中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 杨思悦, 符亚楠, 龙昊, 章翔, 谢珍玉. 2019
- Siyue Yang, Yanan Fu, Hao Long, Xiang Zhang, Zhenyu Xie. 2019
- 珊瑚病原菌株XSBZ03和XSBZ14双重PCR检测方法的建立
- Duplex PCR assay for detection of coral pathogenic strains XSBZ03 and XSBZ14
- 微生物学报, 59(7): 1266-1274
- Acta Microbiologica Sinica, 59(7): 1266-1274
-
文章历史
- 收稿日期:2018-08-27
- 修回日期:2019-01-07
- 网络出版日期:2019-02-19
2. 海南省热带水生生物技术重点实验室, 海南 海口 570228;
3. 南海海洋资源利用国家重点实验室, 海南 海口 570228
2. Key Laboratory of Tropical Aquatic Biotechnology of Hainan Province, Haikou 570228, Hainan Province, China;
3. State Key Laboratory of Marine Resource Utilization in South China Sea, Haikou 570228, Hainan Province, China
处于热带海洋的珊瑚礁具有丰富的生物多样性和重要的生态服务功能,是最重要的生态系统之一[1-2],享有“蓝色沙漠中的绿洲”、“海洋中的热带雨林”等美誉[3]。然而,近20余年来,由于气候和环境变化造成了世界各地的珊瑚礁正以惊人的速度减少,现已有三分之二的珊瑚礁处于灭绝的危险之中[4-7]。珊瑚的白化将直接导致珊瑚礁生态系统结构的瓦解,因此珊瑚白化死亡的原因一直深受人们的关注,其中细菌感染是导致珊瑚白化的重要原因之一[8-10]。研究表明由病原微生物引起的珊瑚疾病已成为全球珊瑚礁的主要威胁[11],当前共有54个国家和地区报道了各类珊瑚疾病3200余例,涉及患病珊瑚达113种,其中80%以上为细菌性病原引起[12-13]。但是有关珊瑚细菌性病原准确快速的检测方法报道较少,仅溶珊瑚弧菌(Vibrio coralliilyticus)、施罗氏弧菌(V. shilonii)等少量珊瑚病原建立了快速检测方法[10, 14-16]。
本研究对象是溶藻弧菌菌株XSBZ03和XSBZ14,它们是从白化的扁枝滨珊瑚中分离得到,并且经过科赫法则验证为扁枝滨珊瑚白化的病原,XSBZ03和XSBZ14对扁枝滨珊瑚的致病性是其他分离菌株所不具备的[17]。扁枝滨珊瑚作为南中国海诸多海域的优势珊瑚种,也是南中国海珊瑚礁人工生态修复最重要的移植物种之一,要预防细菌性病原所导致的南中国海扁枝滨珊瑚白化综合征的发生,首先要对病原进行准确的诊断。本研究从两株病原菌的全基因组序列中分别筛选到的一段特异性高的靶序列,并以之为对象设计合理的引物,拟建立可同时检测XSBZ03和XSBZ14的双重PCR方法,为丰富我国珊瑚疾病快速诊断方法奠定基础。
1 材料和方法 1.1 供试菌株珊瑚病原溶藻弧菌(V. alginolyticus) XSBZ03和XSBZ14;溶藻弧菌ATCC17749,副溶血弧菌(V. Parahaemolyticus) ATCC17802,创伤弧菌(V. vulnificus) ATCC27562,需钠弧菌(V. natriegens) ATCC33788,河流弧菌(V. fluvialis) ATCC33810均购自美国菌种保藏中心;其余37株溶藻弧菌,15株哈维氏弧菌(V. harveyi),2株副溶血弧菌,1株轮虫弧菌(V. rotiferianus),主要由自然海水、养殖海水、病亡的水生生物中分离得到,并已进行16S rRNA菌种鉴定以及各项生理生化指标检测,现由本实验室保存(表 1)。
| No. | Strains | No. | Strains | |
| 1 | V. alginolyticus XSBZ03 | 32 | V. alginolyticus HN07010 | |
| 2 | V. alginolyticus XSBZ14 | 33 | V. alginolyticus HN13001 | |
| 3 | V. alginolyticus HN07011 | 34 | V. alginolyticus PBVAY40119 | |
| 4 | V. alginolyticus HN08809 | 35 | V. alginolyticus HN08202 | |
| 5 | V. alginolyticus HN08304 | 36 | V. alginolyticus PBVAY41120 | |
| 6 | V. alginolyticus HN08307 | 37 | V. alginolyticus PBVAY38117 | |
| 7 | V. alginolyticus HN08805 | 38 | V. alginolyticus PBVAY39118 | |
| 8 | V. alginolyticus HN07005 | 39 | V. alginolyticus PBVAY12023 | |
| 9 | V. alginolyticus XSE381 | 40 | V. alginolyticus PBVAY42121 | |
| 10 | V. alginolyticus HN08803 | 41 | V. harveyi NS131241 | |
| 11 | V. alginolyticus HN08801 | 42 | V. harveyi NS131751 | |
| 12 | V. alginolyticus HN08201 | 43 | V. harveyi NS131651 | |
| 13 | V. alginolyticus PBVAY30031 | 44 | V. harveyi NS131451 | |
| 14 | V. alginolyticus HN08203 | 45 | V. harveyi NS131632 | |
| 15 | V. alginolyticus HN07006 | 46 | V. harveyi NS131631 | |
| 16 | V. alginolyticus HN08811 | 47 | V. harveyi WC13D121 | |
| 17 | V. alginolyticus HN08155 | 48 | V. harveyi HNH11011 | |
| 18 | V. alginolyticus HN07002 | 49 | V. harveyi HNH11001 | |
| 19 | V. alginolyticus TG06003 | 50 | V. harveyi WC13DH21 | |
| 20 | V. alginolyticus HN08306 | 51 | V. harveyi WC13H252 | |
| 21 | V. alginolyticus HN07009 | 52 | V. harveyi HNH11009 | |
| 22 | V. alginolyticus HN08813 | 53 | V. harveyi HNH11013 | |
| 23 | V. alginolyticus E001 | 54 | V. harveyi WC13DH52 | |
| 24 | V. alginolyticus HN08335 | 55 | V. harveyi PBVH78461 | |
| 25 | V. alginolyticus HN08303 | 56 | V. Parahaemolyticus PBVPY07150 | |
| 26 | V. alginolyticus HN07014 | 57 | V. Parahaemolyticus PBVPY06106 | |
| 27 | V. alginolyticus HN08806 | 58 | V. Parahaemolyticus ATCC 17802 | |
| 28 | V. alginolyticus E167 | 59 | V. rotiferianus HN076 | |
| 29 | V. alginolyticus HN08305 | 60 | V. vulnificus ATCC 27562 | |
| 30 | V. alginolyticus HN08156 | 61 | V. natriegens ATCC 33788 | |
| 31 | V. alginolyticus ATCC17749 | 62 | V. fluvialis ATCC 33810 |
1.2 主要试剂和仪器
TCBS培养基(Solarbio),细菌基因组提取试剂盒(TaKaRa),DNA Marker (TaKaRa)、2×AceTaq Master Mix (Vazyme),Multiplex PCR Kit (Vazyme),pMD-19-T载体试剂盒(TaKaRa),PCR仪(Applied Biosystems),DYY-6D型电泳仪(北京六一),全自动凝胶成像系统(Tanon 3500R)。
1.3 引物设计利用Hcluster-sg、Muscle软件对XSBZ03和XSBZ14全基因组序列(注:XSBZ03全基因组序列已在NCBI数据库公布,XSBZ14全基因组序列未公布)进行基因家族的TreeFam聚类分析,结果显示两株菌基因家族共3497个。利用perl脚本对两株菌的单拷贝基因家族的基因序列进行筛选以用于特异引物的设计,其中XSBZ03单拷贝基因序列145条,XSBZ14单拷贝基因序列131条。然后利用NCBI的BLAST功能对每条序列进行特异性分析,结果显示XSBZ03序列S1 (GenBank序列号:CP019959.1;位置:796711–797418)和XSBZ14序列S2 (GenBank序列号:MH702378)具有较高特异性,与其他菌株最高相似性分别为96%和95%。然后针对这两条序列分别设计特异引物(表 2)。
| Primer names | Primer sequences (5ʹ→3ʹ) | Product sizes/bp |
| Z03F1 | TGTCGGCGTTAAGCTCGAAT | 123 |
| Z03R1 | TCAACAAAGGCATTGCCACG | |
| Z14F1 | CTGAGCAGACGCTAGGACT | 244 |
| Z14R1 | TGCGTATTCTCACGAACGAGG |
1.4 细菌培养及其基因组DNA提取
使用TCBS培养基对各菌株纯化培养,挑取单克隆于2216E液体培养基中30 ℃、180 r/min培养18 h。取1.5 mL菌液于2 mL离心管中,采用细菌基因组DNA提取试剂盒说明书进行基因组DNA的提取。
1.5 引物特异性验证单重PCR反应体系为50 μL:2×AceTaq Master Mix 25 μL,上下游引物各2 μL,超纯水19 μL,模板2 μL,反应程序为:94 ℃ 5 min;94 ℃ 30 s,58 ℃ 30 s,72 ℃ 60 s,30个循环;最后72 ℃延伸5 min。产物电泳检测后在凝胶成像系统上拍照。使用引物XSBZ03和XSBZ14分别扩增各参试菌株,对PCR产物用2.5%琼脂糖凝胶(含万分之一goldview)电泳进行检测,电泳时间为30 min,电压120 V。
1.6 双重PCR反应条件优化参考单重PCR反应条件,利用正交实验对影响PCR反应的主要因素温度、循环数、引物浓度进行优化(表 3)。
| T/℃ | Cycle | Final concentration of Z03F1/Z03R1/ (μmol/L) | Final concentration of Z14F2/Z14R2/ (μmol/L) |
| 56 | 30 | 0.16/0.16 | 0.16/0.16 |
| 58 | 32 | 0.20/0.20 | 0.20/0.20 |
| 60 | 34 | 0.24/0.24 | 0.24/0.24 |
| 62 | 36 | 0.28/0.28 | 0.28/0.28 |
| 64 | 38 | 0.32/0.32 | 0.32/0.32 |
| 66 | 40 | 0.36/0.36 | 0.36/0.36 |
1.7 双重PCR特异性检测
以供试菌株的基因组DNA为模板,利用已建立的双重PCR反应体系,检测双重PCR反应体系的特异性。同时以XSBZ03和XSBZ14混合DNA为模板,进行双重PCR检测,产物电泳检测后在凝胶成像系统上拍照。产物回收、纯化后进行连接转化,然后送到华大基因进行测序。
1.8 双重PCR敏感性试验使用NanoDrop 2000超微量分光光度计测定XSBZ03和XSBZ14基因组DNA浓度,使用超纯水对它们进行10倍稀释,分别取3 μL作为模板进行双重PCR,检测双重PCR方法的DNA敏感性。
1.9 海水样品检测采集珊瑚养殖缸中的人工海水。将菌株XSBZ03和XSBZ14分别接种至2216E液体培养基中培养过夜,再用所采集的人工海水对菌株XSBZ03和XSBZ14进行不同浓度的稀释:(1)将稀释后的各稀释液取100 μL分别涂布于TCBS平板上进行菌落计数;(2)同时将各稀释液分别取3 μL作为双重PCR的扩增模板。
2 结果和分析 2.1 单重PCR引物的特异性检测使用上述单重PCR反应体系验证Z03F1/Z03R1、Z14F2/Z14R2的特异性,XSBZ03引物Z03F1/Z03R1特异性验证结果如图 1,仅有XSBZ03扩增出了123 bp的预期产物。XSBZ14引物Z14F2/Z14R2特异性验证结果如图 2,仅有XSBZ14扩增出了244 bp的预期产物。
2.2 双重PCR方法的建立
设计不同的退火温度、引物量、循环次数,对双重PCR反应条件进行优化,综合考虑检测时间、非特异性条带和引物二聚体、检测成本,经过筛选,最佳反应体系:2 × Multiplex Buffer 25 μL;Multiplex DNA Polymerase 1 μL;Z03F1/Z03R1 1 μL (10 μmol /L);Z14F1/Z14R1 1.6 μL (10 μmol/L);模板3 μL;ddH2O添加至50 μL。最佳反应程序:94 ℃ 5 min;94 ℃ 30 s,60 ℃ 30 s,72 ℃ 60 s,36个循环;72 ℃ 5 min。利用优化的双重PCR体系对目标菌株进行扩增可特异性扩增出目的条带,且与预期扩增基因片段大小一致(图 3)。
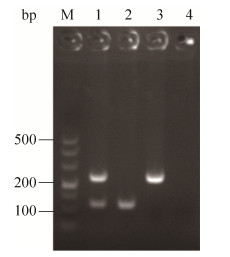
|
| 图 3 双重PCR方法的建立 Figure 3 Development of duplex PCR assay. M: Marker DL500; lane 1: Products of duplex PCR; lane 2: XSBZ03; lane 3: XSBZ14; lane 4: Negative control. |
2.3 双重PCR的特异性检测
结果显示阳性菌株XSBZ03和XSBZ14均扩增出单一目的条带,连接转化后测序结果显示为目标序列。而阴性菌株均未扩增出目的片段(图 4),表明双重PCR方法具有较好的特异性。
2.4 双重PCR敏感性检测
分别以稀释的阳性菌株基因组DNA为模板进行双重PCR扩增,结果显示该方法对XSBZ03基因组DNA检测限为1.7 pg/μL,对XSBZ14基因组DNA检测限为2.0 pg/μL (图 5)。

|
| 图 5 双重PCR敏感性试验 Figure 5 Sensitivity of duplex PCR assay. M: Marker DL500; lane 1–8: 1 to 8 represent the mixed genomic DNA concentration of XSBZ03 and XSBZ14 (The order of XSBZ03 genomic DNA is 1.7 ng/μL, 170 pg/μL, 17 pg/μL, 1.7 pg/μL, 170 fg/μL, 17 fg/μL, 1.7 fg/μL, 170 ag/μL. The order of XSBZ14 genomic DNA concentration is 2 ng/μL, 200 pg/μL, 20 pg/μL, 2 pg/μL, 200 fg/μL, 2 fg/μL, 2 fg/μL, 200 ag/μL); lane 9: Negative control. |
2.5 海水样品检测
于海水样品中分别添加XSBZ03和XSBZ14以制备模拟样品。经平板计数,XSBZ03和XSBZ14初始浓度分别为6×108 CFU/mL、8×108 CFU/mL,然后分别进行10倍梯度稀释,再以不同稀释度作为模板进行双重PCR。凝胶电泳结果显示,在模拟海水样品中,该方法对XSBZ03检测限为6×103 CFU/mL,对XSBZ14检测限为8×103 CFU/mL (图 6)。
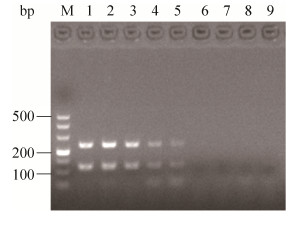
|
| 图 6 双重PCR对海水样品的检测极限 Figure 6 The detection limit of duplex PCR assay from sea water. M: Marker DL500; lane 1–8: 1 to 8 represent the mixed bacterial concentration of XSBZ03 and XSBZ14 (The order of XSBZ03 bacterial concentration is 6×107, 6×106, 6×105, 6×104, 6×103, 6×102, 6×101, 6×100 CFU/mL. The order of XSBZ14 bacterial concentration is 8×107, 8×106, 8×105, 8×104, 8×103, 8×102, 8×101, 8×100 CFU/mL); lane 9: Negative control. |
3 讨论和结论
珊瑚白化对全球珊瑚礁的健康产生严重威胁,严重影响了珊瑚礁生态系统的平衡。其中,由细菌性病原引发的白色综合症(White Syndrome,WT)等疾病的大面积流行已成为导致全球珊瑚白化的主要原因之一[18-20]。溶藻弧菌XSBZ03和XSBZ14作为新发现的珊瑚病原,其感染的宿主范围、流行病学和诊断技术的研究十分有限,因此建立这两株菌的快速检测方法,对于该疾病的诊断以及预防珊瑚白化具有重要意义。
传统的PCR检测方法其设计引物的靶序列多是具有种间特异性的基因片段[10, 13-15],该方法适用于检测遗传型稳定的物种。而溶藻弧菌存在多种不同遗传型,不同遗传型溶藻弧菌菌株间毒力差异较大[20-22],菌株XSBZ03和XSBZ14可使扁枝滨珊瑚白化的能力是其他无毒菌株所不具备的[17]。
本研究利用生物信息学技术结合NCBI数据库对珊瑚病原菌株XSBZ03和XSBZ14的全基因组序列进行比较分析,各筛选出一段具有较高种间及种内特异的靶序列,根据靶序列设计合适引物,对设计的引物进行进一步的筛选组合以便适用于双重PCR,从而利用该方法可同时检测和鉴定珊瑚病原菌株XSBZ03和XSBZ14。该方法可特异识别菌株XSBZ03和XSBZ14,对XSBZ03和XSBZ14基因组DNA的检测极限分别为1.7 pg/μL和2.0 pg/μL。在海水样品模拟检测中,该方法对XSBZ03和XSBZ14的检测极限分别为6×103 CFU/mL和8×103 CFU/mL。
本研究建立的双重PCR方法可检测菌株XSBZ03和XSBZ14,并区分常见的海洋菌株,具有快速简便、特异性强、敏感性高等优点,可用于XSBZ03和XSBZ14特异性快速检测及其分子流行病学调查,在珊瑚疾病诊断领域尚属首例,对珊瑚疾病的诊断和预防有重要意义。
| [1] |
Chen TR, Yu KF, Shi Q, Li S, Price GJ, Wang R, Zhao MX, Chen TG, Zhao JX. Twenty-five years of change in scleractinian coral communities of Daya Bay (northern South China Sea) and its response to the 2008 AD extreme cold climate event. Chinese Science Bulletin, 2009, 54(6): 812-820.
(in Chinese) 陈天然, 余克服, 施祺, 李淑, Price GJ, 王嵘, 赵美霞, 陈特固, 赵建新. 大亚湾石珊瑚群落近25年的变化及其对2008年极端低温事件的响应. 科学通报, 2009, 54(6): 812-820. |
| [2] | Moberg F, Folke C. Ecological goods and services of coral reef ecosystems. Ecological Economics, 1999, 29(2): 215-233. DOI:10.1016/S0921-8009(99)00009-9 |
| [3] |
Wang LR, Zhao HT. The general characteristics of the coral reef ecosystem. Chinese Journal of Ecology, 2001, 20(6): 41-45.
(in Chinese) 王丽荣, 赵焕庭. 珊瑚礁生态系的一般特点. 生态学杂志, 2001, 20(6): 41-45. DOI:10.3321/j.issn:1000-4890.2001.06.011 |
| [4] | Glynn PW. Coral reef bleaching: ecological perspectives. Coral Reefs, 1993, 12(1): 1-17. DOI:10.1007/BF00303779 |
| [5] | Loya Y. The coral reefs of Eilat-past, present and future: three decades of coral community structure studies//Rosenberg E, Loya Y. Coral Health and Disease. Berlin: Springer, 2004: 1-34. |
| [6] | Savini A, Vertino A, Marchese F, Beuck L, Freiwald A. Mapping cold-water coral habitats at different scales within the northern Ionian sea (central mediterranean): an assessment of coral coverage and associated vulnerability. PLoS One, 2014, 9(1): e87108. DOI:10.1371/journal.pone.0087108 |
| [7] | Schleyer MH, Kruger A, Celliers L. Long-term community changes on a high-latitude coral reef in the Greater St Lucia Wetland Park, South Africa. Marine Pollution Bulletin, 2008, 56(3): 493-502. DOI:10.1016/j.marpolbul.2007.11.011 |
| [8] | Munn CB. The role of vibrios in diseases of corals. Microbiology Spectrum, 2015, 3(4). |
| [9] |
Li S, Yu KF. Recent development in coral reef bleaching research. Acta Ecologica Sinica, 2007, 27(5): 2059-2069.
(in Chinese) 李淑, 余克服. 珊瑚礁白化研究进展. 生态学报, 2007, 27(5): 2059-2069. DOI:10.3321/j.issn:1000-0933.2007.05.047 |
| [10] |
Yang YY, Chen C, Luo P, Ding XQ, Xie M. Detection of Vibrio shilonii by using Loop-mediated isothermal amplification (LAMP). Microbiology China, 2018, 45(9): 1871-1880.
(in Chinese) 杨艺滢, 陈偿, 罗鹏, 丁雄祺, 谢媚. 环介导等温扩增技术快速检测施罗氏弧菌(Vibrio shilonii). 微生物学通报, 2018, 45(9): 1871-1880. |
| [11] | Tout J, Siboni N, Messer LF, Garren M, Stocker R, Webster NS, Ralph PJ, Seymour JR. Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Frontiers in Microbiology, 2015, 6: 432. |
| [12] | Willis BL, Page CA, Dinsdale EA. Coral disease on the great barrier reef//Rosenberg E, Loya Y. Coral Health and Disease. Berlin: Springer, 2004: 69-104. |
| [13] | Nozawa Y, Tokeshi M, Nojima S. Reproduction and recruitment of scleractinian corals in a high-latitude coral community, Amakusa, southwestern Japan. Marine Biology, 2006, 149(5): 1047-1058. DOI:10.1007/s00227-006-0285-5 |
| [14] | Pollock FJ, Morris PJ, Willis BL, Bourne DG. Detection and quantification of the coral pathogen Vibrio coralliilyticus by real-time PCR with TaqMan fluorescent probes. Applied and Environmental Microbiology, 2010, 76(15): 5282-5286. DOI:10.1128/AEM.00330-10 |
| [15] | Wilson B, Muirhead A, Bazanella M, Huete-Stauffer C, Vezzulli L, Bourne DG. An improved detection and quantification method for the coral pathogen Vibrio coralliilyticus. PLoS One, 2013, 8(12): e81800. DOI:10.1371/journal.pone.0081800 |
| [16] | Li HY, Zhang X, Long H, Hu CQ, Zhou YC, Wang SF, Ke SW, Xie ZY. Vibrio alginolyticus 16S-23S intergenic spacer region analysis, and PCR assay for identification of coral pathogenic strain XSBZ03. Diseases of Aquatic Organisms, 2018, 129(1): 71-83. DOI:10.3354/dao03233 |
| [17] | Xie ZY, Ke SW, Hu CQ, Zhu ZX, Wang SF, Zhou YC. First characterization of bacterial pathogen, Vibrio alginolyticus, for Porites andrewsi White syndrome in the South China Sea. PLoS One, 2013, 8(9): e75425. DOI:10.1371/journal.pone.0075425 |
| [18] | Arboleda MDM, Reichardt WT. Vibrio sp. causing Porites ulcerative white spot disease. Diseases of Aquatic Organisms, 2010, 90(2): 93-104. DOI:10.3354/dao02222 |
| [19] | Ben-Haim Y, Rosenberg E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Marine Biology, 2002, 141(1): 47-55. |
| [20] | Green E, Bruckner AW. The significance of coral disease epizootiology for coral reef conservation. Biological Conservation, 2000, 96(3): 347-361. DOI:10.1016/S0006-3207(00)00073-2 |
| [21] | George MR, John KR, Iyappan T, Jeyaseelan MJP. Genetic heterogeneity among Vibrio alginolyticus isolated from shrimp farms by PCR fingerprinting. Letters in Applied Microbiology, 2005, 40(5): 369-372. DOI:10.1111/lam.2005.40.issue-5 |
| [22] | Ren CH, Hu CQ, Luo P, Chen C, Jiang X, Wang QB. Genotyping of Vibrio alginolyticus isolates from Daya Bay by infrequent-restriction-site PCR and pulsed-field gel electrophoresis. Molecular and Cellular Probes, 2008, 22(4): 267-271. DOI:10.1016/j.mcp.2008.05.003 |
 2019, Vol. 59
2019, Vol. 59