原CH3-S-CoM生成甲烷和异质二硫化物(CoB-S-S-CoM),即2CH3-S-CoM+2HS-CoB→2CH4+2CoB-S-S-CoM[7, 25, 28]。Mmc不能合成细胞色素和甲烷吩嗪,但同时含有两种异质二硫化物还原酶(HdrABC和HdrD)。异质二硫化物还原酶HdrD,缺失了能接受来自甲烷吩嗪电子的HdrE亚基,但仍具有还原CoB-S-S-CoM的活性[34]。CoB-S-S-CoM能同时被两种不同的异质二硫化物还原酶(HdrABC和HdrD)还原,重新释放出HS-CoM和HS-CoB[25, 33],即2CoB-S-S-CoM→2HS-CoB+2HS-CoM。
5 能量生成途径在马氏甲烷八叠球菌(Methanosarcina mazei)中,F420H2脱氢酶(Fpo)由13个亚基组成(FpoABCDHIJKLMNOF),能与细胞膜相结合(亚基FpoLMNHKJA为跨膜蛋白),参与辅酶F420还原态(F420H2)的氧化,从而形成H+梯度[35]。Mmc存在一种类似Fpo的复合体,但缺少能与辅酶F420作用的亚基FpoF和与甲烷吩嗪作用的亚基FpoO,无法氧化F420H2。但这种类似Fpo的复合体能与HdrD相结合,形成Fpo/HdrD复合体,通过电子歧化催化还原态的铁氧化还原蛋白(Fdred)的生成和CoM-S-S-CoB的还原[30]。此外,在已有的Mmc菌株的基因组中,都发现存在编码H+/Na+反向转运体和AoA1-ATP合成酶的基因[25]。AoA1-ATP合成酶可以利用钠离子梯度或氢离子梯度合成ATP,但Mmc中存在Fpo/HdrD复合体,表明其更可能靠氢离子梯度推动能量的生成。
Mmc可通过MvhADG/HdrABC复合体和Fpo/HdrD复合体共同参与能量的合成。首先,细胞外的氢进入细胞质内,通过MvhADG/HdrABC还原CoM-S-S-CoB和Fdox,产生Fdred,即:2H2+Fdox+CoM-S-S-CoB→Fdred+HS-CoM+HS-CoB+2H+。随后,Fpo复合体氧化Fdred,推动形成跨膜氢离子梯度,同时将电子传递到HdrD还原CoM-S-S-CoB,即:Fdred+2H++CoM-S-S-CoB→ Fdox+HS-CoM+HS-CoB。最后,AoA1-ATP合成酶利用氢离子梯度合成ATP[25, 28, 30, 32]。然而,电子从Fdred到Fpo复合体,再从Fpo复合体到异二硫化物还原酶HdrD的转移过程仍不清楚。
综上所述,Mmc缺少了将CO2还原为CH3-S- CoM的完整代谢途径,但可以通过MvhADG/ HdrABC复合体和Fpo/HdrD复合体的协同作用来弥补这一代谢的缺陷(图 2)。
|
| 图 2 Mmc的甲烷生成途径和能量代谢途径 Figure 2 ethane formation and energy metabolism pathway of Mmc strains. The red color indicates that Mmc does not contain the genes required for production of coenzyme M. MeOH: methanol; MMA: monomethylamine; DMA: dimethylamine; TMA: trimethylamine; Fd: ferredoxin. |
6 基因组特性
目前,已有6个Mmc菌株的基因组完成图发表,分别为Ca. M. alvus[23]、Ca. M. intestinalis[24]、Ca. M. termitum[25]、BRNA1(CP002916)、ISO4-G1[27]和ISO4-H5[28]。此外,还有4个菌株的基因组草图发表,分别为M. luminyensis[36]、RumEn M1[7]、RumEn M2[7]和Ca. Methanomethylophilus[26] (表 2)。
| Clades | Mmc strains | GenBank accession number |
Genome size/Mb |
No. of genes |
No. of tRNA genes |
No. of rRNA genes | GC content/% |
| Environmental | M. luminyensis [36] | CAJE01000001 | 2.62 | 2630 | 45 | 4(2 5S, 1 16S, 1 23S) | 60.5 |
| Ca. M. intestinalis [24] | CP005934 | 1.93 | 1866 | 44 | 4(2 5S, 1 16S, 1 23S) | 41.3 | |
| RumEn M1[7] | LJKK00000000 | 2.21 | 2322 | 44 | 4(2 5S, 1 16S, 1 23S) | 62.5 | |
| Gastrointestinal tract |
RumEn M2[7] | LJKL00000000 | 1.28 | 1419 | 45 | 4(2 5S, 1 16S, 1 23S) | 54.8 |
| Ca. ethanomethylophilus [26] | LOPS00000000 | 1.72 | 1809 | 37 | 5(3 5S, 1 16S, 1 23S) | 60.4 | |
| Ca. M. alvus [23] | CP004049 | 1.67 | 1707 | 45 | 4(2 5S, 1 16S, 1 23S) | 55.6 | |
| Ca. M. termitum [25] | CP010070 | 1.49 | 1458 | 46 | 3(1 5S, 1 16S, 1 23S) | 49.2 | |
| BRNA1(Unpublished) | CP002916 | 1.46 | 1577 | 45 | 4(2 5S, 1 16S, 1 23S) | 58.3 | |
| ISO4-H5[28] | CP014214 | 1.94 | 1877 | 47 | 7(3 5S, 2 16S, 2 23S) | 54.0 | |
| ISO4-G1[27] | CP013703 | 1.59 | 1549 | 45 | 4(2 5S, 1 16S, 1 23S) | 55.5 |
这些基因组不含质粒,仅由一个环状的染色体组成,大小介于1.28 Mb (RumEn M2)和2.64 Mb (M. luminyensis)之间。同一属的Ca. M. intestinalis (1.93 Mbp)和M. luminyensis (2.64 Mbp)两个菌株的基因组大小有较大差异,具有明显的不均匀性。Mmc的基因组大小与基因数量成正比,且基因组编码密度非常接近,大约每个基因1000 bp。不同菌株之间的GC含量差异非常大,但与遗传距离和环境温度没有相关性。
在大多数甲烷菌中,rRNA基因由操纵子组成。Mmc的rRNA基因由独立的5S、16S和23S rRNA基因组成,至少包含了一个16S和一个23S rRNA基因的单拷贝。大多数菌株具有两个非邻接拷贝的5S rRNA基因,但Ca. Methanomethylophilus具有3个拷贝的5S rRNA基因,在Ca. M. termitum中只有1个拷贝的5S rRNA基因。不同基因组之间的tRNA基因数量存在较大的差异。Ca. M. termitum有44个tRNA基因,Ca. M. alvus有47个,ISO4-G1有45个,而Ca. Methanomethylophilus 只有37个。
大多数Mmc基因组都含有转运20种氨基酸的tRNA (M. luminyensis缺少转运赖氨酸的tRNA,RumEn M1缺少转运色氨酸的tRNA),但都没有转运硒代半胱氨酸的tRNA[7, 25-26, 28-30]。吡咯赖氨酸(Pyrrolysine,Pyl),是目前已知的第22个参与蛋白质生物合成的氨基酸,存在于Methanosarcinaceae的甲基转移酶和少数几种细菌中,是合成甲基转移酶的必需氨基酸[37]。研究发现,Mmc基因组中存在编码吡咯赖氨酸必需的pylT、pylS、pylB、pylC和pylD基因簇[28, 38]。暗示,吡咯赖氨酸的合成可能是产甲烷古菌演变的重要标志。
7 基因组与环境适应性“肠道簇”的基因组大小介于1.28 Mb和1.94 Mb之间。相比于“环境簇”,“肠道簇”的基因组大幅简化,这种基因组上的差异可能与环境的适应性相关。Ca. M. termitum在进化关系上属于“肠道簇”,Lang等[25]发现它不能利用三甲胺,这一底物利用缺陷导致Ca. M. termitum不适于在营养物质匮乏或变化较大的环境中生存。与其他自然环境相比,肠道内营养物质较为丰富,较易获得营养物质。
Mmc基因组中存在编码过氧化氢酶、红素氧还蛋白、过氧化物还原酶、赤鲜素蛋白的基因,这些基因能够增强Mmc在有氧环境中的抗氧化能力。“环境簇”的M. luminyensis含有8个拷贝的过氧化物还原酶基因,而“肠道簇”的Ca. M. alvus只含有2个拷贝[30];“环境簇”的RumEn M1含有4个拷贝的过氧化物还原酶基因和4个拷贝的赤鲜素蛋白基因,而“肠道簇”的RumEn M2仅含有2拷贝的过氧化物还原酶基因和2拷贝的赤鲜素蛋白基因[7]。这些差异表明,“环境簇”的Mmc具有更强的抗氧化能力,更容易适应复杂多变的环境。
尽管M. luminyensis、Ca. M. alvus和Ca. M. intestinalis都来源于人肠道,但只有Ca. M. alvus含有编码胆酰甘氨酸水解酶(CBAH)的基因[30]。胆酰甘氨酸水解酶能够增强甲烷菌在消化道中对胆盐的抵抗力。M. smithii和M. stadtmanae是人肠道中的另外两种优势甲烷菌,它们的基因组中也含有这种基因[39-40]。RumEn M1和RumEn M2的基因组中也缺失编码CBAH的基因[7]。在反刍动物肠道中,胆盐直接分泌进入小肠[41],瘤胃中的微生物可能因没有环境胁迫的压力而缺失这一基因。但RumEn M1和RumEn M2的基因组都含有一种甜菜碱ABC转运体,这种转运体可以通过积累甜菜碱调节渗透压,从而应对瘤胃的高盐环境[7]。
此外,“环境簇”的M. luminyensis和Ca. M. intestinalis能适应消化道环境,可能还在于其编码的COG0790蛋白中存在的一个保守的氨基酸结构域[30]。此结构域存在于细菌和真核生物的蛋白质中,但在古菌中一直未被发现,该结构域的存在能够使微生物适应消化道环境[42]。Ca. M. alvus的基因组中,含有28个编码COG0790蛋白的基因,而Ca. M. intestinalis有6个,M. luminyensis只有1个,这表明不同菌株适应消化道环境的能力存在差异。
8 在瘤胃中的丰度和多样性越来越多的证据表明Mmc是瘤胃中仅次于甲烷短杆菌菌群的第二大菌群[2, 43],Janssen和Kirs[15]发现Mmc占瘤胃古菌的15.8%。Seedorf等[19]研究发现在新西兰放牧反刍动物的瘤胃中,Mmc占总甲烷菌的平均比例是10.4%。Kittelmann等报道绵羊、牛和驯鹿瘤胃中的Mmc占总甲烷菌的18.1%。Huang等研究发现青藏高原上的牦牛和黄牛的瘤胃中,Mmc的比例分别高达80.9%和62.9%。
在日粮中使用富含胆碱、甜菜碱的糖蜜和小麦加工副产品等饲料原料时,或者直接添加胆碱和甜菜碱时,瘤胃中Mmc的丰度会增加[44]。本实验室研究发现,饲喂全草料日粮山羊瘤胃内容物中的Mmc丰度显著高于饲喂高精料日粮山羊[29]。这种差异的产生可能是由于高精料日粮降低了瘤胃内的pH值,从而抑制Mmc的生长。研究还发现在中国山羊的瘤胃中,Mmc主要由Group 8、Group 9和Group 10组成[45];而在新西兰绵羊和牛的瘤胃中,Mmc主要由Group 10、Group 11和Group 12组成[17]。
9 展望Mmc分布广泛,作为一个新建立的甲烷菌目,缺乏模式菌株,系统进化分类尚未健全,严重制约了瘤胃甲烷菌以及其他环境中甲烷菌菌群的研究,因此,从不同环境中大量分离培养新的Mmc菌株,了解它们的生理生化和基因组特性是必需的工作。长期以来对瘤胃甲烷生成的认识是“瘤胃甲烷主要来源于氢气还原二氧化碳”。然而,Mmc作为瘤胃中第二大甲烷菌菌群,却利用甲基化合物生成甲烷,这一代谢途径对瘤胃甲烷生成的贡献尚不清楚。全面深入地了解Mmc,有助于高效的瘤胃甲烷减排策略的提出。
| [1] | Garcia JL, Patel BKC, Ollivier B. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe, 2000, 6(4): 205-226. DOI:10.1006/anae.2000.0345 |
| [2] | Wright ADG, Auckland CH, Lynn DH. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Applied Environmental Microbiology, 2007, 73(13): 4206-4210. DOI:10.1128/AEM.00103-07 |
| [3] | Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature, 1992, 356(6365): 148-149. DOI:10.1038/356148a0 |
| [4] | Großkopf R, Stubner S, Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Applied Environmental Microbiology, 1998, 64(12): 4983-4989. DOI:10.1128/AEM.64.12.4983-4989.1998 |
| [5] | Jurgens G, Glöckner FO, Amann R, Saano A, Montonen L, Likolammi M, Münster U. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiology Ecology, 2000, 34(1): 45-56. |
| [6] | Huang LN, Zhou H, Chen YQ, Luo S, Lan CY, Qu LH. Diversity and structure of the archaeal community in the leachate of a full-scale recirculating landfill as examined by direct 16S rRNA gene sequence retrieval. FEMS Microbiology Letters, 2002, 214(2): 235-240. DOI:10.1111/j.1574-6968.2002.tb11353.x |
| [7] | Söllinger A, Schwab C, Weinmaier T, Loy A, Tveit AT, Schleper C, Urich T. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiology Ecology, 2016, 92(1): fiv149. DOI:10.1093/femsec/fiv149 |
| [8] | Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki KI, Igarashi Y, Haruta S. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes and Enviroments, 2013, 28(2): 244-250. DOI:10.1264/jsme2.ME12189 |
| [9] | Luo YH, Chen H, Yu B, He J, Zheng P, Mao XB, Tian G, Yu J, Huang ZQ, Luo JQ, Chen DW. Dietary pea fiber increases diversity of colonic methanogens of pigs with a shift from Methanobrevibacter to Methanomassiliicoccus-like genus and change in numbers of three hydrogenotrophs. BMC Microbiology, 2017, 17(1): 17. DOI:10.1186/s12866-016-0919-9 |
| [10] | Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiology Ecology, 2007, 62(3): 313-322. DOI:10.1111/j.1574-6941.2007.00394.x |
| [11] | Wright ADG, Williams AJ, Winder B, Christophersen CT, Rodgers SL, Smith KD. Molecular diversity of rumen methanogens from sheep in western Australia. Applied and Environmental Microbiology, 2004, 70(3): 1263-1270. DOI:10.1128/AEM.70.3.1263-1270.2004 |
| [12] | Evans PN, Hinds LA, Sly LI, McSweeney CS, Morrison M, Wright ADG. Community composition and density of methanogens in the foregut of the tammar wallaby (Macropus eugenii). Applied and Environmental Microbiology, 2009, 75(8): 2598-2602. DOI:10.1128/AEM.02436-08 |
| [13] | Miyata R, Noda N, Tamaki H, Kinjyo K, Aoyagi H, Uchiyama H, Tanaka H. Phylogenetic relationship of symbiotic archaea in the gut of the higher termite Nasutitermes takasagoensis fed with various carbon sources. Microbes and Environments, 2007, 22(2): 157-164. DOI:10.1264/jsme2.22.157 |
| [14] | Egert M, Wagner B, Lemke T, Brune A, Friedrich MW. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Applied Environment Microbiology, 2003, 69(11): 6659-6668. DOI:10.1128/AEM.69.11.6659-6668.2003 |
| [15] | Janssen PH, Kirs M. Structure of the archaeal community of the rumen. Applied and Environmental Microbiology, 2008, 74(12): 3619-3625. DOI:10.1128/AEM.02812-07 |
| [16] | Paul K, Nonoh JO, Mikulski L, Brune A. "Methanoplasmatales, " Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Applied and Environmental Microbiology, 2012, 78(23): 8245-8253. DOI:10.1128/AEM.02193-12 |
| [17] | Seedorf H, Kittelmann S, Janssen PH. Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Applied and Environmental Microbiology, 2015, 81(3): 986-995. DOI:10.1128/AEM.03018-14 |
| [18] | Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One, 2013, 8(2): e47879. DOI:10.1371/journal.pone.0047879 |
| [19] | Seedorf H, Kittelmann S, Henderson G, Janseen PH. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ, 2014, 2: e494. DOI:10.7717/peerj.494 |
| [20] | Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF. Archaea and the human gut: new beginning of an old story. World Journal of Gastroenterology, 2014, 20(43): 16062-16078. DOI:10.3748/wjg.v20.i43.16062 |
| [21] | Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(8): 1902-1907. |
| [22] | Xie F, Zhang LL, Jin W, Meng ZX, Cheng YF, Wang J, Zhu WY. Methane emission, rumen fermentation, and microbial community response to a nitrooxy compound in low-quality forage fed hu sheep. Current Microbiology, 2019, 76(4): 435-441. DOI:10.1007/s00284-019-01644-5 |
| [23] | Borrel G, Harris HMB, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E, Peyret P, Gribaldo S, O'Toole PW, Brugere JF. Genome sequence of "Candidatus Methanomethylophilus alvus" Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. Journal of Bacteriology, 2012, 194(24): 6944-6945. DOI:10.1128/JB.01867-12 |
| [24] | Borrel G, Harris HMB, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O'Toole PW, Brugère JF. Genome sequence of "Candidatus Methanomassiliicoccus intestinalis" Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announcements, 2013, 1(4): e00453-13. |
| [25] | Lang K, Schuldes J, Klingl A, Poehlein A, Daniel R, Brune A. New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of "Candidatus Methanoplasma termitum". Applied and Environmental Microbiology, 2015, 81(4): 1338-1352. DOI:10.1128/AEM.03389-14 |
| [26] | Noel SJ, Hojberg O, Urich T, Poulsen M. Draft genome sequence of "Candidatus Methanomethylophilus" sp. 1R26, enriched from bovine rumen, a methanogenic archaeon belonging to the Methanomassiliicoccales order. Genome Announcements, 2016, 4(1): e01734-15. |
| [27] | Kelly WJ, Li D, Lambie SC, Jeyanathan J, Cox F, Li Y, Attwood GT, Altermann E, Leahy SC. Complete genome sequence of methanogenic archaeon ISO4-G1, a member of the Methanomassiliicoccales, isolated from a sheep rumen. Genome Announcements, 2016, 4(2): e00221-16. |
| [28] | Li Y, Leahy SC, Jeyanathan J, Henderson G, Cox F, Altermann E, Kelly WJ, Lambie SC, Janssen PH, Rakonjac J, Attwood GT. The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales. Standards in Genomic Sciences, 2016, 11(1): 59. DOI:10.1186/s40793-016-0183-5 |
| [29] | Jin W, Cheng YF, Mao SY, Zhu WY. Discovery of a novel rumen methanogen in the anaerobic fungal culture and its distribution in the rumen as revealed by real-time PCR. BMC Microbiology, 2014, 14: 104. DOI:10.1186/1471-2180-14-104 |
| [30] | Borrel G, Parisot N, Harris HMB, Peyretaillade E, Gaci N, Tottey W, Bardot O, Raymann K, Gribaldo S, Peyret P, O'Toole PW, Brugère JF. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics, 2014, 15: 679. DOI:10.1186/1471-2164-15-679 |
| [31] | Burke SA, Krzycki JA. Reconstitution of monomethylamine: coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. Journal of Biological Chemistry, 1997, 272(26): 16570-16577. DOI:10.1074/jbc.272.26.16570 |
| [32] | Kröninger L, Berger S, Welte C, Deppenmeier U. Evidence for the involvement of two heterodisulfide reductases in the energy-conserving system of Methanomassiliicoccus luminyensis. The FEBS Journal, 2016, 283(3): 472-483. DOI:10.1111/febs.13594 |
| [33] | Ferguson Jr DJ, Krzycki JA, Grahame DA. Specific roles of methylcobamide: Coenzyme M methyltransferase isozymes in metabolism of methanol and methylamines in Methanosarcina barkeri. Journal of Biological Chemistry, 1996, 271(9): 5189-5194. DOI:10.1074/jbc.271.9.5189 |
| [34] | Ferguson Jr DJ, Krzycki JA. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase Ⅱ from Methanosarcina barkeri. Journal of Bacteriology, 1997, 179(3): 846-852. DOI:10.1128/JB.179.3.846-852.1997 |
| [35] | Bäumer S, Ide T, Jacobi C, Johann A, Gottschalk G, Deppenmeier U. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. Journal of Biological Chemistry, 2000, 275(24): 17968-17973. DOI:10.1074/jbc.M000650200 |
| [36] | Gorlas A, Robert C, Gimenez G, Drancourt M, Raoult D. Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated archaea species. Journal of Bacteriology, 2012, 194(17): 4745-4745. DOI:10.1128/JB.00956-12 |
| [37] | Prat L, Heinemann IU, Aerni HR, Rinehart J, O'Donoghue P, Soll D. Carbon source-dependent expansion of the genetic code in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(51): 21070-21075. DOI:10.1073/pnas.1218613110 |
| [38] | Borrel G, Gaci N, Peyret P, O'Toole PW, Gribaldo S, Brugère JF. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea, 2014, 2014: 374146. |
| [39] | Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(25): 10643-10648. DOI:10.1073/pnas.0704189104 |
| [40] | Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. Journal of Bacteriology, 2006, 188(2): 642-658. DOI:10.1128/JB.188.2.642-658.2006 |
| [41] | Bauman DE, Lock AL. Concepts in lipid digestion and metabolism in dairy cows//Proceedings of Tri-State Dairy Nutrition Conference. Port Wayne, Indiana: The Oiho State University, 2006: 1-14. |
| [42] | Mittl PRE, Schneider-Brachert W. Sel1-like repeat proteins in signal transduction. Cellular Signalling, 2007, 19(1): 20-31. |
| [43] | Henderson G, Cox F, Ganesh S, Jonker A, Young W, Global Rumen Census Collaborators, Janssen PH. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Scientific Reports, 2015, 5: 14567. DOI:10.1038/srep14567 |
| [44] | St-Pierre B, Cersosimo LM, Ishaq SL, Wright AD. Toward the identification of methanogenic archaeal groups as targets of methane mitigation in livestock animals. Frontiers in Microbiology, 2015, 6: 776. |
| [45] | Jin W, Cheng YF, Zhu WY. The community structure of Methanomassiliicoccales in the rumen of Chinese goats and its response to a high-grain diet. Journal of Animal Science and Biotechnology, 2017, 8: 47. DOI:10.1186/s40104-017-0178-0 |
 2020, Vol. 60
2020, Vol. 60