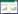中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Jiaqi Wang, Qi Zhang, Bei Jiang, Mengxia Lv, Donghua Jiang. 2019
- 王嘉琦, 张琪, 江北, 吕梦霞, 蒋冬花. 2019
- Deletion of PigE, a pigment biosynthesis-related gene, upregulates the varieties and yields of yellow pigments in Monascus purpureus Mp-21
- 色素生物合成相关PigE基因的缺失对紫色红曲霉Mp-21黄色素种类和产量的影响
- Acta Microbiologica Sinica, 59(8): 1547-1560
- 微生物学报, 59(8): 1547-1560
-
文章历史
- 收稿日期:2018-10-11
- 修回日期:2019-01-07
- 网络出版日期:2019-03-13
Monascus spp. have been widely used for more than one thousand years in China as an important edible and medicinal microorganism[1-4]. It has aroused more attention due to the production of various metabolisms, such as MPs, monacolins, γ-aminobutyric acid and hydrolytic enzymes, displaying a variety of biological activities[5-8]. MPs belong chemically to the group of polyketides, which were usually divided into three groups based on the different characteristic maximum wavelength: red (490-530 nm), orange (460-480 nm) and yellow (330-450 nm)[3, 9-11]. At present, 94 kinds of MPs have been reported, which included 44 kinds of yellow, 8 kinds of orange and 42 kinds of red pigments[3, 12]. The yellow pigments display diverse biological properties including anti-tumor, anti-inflammatory[13], anti-diabetic, anti-hyoxidative stress[14], and anti-cancer activities[15-16].
Specifically, yellow pigments as a major variety of edible pigments have been widely used in the food industry fermentation, such as meat products, pastries or beverages, yellow pigments have accounted for 60% of market demand and may have a wider application than red pigments[17]. In addition to screening high-yielding wild type Monascus strains for industrial fermentation, researchers are currently focusing on improving the yield of yellow pigments by using a variety of methods, including intraspecific protoplast fusion[18], adjusting cultivation mode[19-20] and genetic modification[21-23].
Even though research on the biosynthetic pathway of Monascus pigments began as early as the 1960s using nuclear magnetic resonance analysis[24-26], it is still unclear and controversial. In 2012, a putative 53 kb gene cluster, consisting of genes encoding for polyketide synthase (PKS), fatty acid synthases, dehydrogenase, transporter and regulator, was first reported to be related to the biosynthesis of Monascus pigments in M. ruber M7[27-28]. Recently, the analysis of the genomes of M. pilosus, M. purpureus, and M. ruber via bioinformatics and RT-PCR showed that the MPs gene cluster contains a minimum of 16 genes, include MpigA (nonreducing polyketide synthase, NR-PKS), MpigB (transcription factor), MpigC (dehydrogenase), MpigD (3-O-transacetylase), MpigE (dehydrogenase), MpigF (monoamine oxidase), MpigG (oxidoreductase), MpigH (dehydrogenase), MpigI (transcription factor), MpigJ (fatty acid synthase, α subunit), MpigK (fatty acid synthase, β subunit), MpigL (ankyrin), MpigM (P450-monooxygenase), MpigN/O (monooxygenase), MpigP (unknown function), and MpigQ (transporter)[29]. It's generally accepted hypothesis that the orange pigments are synthesized first and then generated the yellow and red pigments by hydrogenation reaction and amination reaction respectively. Even though many researchers have devoted themselves to studying the MPs biosynthesis pathway, several steps and the identities of related enzymes remain unclear or controversial[30-31].
A number of studies suggest an association between PigE (one of MPs genes) and MPs production. Nevertheless no previous study has investigated the function of PigE gene in the pigment biosynthesis of Monascus purpureus. In this study, the function of PigE gene in the pigment biosynthesis of Monascus purpureus was examined. We constructed gene knockout vector and obtained the △PigE (a PigE gene deletion mutant) based on the homologous recombination principle, followed by analysis of phenotypes, microstructures, growth rates, metabolites and gene expression. The identification and functional characterization of PigE gene will enable more clearly reveal the diversity of Monascus pigments, leading to improved understanding of the potential significances for improving yellow pigments production through the genetic engineering techniques.
1 Materials and methods 1.1 Strains, plasmid, and growth conditionsThe wild-type strain Mp-21 with high yield of Monascus pigments and low production of citrinin and the Agrobacterium tumefaciens AGL-1 strain were used to generate the △PigE in this study. All the gene deleted mutants were selected on potato dextrose agar (PDA) plates containing 50 μg/mL hygromycin B at 28 ℃ for 7 days. For phenotypic characterization, the mycelial mats about 1 cm2 of strain Mp-21 and △PigE were inoculated in PDA, Malt extract agar medium (MEA consisting of malt 2.0%, sucrose 2.0%, peptone 0.1%, agar 2.0%) and Glycerol nitrate agar medium (G25N consisting of NaNO3 0.3%, K2HPO4 0.1%, KCl 0.05%, MgSO4•7H2O 0.05%, yeast extract 0.5%, sucrose 3.0%, glycerol 2.5%, agar 2.0%). The spore suspensions harvested by washing the cultured PDA plates with distilled water were transferred into potato dextrose broth (PDB) for the production of Monascus pigments. For the analysis of citrinin, Monascus strains were inoculated into 100 mL Yeast extract sucrose medium (YES consisting of yeast 4%, sucrose 16%) and incubated at 28 ℃ with continuous shaking at 180 r/min. The plasmid pKD1 with hygromycin B resistance gene (hph) was used for the amplification of resistance selection marker. The plasmid pKO1B showed as Figure 1 was used for the construction of the replacement vector pKOPE.
|
| Figure 1 Overview of the deletion of PigE gene in M. purpureus Mp-21. The deletion strategy based on homologous recombination principle in this study. The homologous recombination mainly created two types of transformants: ectopic insertion mutants and PigE-deleted mutants. |
1.2 DNA extraction and cloning of PigE gene
All the fungal genomic DNA from strain Mp-21 and mutants was isolated from mycelia based on methods in the Omega Fungal DNA kit. For the design of amplification primers, the homologous sequences of pigment metabolism related gene cluster reported in five different Monascus strains were analyzed by the multiple sequences alignment tool in Jellyfish 3.0 software. A pair of primers, PigE-F/PigE-R, was designed for the amplification of PigE gene by Primer Premier 5.0 software (Table 1). The PCR reaction conditions included an initial denaturation for 3 min at 94℃, which was followed by 30 cycles for 30 s at 94℃, 30 s at 50℃, and 90 s at 72 ℃ with a final extension of 10 min at 72 ℃.
| Names | Primer sequences (5′→3′) | Descriptions |
| 5F-F | GGGGTACCCCCGACAGCATCTCCCGTGTTGAAGT | For the amplification of 5′flanking region |
| 5F-R | GCTCCTTCAATATCATCTTCTCTCGCTTTCTTTGGTCGGAGTTATC | |
| 3F-F | TAGAGTAGATGCCGACCGAACAAGAGGAATCCAGTTTCATTAGAG | For the amplification of 3′flanking region |
| 3F-R | GCTCTAGAGCTCTGGCAGTATTTTCGCTTTTCCGC | |
| Phph-F | CGTTATGTTTATCGGCACT | For the amplification of partial hph gene |
| Phph-R | TTGGCGACCTCGTATTGG | Used as probe Phph for Southern blotting |
| hph-F | CGAGAGAAGATGATATTGAAGGAGC | For the amplification of hph gene |
| hph-R | TCTTGTTCGGTCGGCATCTACTCTA | |
| PigE-F | AAAGCACATCTAGGATTTATAG | For the amplification of PigE gene |
| PigE-R | ATTAATCTTCTGGTCAATGCGAAT | |
| GAPDH-F | GTCTATGCGTGTGCCTACTTCC | For the RT-PCR |
| GAPDH-R | GAGTTGAGGGCGATACCAGC | |
| The primers 5F-R and 3F-F were designed with a hph marker tail which showed by a single underline. The sequences underlined by double lines in primers 5F-F and 3F-R represent the restriction sites KpnⅠand XbaⅠrespectively. | ||
1.3 Deletion of the PigE gene
The homologous recombination strategy was applied in this study for the disruption of PigE gene in strain Mp-21. A gene deletion cassette contained 5′-flanking region, 3′-flanking region and hph selectable marker gene was designed by the double-joint PCR. In the first round of PCR, the 694 bp 5′-flanking region (5′-FR) and 752 bp 3′-flanking region (3′-FR) of PigE gene were amplified and served as DNA homologous sequences for recombination event with primers 5F-F and 5F-R, and primers 3F-F and 3F-R, respectively (Table 1). A 1415 bp hph marker gene was amplified from plasmid pKD1 with the primers hph-F and hph-R (Table 1). The three PCR amplifications were all purified using a PCR purification kit and mixed at 1:3:1 (5′-flanking region:hph:3′-flanking region) molar ratio for the second round fusion reaction. The fusion reaction conditions were denaturation at 98 ℃ for 4 min; then 15 cycles consisting of denaturation at 98 ℃ for 30 s, annealing at 55 ℃ for 10 min and extension at 72 ℃ for 4 min; and finally a single extension at 72 ℃ for 10 min. The third round PCR reaction used the product of previous round fusion reaction as template and amplified with primers 5F-F and 3F-R for the construction of PigE-deleted cassette (Figure 2-A).
|
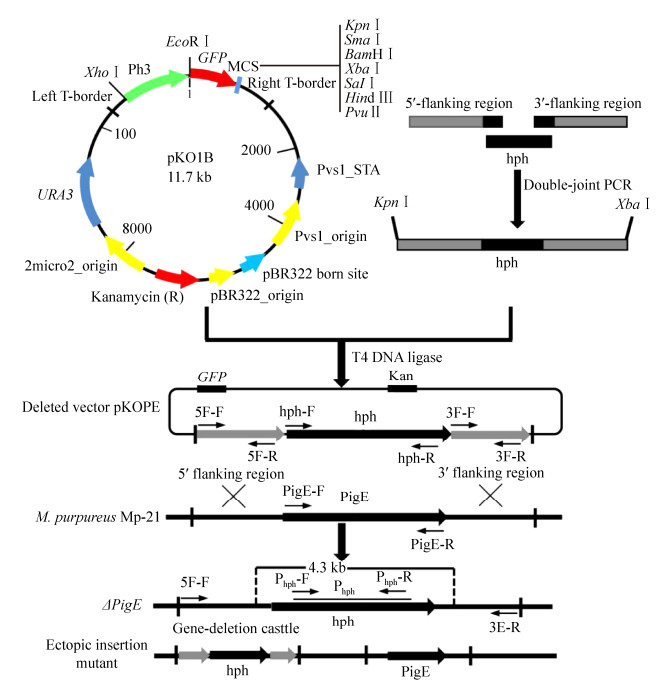
| Figure 2 The verification of PigE deletion mutant (△PigE). A: The construction of PigE deletion cassette. Lane 1: 5′-flanking regions; lane 2: 3′-flanking regions; lane 3: hph gene; lane 4: double-joint PCR production. B: PCR testing for the confirmation of △PigE by six pairs of primers. Lane 1: wild-type strain Mp-21; lane 2: △PigE; lane 3: ectopic insertion mutant. C: Southern blotting analysis of the supposed △PigE by hph probe. Lane 1: wild-type strain Mp-21; lane 2: positive plasmid pKD1; lane 3: △PigE. D: Gene expression in wild-type strain Mp-21 and △PigE determined by RT-PCR. |
After that, the PigE-deleted cassette constructed before and plasmid pKO1B were digested with restriction enzymes KpnⅠand XbaⅠ at 37 ℃ for 3 h. The restriction fragment was inserted into the corresponding sites in pKO1B by T4 DNA ligase to form the replacement vector pKOPE. The Agrobacterium tumefaciens AGL-1 with plasmid pKOPE was constructed by a freeze-thaw method and incubated for transformation at 28 ℃ for 3 d. Agrobacterium tumefaciens-mediated transformation (ATMT) was performed as described by Yang et al.[32] with small modifications. To enhance the transformation efficiency, 1×105 spores of strain Mp-21 were co-cultured with the activated AGL-1 strain in the inducing medium with 0.01% FeSO4 10 μL, 100 g/L MES 10 μL, 0.1 mol/L AS 8 μL. The transformants obtained in the PDA plates containing 50 μg/mL hph were stored in 25% glycerol at -80 ℃ after continuous passage culture.
1.4 PCR testing and Southern hybridization analysisSix pairs of primers, including hph-F/hph-R, PigE-F/PigE-R, 5F-F/3F-R, 5F-F/hph-R, PigE-F/3F-R and 5F-F/PigE-R were used to select and verify the △PigE from the Monascus transformant library (Table 2). For Southern hybridizations, the genomic DNA of strain Mp-21 and supposed △PigE were digested with the BamHⅠrestriction enzyme respectively. The hph fragment probe was amplified from plasmid pKD1 by primers hph-F and hph-R. The experimental methods of southern blot were performed based on the DIG-High Prime DNA Labeling﹠Detection Starter kit Ⅰ (Roche, Mannhein, Germany). The plasmid pKD1 was also digested with the XhoⅠrestriction enzyme as a positive control.
| Primers | Amplified fragments | Length of amplified fragments/bp | ||
| Mp-21 | Ectopic insertion mutant | △PigE | ||
| hph-F/hph-R | hph | 0 | 1415 | 1415 |
| PigE-F/PigE-R | PigE | 1259 | 1259 | 0 |
| 5F-F/3F-R | 5′-FR+hph/PigE+3′-FR | 2705 | 2705 | 2861 |
| 5F-F/hph-R | 5′-FR+hph | 0 | 2109 | 2109 |
| PigE-F/3F-R | PigE+3′-FR | 2011 | 2011 | 0 |
| 5F-F/PigE-R | 5′-FR+PigE | 1953 | 1953 | 0 |
1.5 RT-PCR analysis
The total RNA was extracted from strain Mp-21 and △PigE using TaKaRa RNAiso Plus total RNA kit followed the manufacturer's instructions. The extracted RNA was examined by 1% agarose gel and reverse transcribed into cDNA by Prime Script Reverse Transcriptase. Two pairs of primers, PigE-F/PigE-R and GAPDH-F/GAPDH-R, were used for the amplification of PigE gene and actin GAPDH gene respectively. The reverse-transcription PCR conditions of GAPDH gene were 94 ℃ for 3 min, followed by 30 cycles of 94 ℃ for 30 s, 60 ℃ for 30 s and 72 ℃ for 1 min, and a final extension at 72 ℃ for 10 min.
1.6 Analysis of colony phenotypes and growth rateFor analysis of phenotypic characterization, the wide-type strain Mp-21 and △PigE were incubated in three different culture medium (PDA, MA, G25N) at 28 ℃ for 7 d. The colony diameters of wide-type M. purpureus Mp-21 and △PigE were measured every other day from 4 d to 7 d of cultivation in PDA medium at 30. The same amount of mycelium of wide-type strain Mp-21 and △PigE were punched and inoculated in 250 mL Erlenmeyer flasks containing 100 mL PDB medium at 30 ℃ with 180 r/min. The mycelia filtered from fermentation liquid with gauze were collected and vacuum freeze-dried to measure mycelial dry weight.
1.7 Determination of Monascus pigmentsThe Monascus pigments of the strain Mp-21 and △PigE incubated in three kinds of medium, PDB, MA, G25N, were extracted with 70% (V/V) ethanol and analyzed by a scanning UV-vis spectrophotometer UV-1700 (Shimadzu, Tokyo, Japan) from 300 nm to 600 nm. Thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC) were operated to further analyze the changes of pigment species in △PigE. The pigment extracts of mycelia in PDB were applied on a silica gel plate and separated at room temperature with toluene: ethyl acetate: formic acid of 7:3:1 (V/V/V). The detection of pigments by HPLC was operated as follows: The pigment extracts filtered by 0.45 μm filters were measured in a Waters system (Waters, Milford, MA, USA) fitted with a reverse-phase C18 column (4.6 mm×250 mm, 5 μm). The mobile phase was consisted of methanol (A) and water (B) which was acidified to pH 2.5 with formic acid at 50:50 (V/V) in the beginning. The system was run with a gradient program. The solvent A was increased from 50% to 80% (V/V) over in 75 min, while the solvent B decreased from 50% to 20% (V/V). The column was maintained at 30 ℃ and the flow rate was 0.8 mL/min. Absorbancies were monitored at 510 nm for red pigments and 370 nm for yellow pigments.
The freeze dried mycelia (0.5 g) were dissolved in 10 mL 70% ethanol and incubated at 60 ℃ for 1 h. Then the mixture was centrifuged at 10000 r/min for 10 min and collected supernatant for detection of pigments production. The Monascus pigments extracted from mycelia in PDB were diluted and measured by spectrophotometer at the absorbance of 510 nm and 370 nm which are the maximal absorption of the red and yellow pigments, with 70% ethanol as negative control.
1.8 Detection of citrininCritinin content was measured by HPLC based on the method described by He et al.[33] with some modifications. The wide-type strain Mp-21 and △PigE were cultured in YES medium at 30 ℃ with 180 r/min for 5-13 days. The fermentation filtrate was extracted with equal volume of toluene/ethylacetate/formic acid (7:3:1, V/V/V) to obtain citrinin. Then the extract solution containing citrinin and mobile phase were all passed through the 0.45 μm filter before HPLC detecting. HPLC used a reverse-phase C18 column (4.6 mm×250 mm, 5 μm) at 30. The mobile phase was 75% (V/V) acetonitrile and 25% (V/V) water (pH 2.5, adjusted with orthophosphoric acid), running at 1 mL/min. Ultraviolet absorbance was detected with 2487 UV/Vis Detector (Waters, Milford, MA, USA) at 310 nm wavelength. The citrinin standard solutions with different concentrations were also prepared and detected to draw the standard curve of citrinin.
2 Results 2.1 Sequences analysis of PigEA PigE gene fragment of 1199 bp in length was cloned from genomic DNA of strain Mp-21. The prediction for PigE translation sequence using the GeneMark program (http://exon.gatech.edu/GeneMark/) revealed the putative PigE gene just consisted of a 1029-nt-long open reading frame (ORF) which encoded a protein of 342 amino acids. BLAST searches showed that the PigE gene shared remarkably similar to the pigment biosynthetic gene cluster in Monascus pilosus (93%, KC148521.1) and Mpafr gene which encoded aflatoxin aldehyde reductase in Monacus pilosus (93%, AB206475.1).
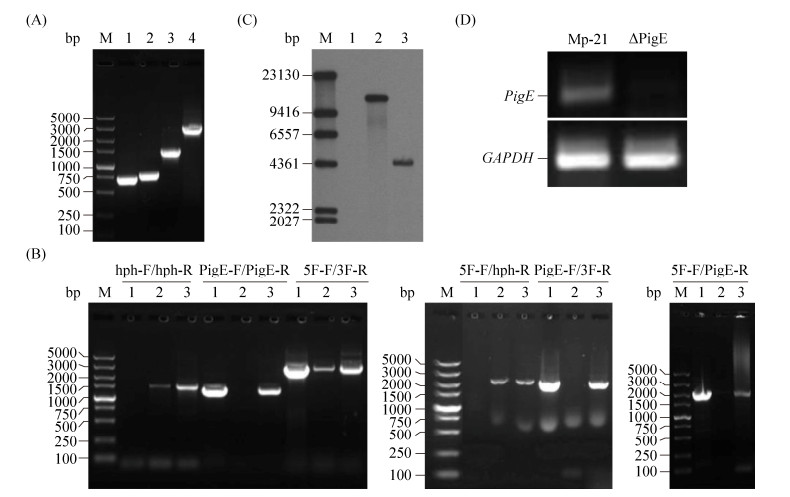
2.2 Selection and verification of △PigEThe △PigE was identified from the transformant library which contained 116 Monascus transformants by incubating on the hph resistance plates, microscopic observation, PCR and Southern hybridization. For the analysis of genetic stability, all the transformants stored before were subcultured for three generations on hph resistance plates and the mutants whose characters changed were discarded (Figure 3-B). Green fluorescent was observed in the mycelia and cleistothecia of △PigE (Figure 3-C). The genomic DNA of transformants was extracted and used as templates for PCR to confirm that the PigE had been replaced by hph successfully (Figure 2-B). The PCR results showed that a 1.4-kb hph gene fragment could be amplified in both putative △PigE and ectopic insertion mutant by primers hph-F and hph-R. The PigE gene was still existed in the genomic DNA of the ectopic insertion mutant, whereas no DNA band was amplified in putative △PigE with primers PigE-F and PigE-R. A 2.8-kb PigE-deleted cassette amplified by primers 5F-F/3F-R in △PigE further proved the homologous events. A probe corresponding to the hph gene coding sequences was designed to verify the single copy of hph in △PigE. Probe Phph yield a 4.3-kb single hybridizing band in a Southern blotting of BamHⅠ-digested genomic DNA of △PigE, compared with no band in wild-type strain Mp-21(Figure 2-C). The PCR and Southern hybridization analysis results all verified the similar conclusion that the PigE gene was deleted in putative △PigE successfully.
|
| Figure 3 The characteristics of M. purpureus Mp-21 transformants. A: The construction of M. purpureus Mp-21 transformant library by ATMT; B: Colony morphology of four M. purpureus Mp-21 transformants; C: The microstructure characteristics of M. purpureus Mp-21 transformants with GFP in confocal laser scanning microscope. |
2.3 Expression analysis of PigE
In this study, the transcription level of PigE in wide-type strain Mp-21 and △PigE was analyzed to investigate the relationship between pigments changes and PigE transcription by RT-PCR. As showed in Figure 2-D, the expression of PigE gene could not be detected in △PigE because of the deletion of PigE gene, while a low level of expression relatively compared with the reference gene GAPDH in the wide-type strain Mp-21. The changes of MPs caused by the little expression of PigE in △PigE suggested that it may be involved in the pigment biosynthetic pathway in Monascus.
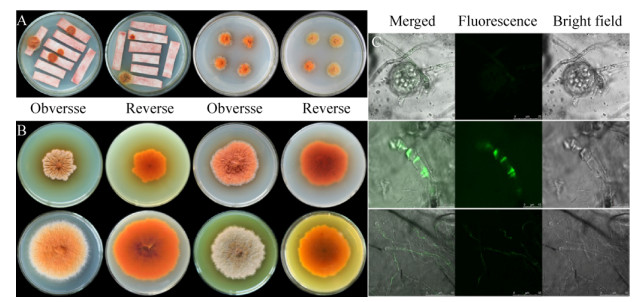
2.4 Characterization and growth rate analysisColony and hyphal phenotypes were observed on different media to investigate the influences of the deletion of PigE in △PigE. Results showed that the colony morphology of △PigE changed obviously, whereas the microstructure characteristics had not significant changes on mycelia, conidia and cleistothecia (Figure 4-A, 4-B). Interestingly, the colors of △PigE colonies on PDA and MA plates were yellow, which were obviously different from the colonies of the wide-type strain Mp-21 in red. This color variation of colony provided a reference for the analysis of Monascus pigments in △PigE. Additionally, compared with the wide-type strain Mp-21, the △PigE produced very few hyphae and was not suitable for growth in MA and G25N media. The determination results of colony diameters and mycelial dry weight showed in Figure 4-C and 4-D supported that the biomass of △PigE changed little in the earlier growth phase, while the decrease was slower compared after culturing for 7 d, with the wide-type strain Mp-21. The △PigE needed more time to adapt to the growing conditions in the earlier growth phase.
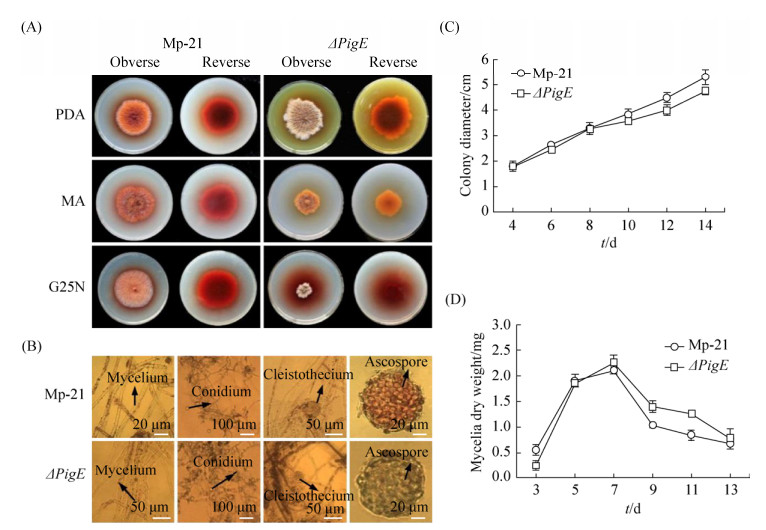
|
| Figure 4 Growth characterization of the wild-type strain Mp-21 and △PigE. A: Colony morphology of the wild-type strain Mp-21 and △PigE incublated at 30 ℃ for 7 d on PDA, MA and G25N; B: Comparison of the wild-type strain Mp-21 and △PigE in microscopic morphological characteristics; C: The colony diameter of the wild-type strain Mp-21 and △PigE incublated on PDA plates at 30 ℃ for different days; D: The mycelial dry weight of the wild-type strain Mp-21 and △PigE on PDB media at 30 ℃ for different days. |
2.5 Analysis of pigments
The pigments extracted from the mycelia of strain Mp-21 and △PigE cultured in three kinds of media (PDB, MA, G25N) were used to analyze the pigment variations (Figure 5-A). Classically, MPs were classified according to OD value: yellow (330-450) orange (460-480) red (490-530). As shown in Figure 5-A, different culture media could lead to obvious changes in MPs yields, which could be further observed from the absorbance value of the extracts of pigments. In the culture medium of PDB, MA and G25N, strain Mp-21 was able to produce three kinds of pigment at the same time (Figure 5-B). Interestingly, no absorbance was detected in the pigment products in the three culture media of △PigE in OD460-530, and the absorbance value in OD330-450 was significantly higher than that of strain Mp-21, which showed that the yellow pigment production of △PigE was significantly higher than that of strain Mp-21, and had lost the ability to produce red pigments. According to the TLC preliminary analysis results, the strain Mp-21 produced an MP complex mixture including two categories of MP compounds (seven kinds of red and two yellow), while the △PigE only yielded yellow pigments (Figure 5-C, YP1 and YP2) without red pigments. The HPLC analysis results further confirmed that the △PigE lost the ability to produce red pigments (Figure 5-D, I a) and yield at least five new yellow pigment compounds (Figure 5-D, Ⅱb).
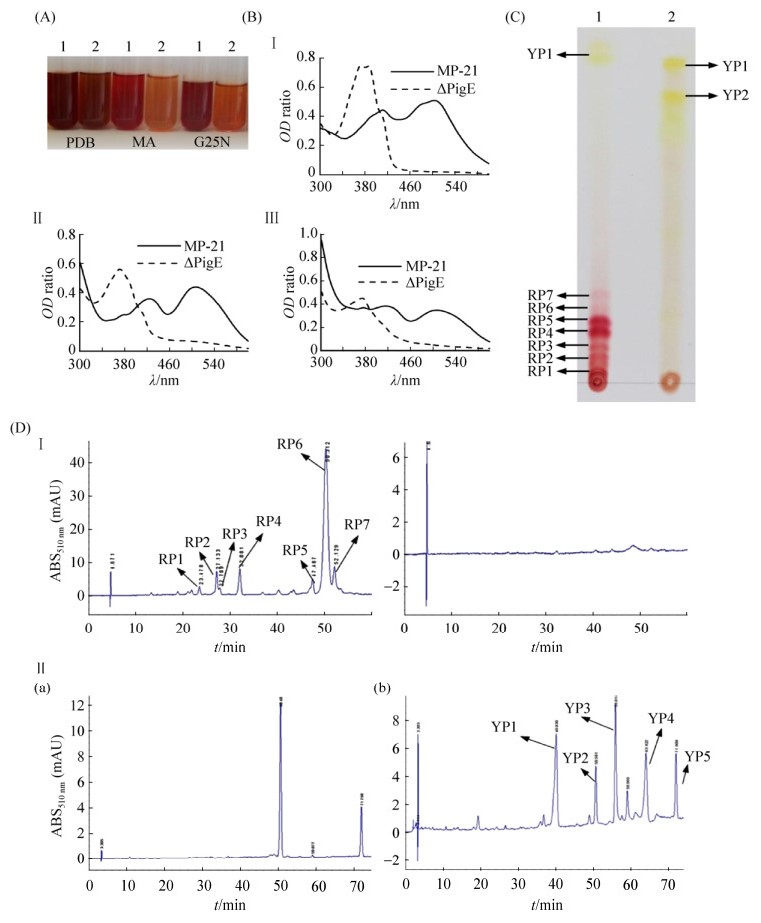
|
| Figure 5 Analysis of Monascus pigments in the wild-type strain Mp-21 and △PigE. A: The Monascus pigments extract from PDB, MA, G25N media respectively in the wild-type strain Mp-21 and △PigE. B: The full wavelength scanning of Monascus pigment extracts from PDB, MA, G25N media respectively in the wild-type strain Mp-21 and △PigE. C: The analysis of Monascus pigment extracts from PDB by TLC. lane 1: strain Mp-21, lane 2: △PigE. D The analysis of pigment varieties by HPLC. a: wild-type strain Mp-21, b: △PigE. |
In this study, the pigment production in strain Mp-21 and △PigE was detected from 5 d to 13 d. The pigments production of △PigE was always much higher and up to 3548.2 U/g after cultivation for 13 d which was about 4.82 times higher than that in the wild-type strain Mp-21 (Figure 6-A).
2.6 Analysis of citrinin productionCitrinin is known for its nephrotoxic activity in mammals, the secretion of citrinin usually accompanies the biosynthesis of MPs[34]. The difference in citrinin production between the wild-type strain Mp-21 and △PigE was detected by HPLC. The citrinin production in △PigE was 1.57 mg/L after fermentation for 13 d in YES medium, which was similar to that of strain Mp-21. Interestingly, The citrinin was produced after culturing for 7 d in △PigE, which was delayed compared with the wild-type strain Mp-21 (Figure 6-B). This phenomenon may be related to the fact that △PigE needed to be longer to adapt to the growth conditions.
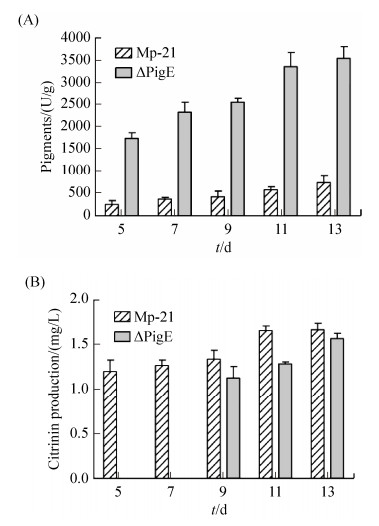
|
| Figure 6 Analysis the productions of pigment and citrinin in the wild-type strain Mp-21 and △PigE. Pigments (A) and citrinin (B) in the wild-type strain M-21 and △PigE after culturing different days. |
3 Discussion
As mentioned above, our finding that the deletion of PigE gene mainly upregulates the varieties and yields of yellow pigments in strain Mp-21 confirms the significant role of PigE gene in pigment biosynthesis. Sequences analysis showed that the PigE gene also encodes an oxidoreductase. In light of the putative biosynthetic pathway of Monascus pigments put forward before, the chemical modification of orange pigments to generate red ones through an aminophilic reaction between orange Monascus pigments and primary amine[35-36]. On the other hand, there may be some oxidoreduction conversion of the polyketide chromophores between yellow and orange pigments or a direct conversion between yellow and orange pigments[20, 37]. The orange pigments monascorubrin and rubropunctatin could be reduced to the yellow pigments ankafavin and monascin, respectively[37]. Our data were consistent with the conclusion that the PigE gene similar to mppC gene may be involved in the conversion of orange and yellow pigments[38]. Unlike the mppE gene encoded a reductive enzyme which controls the conversion reaction of the orange pigments to yellow pigments, the PigE gene catalyzes the reverse reaction[39]. The deletion of PigE blocked the transformation pathway of the yellow pigments to orange pigments, which made the △PigE more favorable to form yellow pigments. As the formation of red pigments requires more complex conditions such as amino acids and suitable pH in culture medium, The △PigE prefers to synthesize yellow pigments first and lost the ability to produce red pigments.
The disruption of PigE had very little effects onto citrinin production, which indicated that the PigE gene was not involved in the citrinin biosynthesis. This study suggests the PigE gene was closely related to the formation of yellow pigments, and it provides a certain contribution to the construction of M. purpureus genetic engineering strain with high yield of yellow pigments.
Acknowledgements
We thank prof. Zhang Chulong (Zhejiang University) for the gift of plasmid vectors used in this study.
| [1] | Dufossé L, Galaup P, Yaron A, Arad SM, Blanc P, Murthy KNC, Ravishankar GA. Microorganisms and microalgae as sources of pigments for food use: a scientific oddity or an industrial reality?. Trends in Food Science & Technology, 2005, 16(9): 389-406. |
| [2] | Shi YC, Pan TM. Beneficial effects of Monascus purpureus NTU 568-fermented products: a review. Applied Microbiology and Biotechnology, 2011, 90(4): 1207-1217. DOI:10.1007/s00253-011-3202-x |
| [3] | Feng YL, Shao YC, Chen FS. Monascus pigments. Applied Microbiology and Biotechnology, 2012, 96(6): 1421-1440. DOI:10.1007/s00253-012-4504-3 |
| [4] | Vendruscolo F, Schmidell W, Moritz DE, Bühler RMM, de Oliveira D, Ninow JL. Isoelectric point of amino acid: importance for Monascus pigment production. Biocatalysis and Agricultural Biotechnology, 2016, 5: 179-185. DOI:10.1016/j.bcab.2015.12.006 |
| [5] | Chen WP, He Y, Zhou YX, Shao YC, Feng YL, Li M, Chen FS. Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics. Comprehensive Reviews in Food Science and Food Safety, 2015, 14(5): 555-567. DOI:10.1111/1541-4337.12145 |
| [6] | Endo A. Monacolin K, a new hypocholesterolemic agent produced by a Monascus species. Journal of Antibiotics, 1979, 32(8): 852-854. DOI:10.7164/antibiotics.32.852 |
| [7] | Jiang DH, Ji H, Ye Y, Hou JH. Studies on screening of higher γ-aminobutyric acid-producing Monascus and optimization of fermentative parameters. European Food Research and Technology, 2011, 232(3): 541-547. DOI:10.1007/s00217-010-1413-5 |
| [8] | Patakova P. Monascus secondary metabolites: production and biological activity. Journal of Industrial Microbiology & Biotechnology, 2013, 40(2): 169-181. |
| [9] | Kang BY, Zhang XH, Wu ZQ, Qi HS, Wang ZL. Effect of pH and nonionic surfactant on profile of intracellular and extracellular Monascus pigments. Process Biochemistry, 2013, 48(5/6): 759-767. |
| [10] | Shi K, Chen G, Pistolozzi M, Xia FG, Wu ZQ. Improved analysis of Monascus pigments based on their pH-sensitive UV-vis absorption and reactivity properties. Food Additives & Contaminants: Part A, 2016, 33(9): 1396-1401. |
| [11] | Jongrungruangchok S, Kittakoop P, Yongsmith B, Bavovada R, Tanasupawat S, Lartpornmatulee N, Thebtaranonth Y. Azaphilone pigments from a yellow mutant of the fungus Monascus kaoliang. Phytochemistry, 2004, 65(18): 2569-2575. DOI:10.1016/j.phytochem.2004.08.032 |
| [12] | Zheng YQ, Xin YW, Guo YH. Study on the fingerprint profile of Monascus products with HPLC-FD, PAD and MS. Food Chemistry, 2009, 113(2): 705-711. DOI:10.1016/j.foodchem.2008.07.105 |
| [13] | Hsu LC, Hsu YW, Liang YH, Kuo YH, Pan TM. Anti-tumor and anti-inflammatory properties of ankaflavin and monaphilone A from Monascus purpureus NTU 568. Journal of Agricultural and Food Chemistry, 2011, 59(4): 1124-1130. DOI:10.1021/jf103652n |
| [14] | Shi YC, Liao VHC, Pan TM. Monascin from red mold dioscorea as a novel antidiabetic and antioxidative stress agent in rats and Caenorhabditis elegans. Free Radical Biology and Medicine, 2012, 52(1): 109-117. DOI:10.1016/j.freeradbiomed.2011.09.034 |
| [15] | Izawa S, Harada N, Watanabe T, Kotokawa N, Yamamoto A, Hayatsu H. Arimoto-Kobayashi S. Inhibitory effects of food-coloring agents derived from Monascus on the mutagenicity of heterocyclic amines. Journal of Agricultural and Food Chemistry, 1997, 45(10): 3980-3984. DOI:10.1021/jf9703821 |
| [16] | Akihisa T, Tokuda H, Ukiya M, Kiyota A, Yasukawa K, Sakamoto N, Kimura Y, Suzuki T, Takayasu J, Nishino H. Anti-tumor-initiating effects of monascin, an azaphilonoid pigment from the extract of Monascus pilosus fermented rice (red-mold rice). Chemistry & Biodiversity, 2005, 2(10): 1305-1309. |
| [17] | Mapari SAS, Thrane U, Meyer AS. Fungal polyketide azaphilone pigments as future natural food colorants?. Cell, 2010, 28(6): 300-307. |
| [18] | Klinsupa W, Phansiri S, Thongpradis P, Yongsmith B, Pothiratana C. Enhancement of yellow pigment production by intraspecific protoplast fusion of Monascus spp. yellow mutant (ade-) and white mutant (prototroph). Journal of Biotechnology, 2016, 217: 62-71. DOI:10.1016/j.jbiotec.2015.11.002 |
| [19] | Hajjaj H, Goma G, François JM. Effect of the cultivation mode on red pigments production from Monascus ruber. International Journal of Food Science and Technology, 2015, 50(8): 1731-1736. DOI:10.1111/ijfs.12803 |
| [20] | Huang T, Wang MH, Shi K, Chen G, Tian XF, Wu ZQ. Metabolism and secretion of yellow pigment under high glucose stress with Monascus ruber. AMB Express, 2017, 7: 79. DOI:10.1186/s13568-017-0382-5 |
| [21] | Xu MJ, Yang ZL, Liang ZZ, Zhou SN. Construction of a Monascus purpureus mutant showing lower citrinin and higher pigment production by replacement of ctnA with pks1 without using vector and resistance gene. Journal of Agricultural and Food Chemistry, 2009, 57(20): 9764-9768. DOI:10.1021/jf9023504 |
| [22] | Liu QP, Cai L, Shao YC, Zhou YX, Li M, Wang XH, Chen FS. Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biology, 2016, 120(3): 297-305. DOI:10.1016/j.funbio.2015.10.008 |
| [23] | Yang YS, Li L, Li X, Shao YC, Chen FS. mrflbA, encoding a putative FlbA, is involved in aerial hyphal development and secondary metabolite production in Monascus ruber M-7. Fungal Biology, 2012, 116(2): 225-233. DOI:10.1016/j.funbio.2011.11.005 |
| [24] | Birch AJ, Cassera A, Fitton P, Holker JSE, Smith H, Thompson GA, Whalley WB. Studies in relation to biosynthesis. Part XXX. Rotiorin, monascin, and rubropunctatin. Journal of the Chemical Society, 1962, 699: 3583-3586. |
| [25] | Kurono M, Nakanishi K, Shindo K, Tada M. Biosynthesis of monascorubrin and monascoflavin. Chemical and Pharmaceutical Bulletin, 1963, 11(3): 359-362. DOI:10.1248/cpb.11.359 |
| [26] | Hadfield JR, Holker JSE, Stanway DN. The biosynthesis of fungal metabolites. Part Ⅱ. The β-oxo-lactone equivalents in rubropunctatin and monascorubrin. Journal of the Chemical Society C: Organic, 1967, 1967: 751-755. |
| [27] | Shao YC, Ding YD, Zhao Y, Yang S, Xie BJ, Chen FS. Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World Journal of Microbiology and Biotechnology, 2009, 25(6): 989-995. DOI:10.1007/s11274-009-9977-6 |
| [28] | Shao YC, Xu L, Chen FS. Genetic diversity analysis of Monascus strains using SRAP and ISSR markers. Mycoscience, 2011, 52(4): 224-233. DOI:10.1007/S10267-010-0087-Y |
| [29] | Liang B, Du XJ, Li P, Sun CC, Wang S. MptriA, an acetyltransferase gene involved in pigment biosynthesis in M. purpureus YY-1. Journal of Agricultural and Food Chemistry, 2018, 66(16): 4129-4138. DOI:10.1021/acs.jafc.8b00661 |
| [30] | Yang Y, Liu B, Du XJ, Li P, Liang B, Cheng XZ, Du LC, Huang D, Wang L, Wang S. Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Scientific Reports, 2015, 5: 8331. DOI:10.1038/srep08331 |
| [31] | Liu QP, Xie NN, He Y, Wang L, Shao YC, Zhao HZ, Chen FS. MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Applied Microbiology and Biotechnology, 2014, 98(1): 285-296. DOI:10.1007/s00253-013-5289-8 |
| [32] | Yang YJ, Lee I. Agrobactrium tumefaciens-mediated transformation of Monascus ruber. Journal of Microbiology and Biotechnology, 2008, 18(4): 754-758. |
| [33] | He Y, Liu QP, Shao YC, Chen FS. ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M7. Applied Microbiology and Biotechnology, 2013, 97(11): 4965-4976. DOI:10.1007/s00253-013-4851-8 |
| [34] | Shimizu T, Kinoshita H, Ishihara S, Sakai K, Nagai S, Nihira T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Applied and Environmental Microbiology, 2005, 71(7): 3453-3457. DOI:10.1128/AEM.71.7.3453-3457.2005 |
| [35] | Jung H, Kim C, Kim K, Shin CS. Color characteristics of Monascus pigments derived by fermentation with various amino acids. Journal of Agricultural and Food Chemistry, 2003, 51(5): 1302-1306. DOI:10.1021/jf0209387 |
| [36] | Xiong X, Zhang XH, Wu ZQ, Wang ZL. Coupled aminophilic reaction and directed metabolic channeling to red Monascus pigments by extractive fermentation in nonionic surfactant micelle aqueous solution. Process Biochemistry, 2015, 50(2): 180-187. DOI:10.1016/j.procbio.2014.12.002 |
| [37] | Hajjaj H, Blanc P, Groussac E, Uribelarrea JL, Goma G, Loubiere P. Kinetic analysis of red pigment and citrinin production by Monascus ruber as a function of organic acid accumulation. Enzyme and Microbial Technology, 2000, 27(8): 619-625. DOI:10.1016/S0141-0229(00)00260-X |
| [38] | Balakrishnan B, Karki S, Chiu SH, Kim HJ, Suh JW, Nam B, Yoo YM, Chen CC, Kwon HJ. Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Applied Microbiology and Biotechnology, 2013, 97(14): 6337-6345. DOI:10.1007/s00253-013-4745-9 |
| [39] | Balakrishnan B, Park SH, Kwon HJ. A reductase gene mppE controls yellow component production in azaphilone polyketide pathway of Monascus. Biotechnology Letters, 2017, 39(1): 163-169. |
 2019, Vol. 59
2019, Vol. 59