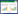中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Lei Chen, Mi Liu, Weilai Sha, Ying Gao, Jiaxin Chen, Jing Zhu. 2019
- 陈磊, 刘咪, 沙未来, 高迎, 陈佳欣, 朱静. 2019
- Diversity of intestinal microbiota of Caracal cat (Caracal caracal) by high-throughput sequence of the bacterial 16S rRNA gene
- 基于16S rRNA基因高通量测序研究狞猫肠道微生物多样性
- Acta Microbiologica Sinica, 58(9): 1685-1694
- 微生物学报, 58(9): 1685-1694
-
文章历史
- 收稿日期:2019-03-11
- 修回日期:2019-05-15
- 网络出版日期:2019-06-10
2. Ji'nan Wildlife Park, Ji'nan 250113, Shandong Province, China
2. 济南野生动物园, 山东 济南 250113
Caracal cats (Caracal caracal) are small felines belonging to Carnivora, Felidae. Caracal cats are found throughout Africa, West Asia, and the northwestern part of South Asia. They occupy a variety of habitats, ranging from savannas to mountain forests. Because of the pressure and threat of human activities and the loss and fragmentation of their habitats, caracal cat population sizes vary considerably between regions[1]. The regional population of caracal cat is also likely to fluctuate based on prey availability[2]. For example, in Africa, caracal cats are common in South Africa and southern Namibia. In central and West Africa, they are largely absent and their densities are apparently lower. In North Africa, caracal cats are considered threatened[3]. However, as caracal cat is an elusive species, it may be more widespread than indicated by records[1].
Intestinal microbes play an important role in intestinal cell proliferation, resistance to pathogens, metabolism of secondary products, and digestion of complex compounds[4-6] and are of great significance to the survival and environmental adaptation of wild animals[7-9]. However, it is difficult to collect intestinal samples from wild animals. Fortunately, fecal samples can be collected non–invasively. Previous research showed that fecal microbes represent the whole intestinal microbial community[10]. Although there are also some reports described that fecal microbes are sometimes different from the intestine microbiota, especially the small intestine microbiota, the effective detection of the fecal microbiota offering a feasible way for the study of intestinal microbes of wildlife, especially in endangered animals.
With increasing researches of the intestinal microbiome of wild animals, the intestinal microorganisms of felids have been investigated by researchers, a number of studies have reported the high correlation between intestinal microbes and the health and survival adaptation of wild felids, and the studies also found that food, sex, living environment, and man-made interference have effects on intestinal microbiota[6, 11-12], but the study of intestinal microbiota in caracal cat has not been reported. Therefore, in this study, we used fecal samples to study the intestinal microbial diversity of caracal cats by high-throughput sequencing. This would provide basic data for further evaluation of the relationship between intestinal microbes and the health and ecological adaption of caracal cats, and provides effective information for the protection of caracal cats.
1 Materials and Methods 1.1 Sample collectionFecal samples from 7 healthy wild adult caracal cats (2 males and 5 females) were used in this study. The samples were collected in Shandong Province from Ji'nan Wildlife Park (1 female and 1 male, designated CCF01 and CCM01) and Weihai Wildlife Park (4 females and 1 male, designated CCF02, CCF03, CCF04, CCF05 and CCM02). The caracal cats, on average of 4 to 5 years old, were half-scattered in an enclosure on the earth floor, weighing about 18 kilograms for males and about 15 kilograms for females, feeding on fresh beef and chicken and drinking tap water. The caracal cats from different parks were fed diets of the same composition. These animals did not suffer from any gastrointestinal disease and had not been administered any anti-inflammatory or antimicrobial drugs in the four months before sample collection. Fresh feces samples were collected within half an hour after defecation before feeding in the morning and preserved in sterilized plastic centrifuge tubes, stored in a mobile refrigerator at 4 ℃, transported to the laboratory, and stored at −80 ℃ until DNA was extracted. All samples were collected in winter of 2017. Fecal samples from the same park were collected at the same day. The experiment was approved by the Animal Protection and Utilization Committee of Qufu Normal University.
1.2 DNA extraction, 16S rRNA gene amplification by PCR, and sequence processingTotal microbial DNA was extracted from the fecal samples using the QIAamp DNA Stool Genomic DNA Extraction Kit (Qiagen, Germany) according to the manufacturer's recommended protocol. We used an ultra-micro spectrophotometer (NanoDrop2000c, Thermo Fisher Scientific, USA) to measure DNA quality and concentration. Primers 341 F (5′-CCTAYGGGRBGCASCAG-3′) and 806 R (5′-GGACTACNNGGGTATCTAAT-3′) were used to amplify the high–mutation region of 16S rRNA V3–V4 by PCR. The volume of the PCR reaction was 30 μL, and the product size was about 410 bp. The PCR reaction included 15 μL of Taq PCR MasterMix, 1.5 μL of each primer, 10 μL of microbial genomic DNA template, and 2 μL of ddH2O. The specific steps of PCR were as follows: 1 cycle of denaturing at 98 ℃ for 1 min, followed by 35 cycles of denaturing at 98 ℃ for 10 s, annealing at 55 ℃ for 30 s, elongation at 72 ℃ for 30 s, and a final extension at 72 ℃ for 5 min. The obtained PCR product was mixed with the same volume of 1×loading buffer (containing SYBR Green) and detected by 2% agarose gel electrophoresis. The samples in the bright bands at 400 bp and 450 bp were mixed at the same density ratio and purified by the QIAquick Gel Extraction Kit (Qiagen). We used the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) to create the library. After Qubit and qPCR quantification, we performed sequencing on the Illumina HiSeq2500 PE250.
2 Bioinformatics AnalysisThe data obtained from the Illumina HiSeq sequencing were spliced and quality controlled. Then, clean tags were obtained, and chimeric filtering was carried out to obtain effective tags that could be used for subsequent analysis[13]. We used Uparse software (v7.0.1001) to cluster effective tags for all samples[14]. The default similarity was 97% for sequential clustering of operational taxonomic units (OTUs). Mothur software and the SILVA 16S rRNA reference database were used to perform species annotation on the typical sequences of selected OTUs[15]. The taxonomic information was obtained and statistically analyzed at each classification level. Qiime software (v1.7.0) was used to calculate the observed species, Chao1, Shannon, Simpson, ACE, Good's coverage, and PD whole tree indices[16]. R software (v2.15.3) was used to generate dilution curves, rank abundance curves, and species cumulative curves and perform intra-group difference analysis of the α-diversity index. The analysis of α-diversity was carried out with the parametric test and non-parametric test. In order to detect the similarity among samples, we compared the shared and specific OTUs among samples by drawing petal maps. Then we used weighted and unweighted Unifrac distance matrices for UPGMA clustering analysis. The UPGMA sample clustering tree was constructed by Qiime software (v1.7.0)[17-18].
3 Results 3.1 Sequencing data and OTU annotationsThe results of this study have been submitted to the SRA database under accession number SRP135784.
A total of 1458741 effective sequences, with an average of 208392 effective sequences and an average length of 433 bp, were obtained from 7 caracal cat fecal samples for OTU clustering and other follow-up analyses. A total of 347 OTUs were obtained by clustering with 97% sequence similarity, with the average number of 233 (Table 1). The end of the rarefaction curve tended to be flat, indicating that the sequencing results effectively reflected the abundance of microbiota in the sample (Figure 1-A). The richness and evenness of species in different samples can be seen in the rank abundance curve (Figure 1-B).
| Sample name | Effective tags | OTUs | Shannon | Simpson | Chao1 | ACE | Goods coverage |
| CCF01 | 66580 | 135 | 4.296 | 0.912 | 138.500 | 138.370 | 1.000 |
| CCF02 | 238804 | 245 | 4.883 | 0.934 | 274.690 | 275.715 | 0.999 |
| CCF03 | 327017 | 235 | 3.846 | 0.884 | 257.455 | 260.036 | 0.999 |
| CCF04 | 238742 | 246 | 4.776 | 0.918 | 274.636 | 272.859 | 0.999 |
| CCF05 | 279820 | 221 | 5.700 | 0.972 | 252.500 | 252.232 | 0.999 |
| CCM01 | 59499 | 120 | 3.863 | 0.863 | 135.111 | 132.478 | 1.000 |
| CCM02 | 248279 | 234 | 4.668 | 0.933 | 255.577 | 254.718 | 0.999 |

|
| Figure 1 Rarefaction (A) and rank abundance curve (B) analyses of caracal fecal microbiota. We perform repeated sampling of the OTU subset to assess whether further sampling may result in additional groups, which is indicated by whether the curve reached the platform value. The y-axis indicates the number of OTUs detected, and the x-axis indicates the number of taxa in the sequence subset analyzed. Rank abundance curves were used to estimate the richness and evenness of taxa present in the samples. The y-axis indicates the relative abundance of OTUs, and the x-axis indicates the number of OTUs according to the relative abundance from large to small. The larger the span curve on the x-axis, the higher the species richness. The smoother the curve on the y-axis, the more even the species distribution. |
3.2 Species annotation and abundance analysis
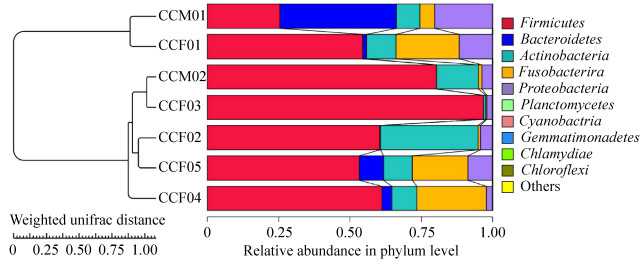
The OTU analysis of all samples revealed caracal cat intestinal microbes belonging to the bacterial domain, including 13 phyla, 26 classes, 43 orders, 75 families, and 119 genera (Figure 2). At the phylum level, Firmicutes demonstrated an absolute advantage with an average relative abundance of 61.70%. The relative abundance of Firmicutes in the sample CCF03 was 96.73%. However, the abundance of Firmicutes highly differed within samples (accounting for about 25.39%–96.73% of all OTUs). Other phyla with high abundance included Actinobacteria (0.71%– 34.15%, average 12.42%), Fusobacteria (0.45%– 24.45%, average 10.53%), Fusobacteroidetes (0.09%– 40.83%, average 7.79%), and Proteobacteria (1.95%– 20.26%, average 7.53%). Bacteroidetes had a higher abundance only in sample CCM01. Other bacteria had a lower abundance. Chloroflexi and Acidobacteria were found only in the sample CCF03. Verrucomicrobia was found in CCF03 and CCF05; Deinococcus–Thermus appeared only in samples CCM01 and CCF03 (Figure 2-A).
|
| Figure 2 Gut bacterial composition at the phylum level in each sample. According to the results of the species notes, we select the gut bacterial species at the phylum level per sample (A) and the relative abundance of the top ten biological species at the family level (B) and the genus level (C), product column maps. |
At the family level, 12 out of 75 detected families had a relative abundance of more than 1%. The most abundant of these was Peptostreptococcaceae (with an average relative abundance of 16.15%) in phylum Firmicutes, Clostridiaceae_I (14.78%), Lachnospiraceae (13.13%), Family_XI (10.03%), Coriobacteriaceae (12.31%) in phylum Actinobacteria, CFT112H7 (8.08%) in phylum Fusobacteria, and Bacteroidaceae (7.41%) in phylum Bacteroidetes. The abundance of each family varied greatly among different samples. For example, the relative abundance of Bacteroidaceae ranged from 0.95% (CCF03) to 40.21% (CCM01), and the relative abundance of Family_XI ranged from 0.46% (CCF01) to 37.74% (CCF03). Streptomycetaceae was found only in CCM01; Thermaceae was only present in CCF03; and Rhodospirillaceae was only present in CCF04 (Figure 2-B).
An average of 15.35% of bacteria in the 7 caracal cat fecal samples were not identified at the genus level. Of the 119 genera identified, 16 had an average relative abundance of more than 1%. The most abundant of these was Collinsella (11.44%), Peptoclostridium (10.91%), Clostridium sensu stricto 1 (10.3%), Peptoniphilus (10.02%), Bacteroides (7.41%), and Peptostreptococcus (5.21%). Among them, Collinsella belongs to Actinobacteria; Peptoclostridium, Clostridium sensu stricto 1, Peptoniphilus, and Peptostreptococcus belong to Firmicutes; and Bacteroides belongs to Bacteroidetes (Figure 2-C).
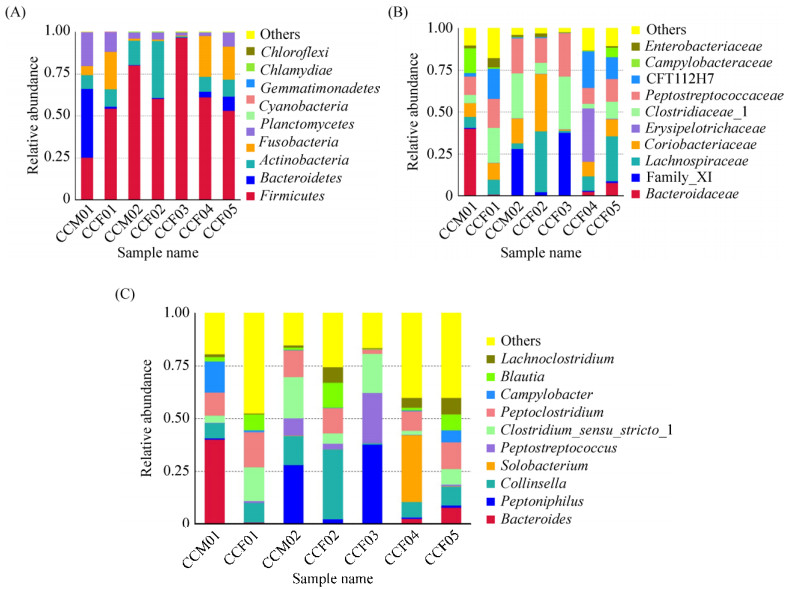
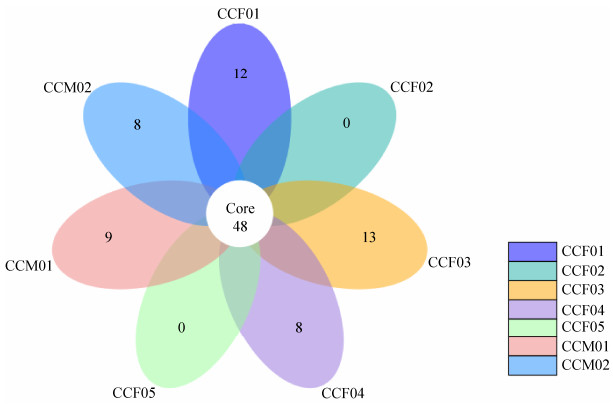
According to the petal map, the number of OTUs shared among samples (about 14% of total OTUs) was much higher than the number of OTUs specific to each individual (Figure 3). The UPGMA clustering analysis showed that CCM01 and CCF01 from Ji'nan Wildlife Park were grouped together, while the remaining 5 samples from Weihai Wildlife Park were grouped together (Figure 4).
|
| Figure 3 The petal map among different samples. Based on the results of OTU cluster, we analyze the common (endemic) OTUs among different samples by product petal map. Each petal on the petal map represents a set of samples. The number in the middle represents the OTUs common to all samples, whereas the number on the petals represents the OTUs unique to the sample. |
|
| Figure 4 UPGMA clustering analysis diagram based on unweighted UniFrac distance matrix. UPGMA clustering analysis is done with weighted Unifrac distance matrix and Unweighted Unifrac distance matrix, and the results are integrated with the relative abundance of species at phylum level. Firstly, the two samples with the smallest distance are gathered together to form a new node (the new sample), whose branch point is at half of the distance between the two samples, and then the average distance between the new "sample" and the other samples is calculated. Then we find out the smallest two samples and cluster them; then we do it again and again until all the samples are gathered together and finally we get a complete clustering tree. |
4 Discussion
More than 1000 species of microbes are colonized in the intestinal tract of humans and mammals, a large proportion of which are colonized at birth and remain stable throughout the life of the animal[9]. This is known as the core microbial community. The core microbiota has an important relationship with the health of the host and plays a vital role in food digestion, nutrient metabolism, intestinal health maintenance and immune protection, adaptation to the environment, and adaptive evolution[19]. According to the standard phyla established in existing reports, the core microbiota of the intestinal tract in catamounts and most carnivores mainly include Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Proteobacteria[19-21]. For example, the Bacteroides/Chlorobi group (about 68%) represents the most abundant intestinal microorganisms in cats (Felis catus), followed by Firmicutes (about 13%) and Proteobacteria (about 6%)[5]. The phyla with the highest abundance in the intestinal tract of snow leopard (Panthera uncia) are Firmicutes (56.5%), Actinobacteria (17.5%), Bacteroidetes (13%) and Proteobacteria (13%)[6]. In this study, the fecal microflora of caracal cats is studied by high-throughput sequencing. Firmicutes shows the highest abundance in the caracal intestinal microbiota. Our study also found that the number of OTUs shared among the samples is much higher than the number of OTUs unique to each sample, which indicates that the core microbiota played an important role in the gut of the caracal cats.
Firmicutes is the dominant phylum in the gastrointestinal tract and feces of humans and many mammals[6], and these microbes can degrade plant fibers into volatile fatty acids that can be used by the host[22]. The food of herbivores is rich in plant fiber, and the high abundance of Firmicutes in herbivores is related to their specific dietary, anatomical, and physiological characteristics[22-26]. The caracal cat is a typical carnivorous animal that mainly hunts small and medium sized mammals, primarily small rodents[1]. In the diet of the caracal cat, the proportion of vegetable food is very small, so there is no direct relationship between the food habit and the high abundance of Firmicutes in the gut of caracal cat. In the digestive tract of other predators, such as snow leopards[6], wolves[8], etc., the abundance of Firmicutes is also high, indicating that Firmicutes might play an important role in the nutrition intake, metabolism and intestinal health of carnivorous animals.
Actinobacteria and Bacteroidetes are the other two core phyla in the fecal samples of caracal cats. The abundance of Bacteroidetes in sample CCM01 is over 40%. Bacteroidetes is commonly abundant among the intestinal microbes in carnivorous animals and is the most abundant intestinal phylum in Felis catus (68%) and Cuon alpinus (21.63%–38.97%)[8, 21]. Bacteroidetes mainly degrades indigestible dietary polysaccharides[9], such as high molecular weight organic matter, and helps the host degrade plant cell wall compounds, including cellulose, pectin, and xylan. It also digests endogenous carbohydrates secreted from the gastrointestinal tract of the host, such as mucin and chondroitin sulfate polysaccharides[27]. In addition, Bacteroidetes has anti-tumor properties and participates in the metabolism and transformation of toxic or mutagenic bile acids[27], which can help maintain the host's intestinal health. In a study of dhole fecal microbes, the researchers believe that the increased abundance of Bacteroidetes in the feces of dholes in captivity is related to the consumption of commercially prepared foods containing adequate carbohydrates and soluble fiber from plant sources[9]. Bacteroidetes shows low abundance in most caracal cat fecal samples. This may be because the diet of the caracal cats is dominated by proteins and lipids, and the content of macromolecular polysaccharides is relatively low.
It has been reported that the increase in Actinobacteria abundance is related to the intake of fatty foods and animal proteins[24-25]. At the same time, Actinobacteria is also an important producer of secondary metabolites, which are important antibiotics. This suggests that the high abundance of Actinobacteria may be associated with high fat and protein intake and intestinal immunity in wild carnivores. In the present study, Actinobacteria shows a high abundance in the intestinal microbiota of the caracal cat, which is similar to the results of the study on intestinal microbiota of snow leopards[6].
With the development of high throughput sequencing technology, the study of intestinal microflora in mammals is getting deeper and deeper. In this paper, high-throughput sequencing is used to detect the intestinal microflora of caracal cats. The results of UPGMA cluster analysis show that the samples from the same sampling site are gathered together. Because of the small sample size involved in this experiment, it is difficult to objectively evaluate the effect of feeding environment on caracal cat intestinal microbiota. However, the results of this study still provide important data for protection, rescue feeding and nutritional ecology research of caracal cats.
Acknowledgements
We thank Ji'nan Wild Animal Park and Weihai Wildlife Park for their assistance in the fecal samples collection.
| [1] | Avgan B, Henschel P, Ghoddousi A. Caracal caracal (errata version published in 2016). The IUCN Red List of Threatened Species, 2016: e. T3847A102424310. |
| [2] | Mallon DP, Budd K. Regional red list status of carnivores in the Arabian Peninsula. Sharjah, UAE: Environment and Protected Areas Authority, 2011. |
| [3] | Stuart C, Stuart T. Caracal caracal//Kingdon JS, Hoffmann M. The Mammals of Africa. Amsterdam, The Netherlands: Academic Press, 2013. |
| [4] | Depauw S, Bosch G, Hesta M, Whitehouse-Tedd K, Hendriks WH, Kaandorp J, Janssens GPJ. Fermentation of animal components in strict carnivores:a comparative study with cheetah fecal inoculum. Journal of Animal Science, 2012, 90(8): 2540-2548. DOI:10.2527/jas.2011-4377 |
| [5] | Tun HM, Brar MS, Khin N, Jun L, Hui RKH, Dowd SE, Leung FCC. Gene-centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. Journal of Microbiological Methods, 2012, 88(3): 369-376. DOI:10.1016/j.mimet.2012.01.001 |
| [6] | Zhang HH, Liu GS, Chen L, Sha WL. Composition and diversity of the bacterial community in snow leopard (Uncia uncia) distal gut. Annals of Microbiology, 2015, 65(2): 703-711. DOI:10.1007/s13213-014-0909-9 |
| [7] | Zhu LF, Wu Q, Dai JY, Zhang SN, Wei FW. Evidence of cellulose metabolism by the giant panda gut microbiome. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(43): 17714-17719. DOI:10.1073/pnas.1017956108 |
| [8] | Chen L, Zhang HH, Liu GS, Sha WL. First report on the bacterial diversity in the distal gut of dholes (Cuon alpinus) by using 16S rRNA gene sequences analysis. Journal of Applied Genetics, 2016, 57(2): 275-283. DOI:10.1007/s13353-015-0319-0 |
| [9] | Wu XY, Zhang HH, Chen J, Shang S, Wei QG, Yan JK, Tu XY. Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3-V4 region of the 16S rRNA gene. Applied Microbiology and Biotechnology, 2016, 100(8): 3577-3586. DOI:10.1007/s00253-015-7257-y |
| [10] | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science, 2005, 308(5728): 1635-1638. DOI:10.1126/science.1110591 |
| [11] | He FP, Liu D, Zhang L, Zhai JC, Ma Y, Xu YC, Jiang GS, Rong K, Ma JZ. Metagenomic analysis of captive Amur tiger faecal microbiome. BMC Veterinary Research, 2018, 14(1): 379. DOI:10.1186/s12917-018-1696-5 |
| [12] | Han SY, Guan Y, Dou HL, Yang HT, Yao M, Ge JP, Feng LM. Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing. PeerJ, 2019, 7: e6684. DOI:10.7717/peerj.6684 |
| [13] | Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research, 2011, 21(3): 494-504. DOI:10.1101/gr.112730.110 |
| [14] | Edgar RC. UPARSE:highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 2013, 10(10): 996-998. DOI:10.1038/nmeth.2604 |
| [15] | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project:improved data processing and web-based tools. Nucleic Acids Research, 2013, 41(D1): D590-D596. |
| [16] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [17] | Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics, 2006, 7: 371. DOI:10.1186/1471-2105-7-371 |
| [18] | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [19] | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature, 2009, 457(7228): 480-484. DOI:10.1038/nature07540 |
| [20] | Li YM, Hu XL, Yang S, Zhou JT, Zhang TX, Qi L, Sun XN, Fan MY, Xu SH, Cha MH, Zhang MS, Lin SB, Liu SQ, Hu DF. Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Frontiers in Microbiology, 2017, 8: 1705. DOI:10.3389/fmicb.2017.01705 |
| [21] | Zhang HH, Chen L. The complete mitochondrial genome of dhole Cuon alpinus:phylogenetic analysis and dating evolutionary divergence within Canidae. Molecular Biology Reports, 2011, 38(3): 1651-1660. DOI:10.1007/s11033-010-0276-y |
| [22] | Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo SJ, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science, 2011, 331(6016): 463-467. DOI:10.1126/science.1200387 |
| [23] | Hu XL, Liu G, Shafer ABA, Wei YT, Zhou JT, Lin SB, Wu HB, Zhou M, Hu DF, Liu SQ. Comparative analysis of the gut microbial communities in forest and alpine musk deer using high-throughput sequencing. Frontiers in Microbiology, 2017, 8: 572. |
| [24] | Guan Y, Yang HT, Han SY, Feng LM, Wang TM, Ge JP. Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express, 2017, 7: 212. DOI:10.1186/s13568-017-0517-8 |
| [25] | He J, Yi L, Hai L, Ming L, Gao WT, Ji R. Characterizing the bacterial microbiota in different gastrointestinal tract segments of the Bactrian camel. Scientific Reports, 2018, 8(1): 654. DOI:10.1038/s41598-017-18298-7 |
| [26] | Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, Ahn J. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One, 2015, 10(4): e0124599. DOI:10.1371/journal.pone.0124599 |
| [27] | Chen J, Zhang HH, Wu XY, Shang S, Yan JK, Chen Y, Zhang HH, Tang XX. Characterization of the gut microbiota in the golden takin (Budorcas taxicolor bedfordi). AMB Express, 2017, 7: 81. DOI:10.1186/s13568-017-0374-5 |
 2019, Vol. 58
2019, Vol. 58