中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 范成莉. 2019
- Chengli Fan. 2019
- 新生隐球菌一个新的产孢相关蛋白Srp1的鉴定与功能分析
- Identification and characterization of a novel sporulation-related protein Srp1 in Cryptococcus neoformans
- 微生物学报, 59(7): 1395-1407
- Acta Microbiologica Sinica, 59(7): 1395-1407
-
文章历史
- 收稿日期:2019-01-24
- 修回日期:2019-02-15
- 网络出版日期:2019-03-07
新生隐球菌是自然界广泛存在的具荚膜的环境腐生菌,其担孢子和干燥的酵母细胞可通过呼吸方式进入人体,引起隐球菌性肺炎和脑膜炎[1]。近年来,新生隐球菌感染的发病率逐年增高,在国外隐球菌感染常与艾滋病并发,成为导致患者死亡的首要原因;在我国新生隐球菌感染的发病率也呈逐年增加的态势并展现出能感染免疫健全人群的特点[2]。据最新统计,新生隐球菌每年能引起全球约22万人感染并导致大约18万人死亡[3],已成为威胁现代人们健康的重要杀手。新生隐球菌属于担子菌,有α和a两个交配型,不同交配型的细胞可交配融合并产生担子菌所特有的双核菌丝。双核菌丝进一步特化为担子,其内部的双核经核融合和减数分裂后在担子顶端产生4条串状的担孢子。新生隐球菌作为一种能够快速生长的酵母型病原真菌,具有明确的性周期,已成为真菌遗传和致病性研究的模式生物之一[4-5]。
有性生殖可促进不同交配型菌株之间基因组合的广泛变异,增加子代适应自然选择的能力,促进有利突变在种群中的传播。在新生隐球菌中,目前已有许多参与有性生殖的相关因子和通路被研究。锌指蛋白是一类广泛存在于真核生物中的重要转录因子,在隐球菌的生长发育中起重要作用。锌指蛋白Znf2、Znf3和Zfp1分别被证明能够调控隐球菌的有性生殖过程,因为上述蛋白编码基因分别敲除后都会阻断新生隐球菌的有性生殖过程,使新生隐球菌丧失产生担孢子的能力[6-9]。RNA结合蛋白也被证明参与调控新生隐球菌的有性生殖过程;RNA结合蛋白Csa1和Csa2是介导减数分裂和担子成熟的重要元件,相关基因敲除后突变体丧失产孢能力[10];而另一RNA结合蛋白Rbp1也调控隐球菌的有性生殖过程[11]。群体感应蛋白Qsp1及其通路重要组分锌指调节因子Cqs2也被证明调控新生隐球菌的有性生殖过程[12]。另外,钙调磷酸酶应答蛋白锌指转录因子Crz1也调控新生隐球菌有性产孢过程[13]。蛋白降解通路也被证明调控新生隐球菌的有性生殖。我们课题组前期研究鉴定了泛素蛋白降解通路E3中泛素连接酶的一个关键蛋白F-box蛋白Fbp1,功能分析发现Fbp1调控新生隐球菌的有性生殖和致病性[14]。初步机理分析发现α和a交配型的fbp1Δ突变体交配形成双核菌丝后其减数分裂过程被阻断,从而导致新生隐球菌丧失产孢能力。
Fbp1作为E3泛素连接酶的关键蛋白负责特异性识别底物蛋白并进行泛素化,因此Fbp1可能通过调控其下游底物来调控新生隐球菌的有性生殖过程。本课题组前期通过iTRAQ蛋白质谱技术鉴定了新生隐球菌野生型H99及fbp1Δ突变体背景下的差异蛋白,发现fbp1Δ背景下一高丰度蛋白CNAG_00626。其可能为Fbp1下游底物,可能参与新生隐球菌的有性生殖或致病性过程,因此我们决定对其进行功能分析。本研究通过克隆获得CNAG_00626的序列,并命名为SRP1 (Sporulation-related protein 1)。通过qRT-PCR分析和构建SRP1-mCherry融合菌株分析了SRP1的时空表达特点。利用Split marker策略构建基因敲除片段,经基因枪转化和筛选验证后得到SRP1基因敲除突变体srp1Δ;利用体外无缝克隆技术构建SRP1基因互补质粒,经转化和筛选验证后得到互补菌株srp1Δ::SRP1。通过对比野生型和srp1Δ突变体菌株在毒力和有性产孢等方面的差异,来研究Srp1蛋白在新生隐球菌致病性和有性生殖中的作用。
1 材料和方法 1.1 材料 1.1.1 菌株、质粒与引物: 本研究所采用的隐球菌菌株详见表 1,其中H99和KN99a分别为隐球菌α和a交配型野生型菌株,其余均为野生型菌株基础上构建的菌株。本研究所用质粒和引物分别详见表 1和表 2。| Strains/plasmids | Genotype/properties | Source/reference |
| C. neoformas | ||
| H99 | MATα | Perfect et al., 1993[17] |
| KN99a | MATa | Nielsen et al., 2003[18] |
| TBL101 | MATα Nop1-mCherry::NAT | Fan et al., 2019[6] |
| TBL102 | MATa Nop1-mCherry::NAT | Fan et al., 2019[6] |
| TBL105 | MATα srp1::NEO | In this study |
| TBL138 | MATa srp1::NEO | In this study |
| TBL218 | MATα srp1::NEO SRP1::NAT | In this study |
| TBL219 | MATa srp1::NEO SRP1::NAT | In this study |
| TBL220 | MATα Srp1-mCherry::NAT | In this study |
| TBL221 | MATa Srp1-mCherry::NAT | In this study |
| TBL222 | MATα srp1Δ::NEO Nop1-mCherry::NAT | In this study |
| TBL223 | MATa srp1Δ::NEO Nop1-mCherry::NAT | In this study |
| Plasmids | ||
| pBSK(–) | pBluescript SK(–) | |
| pJAF1 | Ampr plasmid containing NEO marker gene | Fraser et al., 2003[19] |
| pCN19 | Ampr plasmid harboring GFP under histone H3 promoter | Price et al., 2008[20] |
| pTBL1 | Ampr plasmid containing NAT marker gene | Fan et al., 2019[6] |
| pTBL3 | Ampr plasmid harboring mCherry-GPD1 terminator and NAT marker | Fan et al., 2019[6] |
| pTBL131 | Ampr vector for PSRP1-SRP1-mCherry for Srp1 localization | In this study |
| pTBL132 | Ampr vector for PSRP1-SRP1-NAT for complementation | In this study |
1.1.2 培养基: 新生隐球菌基础培养基为YPD培养基,于30 ℃黑暗条件下培养;菌株交配诱导培养基为MS培养基和V8培养基[15];黑色素形成培养基为Niger Seed培养基,荚膜诱导培养基为DME培养基,配制方法如前所述[16]。 1.1.3 主要仪器: 外源DNA转化:采用美国Bio-Rad公司PDS-1000/He基因枪进行转化;细胞破碎:采用美国MP公司Fastprep-24 5G匀浆仪进行破碎;交配菌丝及荧光观察:分别采用Olympus CX41显微镜和Olympus FV1200激光共聚焦显微镜进行观察;荧光定量PCR:采用德国Roche公司的LightCycler®96实时荧光定量PCR仪进行PCR扩增和数据收集。 1.2 SRP1序列及表达模式分析
SRP1基因及蛋白序列获取数据库为FungiDB (Fungal and Oomycete Genomics Resources),使用BLASTP程序对蛋白数据库进行同源性搜索和对比分析。为了研究SRP1基因在隐球菌交配过程中的表达情况,我们采用实时荧光定量PCR技术检测了SRP1在mRNA水平上的变化情况。样品准备及操作方法如前述[14]。SRP1及内源参照基因GAPDH的扩增引物分别为TL537/TL538和TL217/TL218 (详见表 2)。为了分析SRP1基因的表达模式,以野生型菌株H99基因组DNA为模板利用引物TL877/878扩增一包含SRP1基因上游约1.5 kb基因编码序列(无终止密码子)片段,并通过体外同源重组方式克隆到载体pTBL3中得到pTBL131。pTBL131经Apa I酶切后分别转化到两交配型srp1Δ突变体中,经筛选验证后获得Srp1-mCherry融合荧光表达菌株。分别选取α和a交配型Srp1-mCherry融合荧光表达菌株设置交配实验并利用激光共聚焦荧光显微镜(Olympus,FV1200)观察荧光在隐球菌各发育阶段的定位情况。
| Primers | Targeted genes | Sequence (5′→3′) |
| TL217 | GAPDH qPCR F | TGAGAAGGACCCTGCCAACA |
| TL218 | GAPDH qPCR R | ACTCCGGCTTGTAGGCATCAA |
| TL228 | SRP1 KO F1 | GTAGGCGTTGAAGGTGTCGGTGTC |
| TL229 | SRP1 KO R1 | CTGGCCGTCGTTTTACGGAAGGTGGTGGAATGGAATCAAC |
| TL230 | SRP1 KO F2 | GTCATAGCTGTTTCCTGAGTATGGCCCGTTTGCTCTTG |
| TL231 | SRP1 KO R2 | CACGGCGCTTGCTTTCTCTTAC |
| TL232 | SRP1 KO F3 | ACCCCTTGGATCATTGGCTCTGCT |
| TL233 | SRP1 KO R3 | AACAATGGAGAGGAAAAGACGGAG |
| TL234 | SRP1 KO F4 | TTTCCAAATCCCCATCAATCGTG |
| TL537 | SRP1 qPCR F1 | TGCCAATGCCAATGAAACC |
| TL538 | SRP1 qPCR R1 | AACAGGAGATTCGCCCTTC |
| TL564 | SRP1 Comp F | GATATCGAATTCCTGCAGCCCGGGGGATCCCGTGGACTGCTAGGGGTGATAAGGAT (BamH I) |
| TL565 | SRP1 Comp R | CGGTGGCGGCCGCTCTAGAACTAGTGGATCCAGGACCAAAGTTGAACAGGATAGGAG (BamH I) |
| TL877 | SRP1-mCherry F1 | TTAGTAAACTCGCCCAACATGTCTGGATCCTTTCCAAATCCCCATCAATCGTG (BamH I) |
| TL878 | SRP1-mCherry R1 | CTTGCTCACCATTCTAGAACTAGTGGATCCTTCCTGCTTTCCAACAGGAGATTC (BamH I) |
1.3 SRP1基因的敲除与互补
采用两步PCR法构建基因敲除片段,具体方法如前所述[6]。通过PCR初筛和Southern blotting验证获得SRP1基因敲除α交配型突变体α srp1Δ菌株,并通过交配产孢分离验证方式获得a交配型突变体a srp1Δ菌株。为获得srp1Δ突变体互补菌株,我们以野生型菌株H99基因组DNA为模板,利用引物TL564/TL565扩增一长约3.0 kb的SRP1基因互补片段(含1.5 kb SRP1基因启动子、0.85 kb基因编码序列及0.5 kb终止子序列)并克隆到pTBL1载体中构建互补质粒pTBL132。互补质粒经Apa I线性化后转化到α和a srp1Δ突变体中,经抗性筛选、PCR验证和表型测定,最终得到SRP1基因的互补突变体菌株TBL220/TBL221。
1.4 突变体毒力因子测定将野生型H99、突变体srp1Δ及互补菌株srp1Δ::SRP1菌株接种于YPD液体培养基中于30 ℃培养过夜并收集上述菌株酵母细胞,经ddH2O洗涤和浓缩后分别滴于Niger seed和DME培养基上,于37 ℃黑暗培养2 d,分别检测上述隐球菌黑色素和荚膜形成情况;同时,上述菌株经ddH2O洗涤后调整OD600至2.0,经过一系列10倍稀释后滴于含有不同应激条件的平板上,黑暗培养2–3 d,观察菌株生长情况。
1.5 致病性分析收集30 ℃过夜培养的上述三种菌株酵母细胞,经PBS洗过后调整细胞浓度至2×106 CFU/mL。通过滴鼻感染方式分别将50 μL上述菌株酵母细胞接种至8周龄大C57BL/6小鼠中,具体接种方法及小鼠发病情况判定标准参见Liu等[14]。菌株致病性统计及小鼠存活曲线绘制均采用PRISM 7.0 (GraphPad Software,San Diego,CA)软件进行分析。组织载菌量分析:分离处死小鼠的脑肺脾取其中一半进行匀浆和稀释,取100 μL稀释后的匀浆液涂布于YPD平板(含有青霉素和氯霉素)并置于30 ℃培养,2 d后统计酵母菌落数并计算组织载菌量(CFU/g新鲜器官,Log10);剩余器官经10%福尔马林固定后送往武汉赛维尔生物科技有限公司进行组织切片分析。
1.6 菌株的交配分析分别将过夜培养的新生隐球菌两个不同交配型的野生型(H99 × KN99a)、srp1Δ突变体(α×a)及互补菌株srp1Δ::SRP1(α×a)细胞等量混合后滴于MS培养基上并于25 ℃黑暗诱导培养14 d,用Olympus CX41显微镜观察并记录隐球菌交配菌丝和担孢子形成情况。为了检测srp1Δ突变体菌株交配过程中细胞核变化情况,将含有Nop1-mCherry融合蛋白的表达载体pTBL72[6]分别转化到α和a srp1Δ突变体中,经筛选得到具有细胞核定位的荧光菌株,用于后续的交配实验和荧光观察。
1.7 数据分析方法采用PRISM 7.0软件的Log-rank (Mantel-Cox) test过程对隐球菌接种小鼠后致病性进行差异性分析;采用nonparametric Mann-Whitney test过程对组织载菌量和荚膜大小进行差异性分析(P < 0.01视为差异显著)。
2 结果和分析 2.1 Srp1序列分析根据新生隐球菌H99基因组序列经PCR扩增得到SRP1(CNAG_00626)基因,序列分析显示该基因全长853 bp,含4个外显子(图 1-A);其cDNA全长675 bp,编码一个由224个氨基酸组成的蛋白(图 1-B),分子量为23.6 kDa。然而当我们对该蛋白序列进行保守结构域预测时发现目前没有任何结构域与之匹配;同时我们又通过BLASTP程序对该蛋白进行同源搜索,结果显示与该蛋白同源性较高的蛋白均为假设蛋白(表 3),表明该蛋白可能还未被研究过,因此有必要对其功能进行研究。
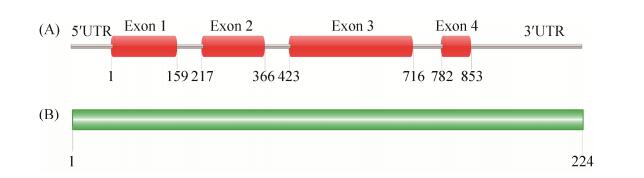
|
| 图 1 产孢相关蛋白Srp1基因及蛋白序列分析 Figure 1 Gene and protein sequence analysis of the sporulation-related protein Srp1. A: The sequence structure of the sporulation-related protein gene SRP1; B: Sequence of the sporulation-related protein Srp1. Non-conserved domains were found in Srp1. |
| Accession | Description | Query cover/% | E-value | Identity/% |
| XP_012046847 | Srp1, Cryptococcus neoformans var. grubii H99 | 100 | 7e–158 | 100 |
| XP_566960 | Hypothetical protein, Cryptococcus neoformans var. neoformans JEC21 | 100 | 5e–130 | 91 |
| KGB75464 | Hypothetical protein, Cryptococcus gattii VGII R265 | 99 | 4e–109 | 81 |
| ODN89942 | Hypothetical protein, Cryptococcus depauperatus CBS 7841 | 99 | 7e–59 | 49 |
| XP_018996930 | Hypothetical protein, Cryptococcus amylolentus CBS 6039 | 100 | 3e–45 | 46 |
| XP_019032557 | Hypothetical protein, Tsuchiyaea wingfieldii CBS 7118 | 70 | 3e–44 | 56 |
| RSH85290 | Hypothetical protein, Saitozyma podzolica | 93 | 1e–07 | 31 |
| PPQ76540 | Hypothetical protein, Gymnopilus dilepis | 87 | 0.005 | 31 |
| KIK43939 | Hypothetical protein, Suillus luteus UH-Slu-Lm8-n1 | 67 | 0.002 | 28 |
2.2 SRP1表达模式分析
为了检测SRP1基因在隐球菌各发育阶段的表达情况,我们采用了qRT-PCR对其进行了检测,结果表明SRP1在隐球菌交配过程中呈诱导表达方式(图 2-A),表明SRP1基因可能在隐球菌交配过程中起重要作用。同时我们也构建了PSRP1-SRP1-mCherry融合表达载体pTBL131,经转化和筛选后得到α和a交配型荧光菌株(TBL220和TBL221),荧光观察结果表明Srp1-mCherry融合蛋白定位于隐球菌的细胞质中(图 2-B);为了检测SRP1在隐球菌各个发育阶段的表达情况,我们将TBL220和TBL221交配后在共聚焦显微镜下观察,发现Srp1-mCherry在隐球菌的酵母细胞、双核菌丝、担子以及担孢子阶段都有表达,且定位于细胞质中(图 2-B)。
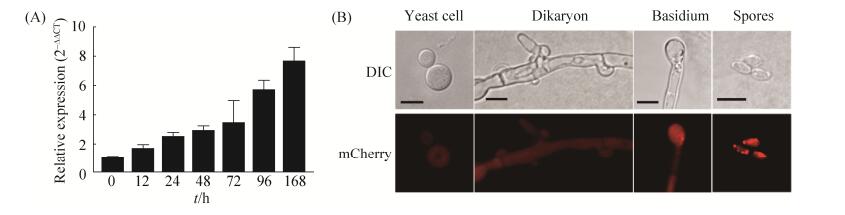
|
| 图 2 SRP1基因的表达模式分析 Figure 2 Expression pattern of the SRP1 gene in C. neoformans. A: Expression of SRP1 during early stages of mating was detected by qRT-PCR. Mating cultures of the wild type strains (H99 × KN99a) were collected from the V8 plates after 12, 24, 48, 72, and 96 h and 7 d incubation for RNA purification and cDNA synthesis. The 2–ΔΔCT method was used to analyze the relative changes in SRP1 expression and GAPDH was used as internal control. The experiment was repeated three times; B: Localization of Srp1-mCherry fusion protein in cryptococcal different developmental stages. The Srp1-mCherry fusion protein was located to the cytoplasm of the yeast cell, mating hyphae, basidium and basidiospores of C. neoformans. Bars, 5 μm. |
2.3 SRP1基因敲除及互补菌株的获得
利用Split marker策略构建SRP1基因敲除片段,通过基因枪将基因敲除片段转化到H99菌株中,经G418(200 mg/L)筛选获得3个转化子,并经Southern blotting验证后确认获得srp1Δ基因敲除突变体菌株(图 3)。构建SRP1基因互补质粒(pTBL132)并通过基因枪转化至srp1Δ突变体中,经NAT抗性筛选、PCR验证和表型测定确认得到4株互补菌株,选取其中1株互补菌株进行后续实验。
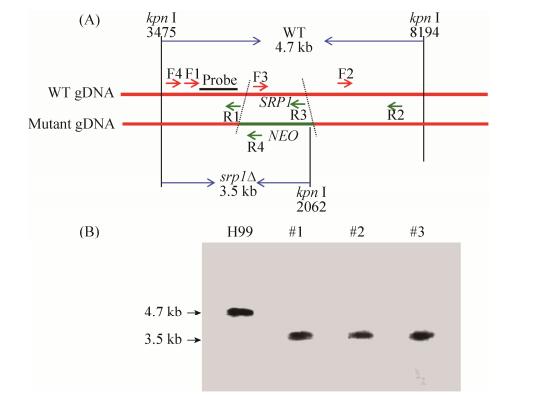
|
| 图 3 SRP1基因敲除突变体的验证 Figure 3 Verification of SRP1 gene knockout mutants. A: The restriction enzyme Kpn I was used to digest the genomic DNAs for Southern blotting; B: Disruption of the wild type SRP1 gene was confirmed by Southern blotting analysis. The Southern blotting was probed with a F1/R1 PCR product specific probe. A fragment of 4.7 kb is present in the wild-type strains while the 3.5 kb band is shown in srp1Δ mutant strains. Number 1–3 are the three G418 resistant transformants used for Southern blotting. |
2.4 SRP1基因敲除不影响隐球菌毒力因子的形成
黑色素、荚膜形成以及能在37 ℃条件下生长是新生隐球菌目前研究较为透彻的经典毒力因子,与新生隐球菌的致病性密切相关。为了研究Srp1在新生隐球菌致病性中的作用,我们首先检测了SRP1基因敲除突变体srp1Δ背景下各毒力因子变化情况。结果发现与野生型菌株H99相比,srp1Δ突变体在Niger seed培养基上能够形成正常的黑色素(图 4-A),在DME培养基上形成正常的荚膜(图 4-B),且在37℃条件生长正常(图 4-C),表明Srp1不参与新生隐球菌上述毒力因子的形成。
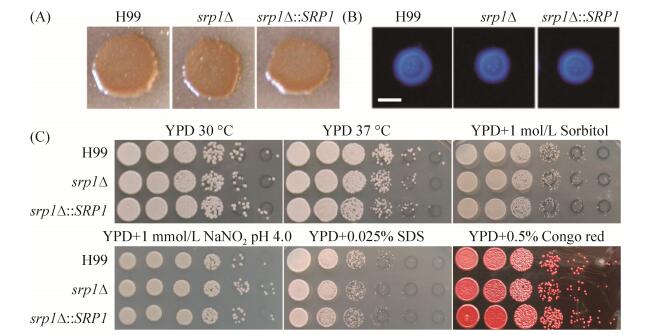
|
| 图 4 SRP1基因敲除不影响新生隐球菌经典毒力因子的形成 Figure 4 Srp1 is dispensable for classical virulence factors development in C. neoformans. A: Melanin production was assayed on Niger seed medium. Melanin levels produced by the strains were observed in photographs after incubation for 2 days at 37 ℃; B: Capsule formation was assayed at 37 ℃ on DME medium. Capsule production was visualized by India ink staining after cells were grown on DME medium for 2 days; Bar, 5 μm. C: Cryptococcal overnight cultures in YPD were diluted to an OD600 of 2.0. Tenfold dilutions were made in water and 5 μL of each dilution was spotted on YPD agar with indicative stresses. The plates were incubated for two to three days at indicated temperature. H99, srp1Δ mutant and srp1Δ complemented strains are indicated on the left and the conditions at the top. |
为了进一步研究Srp1在真菌毒力中的作用,我们又检测了srp1Δ突变体在高渗(YPD+1 mol/L Sorbitol)、硝化应激(YPD+1 mmol/L NaNO2,pH 4.0)、细胞完整性应激(YPD+SDS/Congo red)等条件下的生长情况(图 4-C)。与野生型菌株H99相比,srp1Δ突变体对上述应激条件不敏感,表明Srp1也不调控隐球菌在上述应激条件下生长。
2.5 SRP1敲除不影响隐球菌的致病性由于真菌毒力是一个复杂性状,我们进一步通过C57BL/6小鼠致病性实验检测了Srp1在隐球菌毒力中的作用。当用野生型菌株H99接种时,小鼠在接种后22 d至27 d内死亡,而用srp1Δ接种时小鼠则在接种后20 d到28 d内死亡,两者致病性无差异(图 5-A)。组织载菌量(CFU)分析发现两者感染小鼠脑肺脾的组织载菌量分别在107、109和107,两者之间无明显差异,表明SRP1基因的敲除并没有影响新生隐球菌的致病性。隐球菌感染小鼠后的病理组织切片结果也显示小鼠感染野生型菌株H99和srp1Δ突变体后其肺脑脾病变严重,病变部位充满大量含有荚膜增大的酵母细胞(图 5-C)。
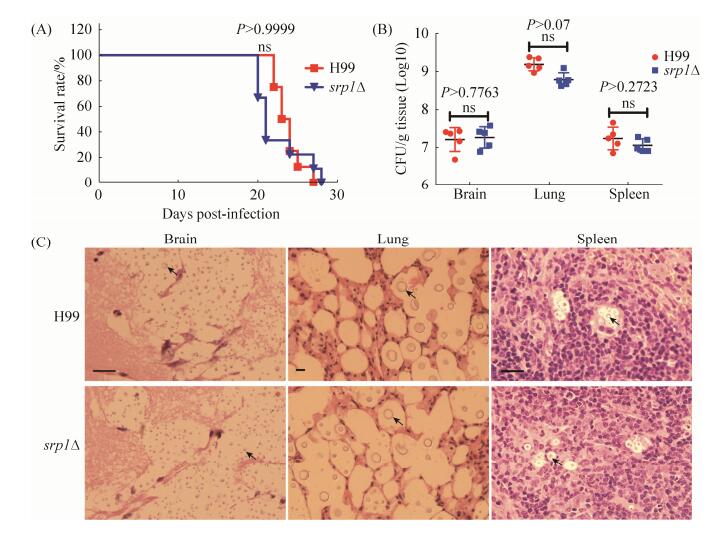
|
| 图 5 SRP1基因敲除不影响新生隐球菌的致病性 Figure 5 Srp1 is not required for pathogenicity in C. neoformans. A: Groups of 10 female C57 BL/6 mice were infected intranasally with 1×105 cells of wild-type H99 and srp1Δ mutant cells, and progression to severe morbidity was monitored for 30 days. There was no significant difference between srp1Δ mutant and the wild-type strain, H99 in virulence. B: Brains, Lungs and spleens were harvested and homogenized, and serial dilutions were plated for CFU determinations. Error bars indicate standard deviations for five mice per treatment group. ns: not significant. C: Histopathological analysis of C. neoformans-infected brain, lung and spleen tissues. Brain, lung and spleen tissues from H99 and srp1Δ infected mice were isolated, fixed and stained with H & E. Arrows indicates strained cryptococcal cells. Bars, 25 μm. |
2.6 Srp1调控新生隐球菌有性产孢过程
新生隐球菌是担子菌,有α和a两个交配型,两个交配型的细胞可交配形成双核菌丝并产生担孢子。为了检测Srp1在新生隐球菌有性生殖中的作用,我们等量混合srp1Δ突变体α和a交配型酵母细胞并滴于MS诱导培养基上,25 ℃黑暗条件下诱导培养14 d后显微镜下观察交配菌丝和担孢子发育情况。结果显示srp1Δ突变体能形成正常的交配菌丝和担子,但不能产生担孢子(图 6),表明Srp1调控隐球菌的有性产孢过程。
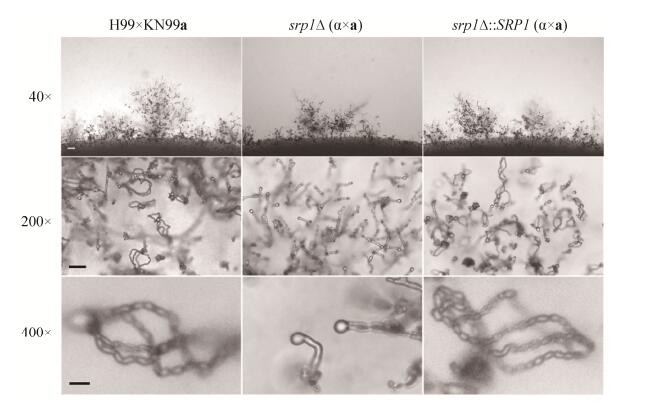
|
| 图 6 Srp1调控隐球菌的有性生殖 Figure 6 Srp1 is required for sexual reproduction in C. neoformans. Mating phenotypes for a wild-type cross between H99 and KN99a, the srp1Δ mutant bilateral cross between TBL105 and TBL138, and srp1Δ complemented strains cross between TBL218 and TBL219. All mating patches were spotted on MS medium and incubated for 14 days in the dark at 25 ℃. The top row shows hyphal growth on the edge of mating patches at ×40 magnification. bar=100 μm. The middle and the bottom rows show the basidium and spore chain morphology at ×200 (bar=25 μm) and ×400 magnification (Bar=10 μm), respectively. |
为了检测srp1Δ突变体细胞核在有性生殖各发育阶段的变化情况,我们分别构建了野生型和srp1Δ突变体α和a交配型背景下细胞核定位荧光菌株并设置交配实验。荧光显微镜观察发现野生型菌株(H99和KN99a)和srp1Δ突变体菌株(α和a交配型)的酵母细胞及交配菌丝中分别存在1个和2个细胞核(图 7),而交配后形成的新鲜担子内存在1个融合的细胞核(图 7),这表明野生型和srp1Δ突变体菌株都能够正常交配产生双核菌丝并能够形成担子和进行正常的细胞核融合。然而,srp1Δ突变体成熟担子中仍只观察到1个细胞核,而野生型菌株产生的成熟担子中则可以观察到4个细胞核,表明srp1Δ突变体菌株成熟担子中的细胞核融合后不能够正常进行减数分裂,导致只有1个细胞核存在。以上结果表明Srp1调控新生隐球菌有性生殖的减数分裂过程,Srp1缺失导致srp1Δ突变体菌株交配后不能正常进行减数分裂从而丧失产生担孢子能力。

|
| 图 7 srp1Δ突变体交配过程中细胞核变化情况 Figure 7 Fungal nuclei development assay in yeast cell, mating hyphae and basidia of the wild-type and srp1Δ mutant strains. Mating cultures were harvested from MS plates after incubation for 7 or 14 days at 25 ℃ in the dark and examined under inverted confocal laser scanning microscope (Olympus, FV1200). Bars, 5 μm. |
3 讨论
新生隐球菌是自然界广泛存在的具荚膜的酵母型病原真菌,能感染免疫缺陷及正常人群引起隐球菌性肺炎和脑膜炎,导致每年超过18万人的死亡[3]。新生隐球菌具有α和a两个交配型,可通过交配进行有性生殖并产生担孢子[1]。有性生殖可以促进隐球菌不同菌株之间遗传物质交换,利于子代细胞适应新的环境,也会导致高致病性和耐药性菌株的出现[21],这对于新生隐球菌致病力进化有积极意义。本研究鉴定了一新的产孢相关蛋白Srp1,功能分析结果显示Srp1能够影响新生隐球菌的有性生殖过程,SRP1基因敲除能导致隐球菌丧失产生担孢子的能力,表明Srp1在隐球菌有性生殖过程中起重要作用。
Srp1为一新的产孢相关蛋白。我们研究组前期通过iTRAQ蛋白组学方法鉴定fbp1Δ突变体背景下高丰度蛋白时发现一未被研究的蛋白CNAG_00626,该蛋白在fbp1Δ突变体背景下的丰度与野生型的丰度比值(fbp1Δ/H99)为3.0265681,为fbp1Δ突变体背景下最高丰度蛋白。我们首先对Srp1进行了序列分析,经过同源比对和保守结构域分析后发现Srp1不含有目前已知的保守结构域;与其同源性较高的蛋白都是假设蛋白,功能还未被研究,因此Srp1为一未被研究蛋白,对其进行功能研究十分有意义。
Srp1调控新生隐球菌的减数分裂过程。为了研究Srp1的功能,我们对其进行了基因敲除,结果表明新生隐球菌产孢相关蛋白基因SRP1敲除后其突变体交配时可正常进行细胞融合并形成双核菌丝和担子,但丧失产生担孢子的能力,推测可能原因是突变体交配后其有性生殖过程被阻断,导致其不能产生担孢子。至于是否是有性生殖过程被阻断,是有性生殖的细胞核融合过程被阻断,还是融合细胞核的减数分裂过程被阻断尚不明确。为进一步探究Srp1调控隐球菌有性产孢的机理,我们构建了核定位蛋白Nop1和红色荧光蛋白mCherry融合表达载体用以示踪新生隐球菌有性生殖过程中细胞核变化情况。表达载体经转化和筛选后分别获得野生型和srp1Δ突变体α和a交配型核定位荧光菌株。交配实验及细胞核发育结果表明srp1Δ突变体交配后形成的年轻担子里面含有2个细胞核,可进一步融合形成1个细胞核,但不能继续分裂形成4个细胞核,表明Srp1缺失阻断了减数分裂过程,从而导致srp1Δ突变体丧失产生担孢子的能力。至于Srp1如何调控隐球菌的减数分裂过程,是间接还是直接,目前还不得而知。因此Srp1调控隐球菌有性生殖的分子机制还有待于进一步研究。
Srp1是否是Fbp1下游底物还有待于进一步验证。Fbp1是新生隐球菌E3泛素连接酶关键蛋白,负责特异性地识别和泛素化底物蛋白并进行降解,其基因敲除突变体fbp1Δ完全丧失致病性和产孢能力[14, 22]。DAPI染色结果显示fbp1Δ突变体交配后其成熟担子中的2个细胞核融合形成1个细胞核后不再进行减数分裂,从而导致隐球菌丧失产孢能力。我们前期的质谱工作显示Srp1在泛素连接酶关键蛋白Fbp1缺失突变体背景下高丰度表达,表明Srp1可能是Fbp1的一下游底物。功能分析表明srp1Δ突变体丧失产生担孢子的能力也是由于细胞核融合后减数分裂过程受到阻断,表型与fbp1Δ一致,因此Fbp1有可能通过调控Srp1来调控新生隐球菌的有性生殖过程。至于Fbp1与Srp1是否存在相互作用以及Srp1是否是Fbp1的下游底物还有待于进一步验证。
产孢相关蛋白Srp1通过调控减数分裂过程而影响新生隐球菌的有性产孢,在新生隐球菌有性生殖进程中发挥着关键作用。对该蛋白进行功能和调控机理研究不仅为新生隐球菌有性生殖的调控及机制阐述提供数据支撑,而且也为后续新生隐球菌有性生殖精确调控机理的解析提供理论基础和技术支持。
| [1] | Lin XR, Heitman J. The biology of the Cryptococcus neoformans species complex. Annual Review of Microbiology, 2006, 60: 69-105. DOI:10.1146/annurev.micro.60.080805.142102 |
| [2] | Fang W, Fa ZZ, Liao WQ. Epidemiology of Cryptococcus and cryptococcosis in China. Fungal Genetics and Biology, 2015, 78: 7-15. DOI:10.1016/j.fgb.2014.10.017 |
| [3] | Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. The Lancet Infectious Diseases, 2017, 17(8): 873-881. DOI:10.1016/S1473-3099(17)30243-8 |
| [4] | Carroll SF, Guillot L, Qureshi ST. Mammalian model hosts of cryptococcal infection. Comparative Medicine, 2007, 57(1): 9-17. |
| [5] | Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annual Review of Genetics, 2002, 36: 557-615. DOI:10.1146/annurev.genet.36.052402.152652 |
| [6] | Fan CL, Han LT, Jiang ST, Chang AN, Zhou ZY, Liu TB. The Cys2His2 zinc finger protein Zfp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Fungal Genetics and Biology, 2019, 124: 59-72. |
| [7] | Feretzaki M, Heitman J. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genetics, 2013, 9(8): e1003688. |
| [8] | Gyawali R, Zhao YB, Lin JF, Fan YM, Xu XP, Upadhyay S, Lin XR. Pheromone independent unisexual development in Cryptococcus neoformans. PLoS Genetics, 2017, 13(5): e1006772. DOI:10.1371/journal.pgen.1006772 |
| [9] | Lin XR, Jackson JC, Feretzaki M, Xue CY, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genetics, 2010, 6(5): e1000953. |
| [10] | Liu LX, He GJ, Chen L, Zheng J, Chen YY, Shen L, Tian XY, Li EW, Yang EC, Liao GJ, Wang LQ. Genetic basis for coordination of meiosis and sexual structure maturation in Cryptococcus neoformans. eLife, 2018, 7: e38683. DOI:10.7554/eLife.38683 |
| [11] |
Fan CL, Yu QK, Liu TB. Cloning and functional analysis of RBP1 gene in Cryptococcus neoformans. Mycosystema, 2018, 37(11): 1466-1478.
(in Chinese) 范成莉, 余启昆, 刘同宝. 新生隐球菌RBP1基因克隆与功能分析. 菌物学报, 2018, 37(11): 1466-1478. |
| [12] | Tian XY, He GJ, Hu PJ, Chen L, Tao CY, Cui YL, Shen L, Ke WX, Xu HJ, Zhao YB, Xu QJ, Bai FY, Wu B, Yang EC, Lin XR, Wang LQ. Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nature Microbiology, 2018, 3(6): 698-707. DOI:10.1038/s41564-018-0160-4 |
| [13] | Fu C, Donadio N, Cardenas ME, Heitman J. Dissecting the roles of the calcineurin pathway in unisexual reproduction, stress responses, and virulence in Cryptococcus deneoformans. Genetics, 2018, 208(2): 639-653. DOI:10.1534/genetics.117.300422 |
| [14] | Liu TB, Wang YN, Stukes S, Chen Q, Casadevall A, Xue CY. The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryotic Cell, 2011, 10(6): 791-802. DOI:10.1128/EC.00004-11 |
| [15] | Xue CY, Tada Y, Dong XN, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host & Microbe, 2007, 1(4): 263-273. |
| [16] | Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes & Development, 1997, 11(23): 3206-3217. |
| [17] | Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL. Karyotyping of Cryptococcus neoformans as an epidemiological tool. Journal of Clinical Microbiology, 1993, 31(12): 3305-3309. |
| [18] | Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infection and Immunity, 2003, 71(9): 4831-4841. DOI:10.1128/IAI.71.9.4831-4841.2003 |
| [19] | Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryotic Cell, 2003, 2(5): 1036-1045. DOI:10.1128/EC.2.5.1036-1045.2003 |
| [20] | Price MS, Nichols CB, Alspaugh JA. The Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infection and Immunity, 2008, 76(12): 5729-5737. DOI:10.1128/IAI.00896-08 |
| [21] | Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature, 2005, 437(7063): 1360-1364. |
| [22] | Masso-Silva J, Espinosa V, Liu TB, Wang YN, Xue CY, Rivera A. The F-Box protein Fbp1 shapes the immunogenic potential of Cryptococcus neoformans. mBio, 2018, 9(1): e01828-17. |
 2019, Vol. 59
2019, Vol. 59




