中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 何晴, 王尚, 邓晔. 2019
- Qing He, hang Wang, Ye Deng. 2019
- 分子生物学技术在热泉地质微生物学研究中的应用
- Molecular biological technologies applied in geo-microbiology of terrestrial geothermal environments
- 微生物学报, 59(6): 996-1011
- Acta Microbiologica Sinica, 59(6): 996-1011
-
文章历史
- 收稿日期:2018-09-30
- 修回日期:2018-11-14
- 网络出版日期:2018-12-05
2. 中国科学院大学资源与环境学院, 北京 100049
2. College of Resources and Environment, University of Chinese Academy of Sciences, Beijing 100049, China
全球广布的陆地热泉多分布在构造运动活跃的地热带上,它是连接深部和地表环境的桥梁,也是我们追寻生命起源、探索地外生命的一扇窗口[1-2]。热泉具有多样的水化学类型、温度和pH变化梯度大、富含矿物质元素等特点,特殊的地球化学条件导致其孕育着大量的生命“暗物质”[3],这些生命获得赖以生存的能量的过程伴随着一系列独特的生物地球化学反应[4],推动着生命与环境的相互作用和协同演化。将其作为一个有机生命-无机环境相互作用的复杂地质系统加以研究,对揭示生命-极端环境相互作用机制有重要意义。
分子生物学技术如组学技术、探针技术和同位素标记技术等的应用,在揭开热泉生命“暗物质”神秘面纱和解析其与地球环境相互作用等方面发挥了关键作用[5-9],尤其在生物地球化学循环方面产生了一系列的新认识。以碳循环为例,在酸性地热土壤中发现了能利用H2进行自养生长的甲烷氧化菌Methylacidiphilum sp. RTK17.1,这是首次通过实验证实甲烷氧化菌能够同时进行自养和异养生长[10];此外,Science杂志上刊登了两篇关于嗜热微生物Thermosulfidibacter takaii ABI70S6T和Desulfurella acetivorans通过柠檬酸合酶的逆反应进行碳固定的新途径[11-12]。这些最新发现使人们对高温热泉的碳循环过程有了更深的认识,同时揭示了嗜热微生物蕴含的特殊代谢能力。本文将对热泉地质微生物学研究中常用的分子生物学技术进行梳理,阐述其在热泉微生物多样性分析和功能活性挖掘验证方面的应用及最新研究进展,并对存在问题和未来的研究方向进行剖析与展望。
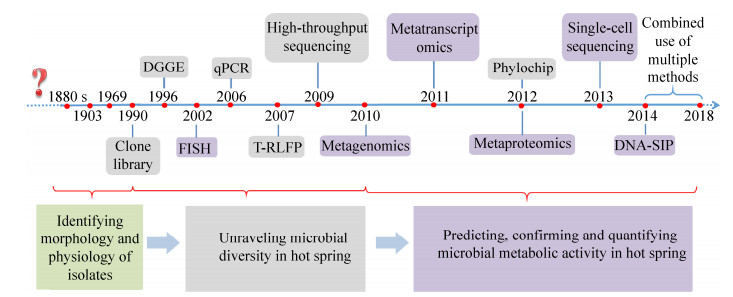
1 分子生物学技术应用于热泉地质微生物学研究的发展历程学者们对热泉生命的关注已有一百多年的历史,从发现到确认热泉中有生命经历了十几年(1880–1903),此后的60多年学者们致力于光合藻类的研究,一直认为光合作用的上限温度是生命的极限。直到Thermus aquaticus的成功分离培养才掀起了对其他嗜热微生物的研究浪潮。这一时期对热泉极端微生物的认识主要来自于分离培养和显微镜观察,然而在实验室重建极端的生长环境条件是很困难的,所以导致对嗜热微生物的认识具有片面性和选择性。
分子生物学手段的应用改善了这种困境,学者们最初通过5S rRNA来检测热泉微生物多样性[13],但由于片段短(120 bp)和电泳分辨率低不能应用于复杂环境。得益于基于16S rRNA基因的分子手段的开发和利用,热泉微生物多样性得到了很大的拓展[14-15]。传统分子生物学手段如克隆文库(Clone library)、变性梯度凝胶电泳分析(DGGE)和末端限制性酶切片段长度多态性分析(T-RFLP),分别于1990年[16]、1996年[17]、2007年[18]首次应用到热泉微生物多样性的研究中。近几年的技术以通量高为特点,如新一代高通量测序技术(High-throughput sequencing)[6]和系统发育芯片(Phylochip)[19]。高通量测序技术的广泛应用是热泉地质微生物学研究的革命性转折点,使学者们能够全面了解嗜热微生物群落结构的多样性及深入解析热泉极端微生物的生态功能,为探索地质历史时期的微生物生命过程提供了良好的资源。
解析热泉微生物多样性是认识热泉微生物在地质环境中功能的基础,基础夯实才能进行更深入的挖掘热泉微生物功能的研究。目前基于组学的技术,如宏基因组(Metageonomics)、单细胞基因组(Single-cell genome)、宏转录组测序(Metatrascriptomics)和蛋白质组学(Proteomics)[7, 28-30]可以预测热泉微生物的代谢能力并证实相关基因产物的存在,而基于荧光原位杂交(Fluorescence in situ hybridization,FISH)的技术[9, 31]和基于同位素标记(Isotope-probing)的技术[8-9, 23, 31-33]却能验证基因产物的功能或直接针对物种进行代谢活力的检测和量化,真正实现生命-地球化学过程相关联。这些技术的应用为解析嗜热微生物在元素循环、生物成矿机制等方面的作用提供了有效手段。综上所述,热泉微生物群落结构和功能解析技术经历了由片面向全面、由低通量向高通量、由低分辨水平向高分辨水平的发展过程(图 1)。技术的进步推动了研究的深入,更丰富了热泉地质微生物学研究的内容,拓展了人们对热泉微生物与地球环境相互作用关系的认识。
|
| 图 1 不同分子检测技术应用于热泉地质微生物研究的发展史 Figure 1 The development history of different molecular detection technologies applied in geomicrobiology research of terrestrial geothermal environments[6-7, 17-25]. In 1880s, there were reports of bacteria that could survive at high temperature[26], until 1903 there was first report confirming the presence of microorganisms in hot spring[26]; in 1969 Thermus aquaticus was isolated from hot spring[27] |
2 解析热泉微生物多样性的主要研究方法和进展 2.1 传统分子生物学方法
基于分子标记基因的分子生物学技术使人们对热泉微生物的认知从表型上升到基因水平。传统分子生物学方法在热泉微生物多样性研究中占据了很重要的地位,其主要通过16S rRNA基因或者功能基因(如氨氧化基因amoA等)的扩增来检测微生物整体群落或者功能类群的多样性及系统发育特征[25, 34-39]。目前在基于16S rRNA基因的微生物群落多样性研究中,传统方法已经被高通量测序所取代,但针对功能基因的研究,仍以传统分子生物学方法为主。大量的研究表明热泉中富含参与氮循环和硫循环过程的功能基因[40-46],然而功能基因的可检测性并不代表其在当前环境的顺利表达以合成相应的蛋白酶系,需结合其他方法如同位素标记技术来验证其代谢活性。此外,对于某些特有的微生物类群,可以设计特异性探针和引物分别通过FISH或定量PCR技术(qPCR)来鉴别其分布和丰度[36]。
2.2 新一代高通量分子生物学方法目前用于研究热泉微生物群落结构和物种多样性的高通量技术方法主要有两种,即高通量测序技术和基因芯片技术[47]。高通量测序方法主要可以分为扩增子测序和鸟枪法测序,这部分内容针对扩增子测序的应用进行总结。与传统方法类似,扩增子高通量测序也需要扩增分子标记基因(如16S rRNA基因、ITS基因和amoA基因等),随后对扩增产物进行测序来研究目标微生物群落结构及多样性,但由于其测序深度大,获得的多样性远高于传统方法。虽然测序结果同样会受到引物的偏好性的影响[48],其在解析热泉微生物群落多样性及挖掘群落分布控制因素方面的巨大贡献仍是不容忽视的。Miller等[6]于2009年首次利用454焦磷酸测序对美国黄石国家公园的两个碱性热泉微生物群落进行了研究,发现微生物群落结构沿温度梯度变化明显。随后该技术被多国学者用来解析热泉原位或富集产物的微生物多样性和群落组成[49-57]。上述研究大多集中在热泉原核微生物,热泉真菌和真核微生物的研究则明显滞后。目前,只有少量研究运用高通量测序技术分析了热泉真菌和真核生物的多样性。Liu等[58-59]通过基于ITS基因的Illumina Hiseq测序发现云南腾冲热海高温热泉蕴藏着丰富的真菌物种资源,其群落分布的控制因素与原核生物相似,主要有温度、pH和水化学离子如Fe2+和NH4+等;Oliverio等[60]通过基于18S rRNA基因的Illumina Miseq测序技术对新西兰Taupō火山区160个热泉的原生生物进行研究,发现温度和pH对多样性和群落结构有显著影响。
本课题组开展热泉相关研究工作较早,经历了从传统克隆文库到高通量测序的转变。同样是西藏谷露热泉,高通量技术获得的群落多样性比传统方法更高,得到的群落结构随温度的变化规律更明显[49]。在原核生物群落结构与环境因子的相关性方面得到了与其他学者一致的结果:微生物群落结构主要受温度和pH的影响,其次是水化学条件[52, 57, 61]。然而当研究载体同时包括水相和沉积相时,发现二者的微生物群落具有巨大差异[50, 62-64],极有可能是因为沉积物的异质性为微生物提供了更多的生态位,而且黄铁矿、碳酸盐、硅酸盐等矿物溶解释放的离子可以为微生物提供更多的营养[62-64]。此外,本课题组相关研究发现热泉微生物群落有明显的季节特征[50, 65]。与旱季相比,雨季热泉中营养物质如钾、总有机碳、铵、钙、钠和总氮等的补给增加,细菌多样性也随之变高,且微生物群落分布模式由高度有序变得杂乱无序[65]。我们还发现这种变化是个渐变的过程,刚进入雨季(6月份)时的群落与旱季(1月份)相比稍有变化,而到降雨量最大的时期(8月份)微生物群落变化最为剧烈[50]。进一步研究发现,通过LSA (Local similarity analysis)算法构建的微生物共变网络结构的变化也有相似的变化趋势,8月份微生物网络结构相较1月份和6月份发生明显改变,虽然此时群落多样性最高,但其网络结构最简单,关联度减少(1月份:节点数=95,连接数=465;6月份:节点数=99,连接数=408;8月份:节点数=82,连接数=123),表明在资源充足的情况下,不同的生态功能群落的生态位不重叠,彼此关联较弱。我们推测热泉微生物群落的季节变化与微生物对有机质的利用能力有关,但还需要通过实验来验证。
目前对热泉微生物多样性,尤其是基于16S rRNA基因的原核微生物的研究已经较为透彻,这为开展热泉微生物功能多样性的研究奠定了夯实的基础。以上这些手段只能解析微生物群落组成,却不能用来确定微生物在环境中的功能性状,而只有明确了群落组成和功能的对应关系才能更好地预测微生物响应环境变化的演替规律以及定向调控微生物在生态系统中的功能。
3 挖掘热泉微生物功能和代谢活性的主要研究方法和进展 3.1 宏基因组测序预测代谢潜力宏基因组可以在DNA水平上预测微生物潜在的代谢功能,通过基因组信息发现新类群和新的代谢途径(表 1)。Inskeep等[66]通过宏基因组鸟枪测序技术对美国黄石公园高温热泉微生物的群落结构和功能进行研究,发现热泉环境条件的异质性同时影响群落多样性和其功能多样性。Hua等[67]通过对云南腾冲热泉样品进行宏基因组测序分析,组装了6个平均大小为1.4 Mbp接近完整的Agiarchaeata基因组,并且对其代谢通路进行构建,显示Agiarchaeata是严格厌氧或兼性厌氧菌,能够通过氧化硫化氢进行化能自养生长,并且水平基因转移的发生导致其从细菌中获得了异化硫酸盐还原基因和一氧化碳氧化基因。
| Research point | Combined method | Major discovery |
| Metagenomics | ||
| Predicting potential metabolic functions | Six near-complete Agiarchaeata genomes with an average size of 1.4 Mbp have been assembled and the metabolic pathway of Agiarchaeata has been constructed[67]. | |
| Virus study | Single-cell sequencing | First report of Hydrogenobaculum-infected virus and an assessment of genome conservation and evolution of the Ampullaviridae family as well as Sulfolobus Monocaudavirus 1 (SMV1) -related viruses[71-72]. |
| Single-cell sequencing | ||
| Identifying microbial dark matter | Shotgun sequencing | Proposed Atribacteria (previous OP9)[7] Proposed Kryptonia[30] Proposed Calescamantes (EM19), Fervidibacteria (OctSPA-106) and Aminicenantes (OP8)[28]. |
| Shotgun sequencing Metatranscriptome FISH | The phylogeny and metabolic potential of the candidate archaeal phylum ‘Aigarchaeota’ has been deduced[29]. | |
此外,宏基因组测序自新一代测序应用以来便成为最高效的研究病毒的方法,宏基因组装可以反映完整病毒组信息,涵盖丰富的进化、遗传和功能等信息,帮助发现新型病毒,弥补了以往病毒发现及分类中存在的大量间隙(表 1)。例如,通过宏基因组序列组装发现高温热泉病毒的多样性与中温环境相当[68],还发现了首例侵染古菌的单链RNA病毒[69]。相比较热泉古菌病毒,热泉细菌病毒和真菌病毒的研究相对较少,最新对陆地热泉细菌病毒的综述指出目前NCBI只能检索到16条嗜热噬菌体的基因组,分属于5个病毒科[70]。Gudbergsdóttir等[71]从6个环境宏基因组中组装得到了10个完整的病毒基因组,其中一个疑似是细菌Hydrogenobaculum的病毒基因组,这是首次获 得该细菌的病毒序列,并且通过CRISPR分析方法发现了多样性极高的CRISPR基因组和CRISPR间隔序列[71];Munson-McGee等[72]通过单细胞测序结合宏基因组数据发现美国黄石国家公园热泉中普遍存在病毒-宿主相互作用体系,并且大多数的细胞中至少含有两种病毒,体现了热泉病毒的多样性。针对热泉病毒的研究仍处于起始阶段,关于热泉病毒多样性的影响因素、病毒-宿主-环境的相互作用关系等问题还亟待回答。
3.2 单细胞测序挖掘微生物代谢潜力单细胞测序可以在单细胞水平上获取微生物基因组信息,用于探索环境中物种丰度极少的未培养微生物,或是发现未被宏基因组及其他技术检测到的新型功能基因,以及研究微生物个体的生理特性、物质能量代谢和生态功能等[73]。微生物单细胞基因组技术包括单细胞获取、全基因组扩增、全基因组测序以及数据分析等步骤[73]。Rinke等[28]利用单细胞基因组测序首次从热泉中分别获得未培养类群EM19、OctSPA-106以及OP8的基因组,并将它们命名为Calescamantes、Fervidibacteria和Aminicenantes。Munson-McGee等[74]在美国黄石公园高温酸性热泉中得到了Nanoarchaeota的单细胞基因组,并且系统发育分析显示其在进化上与中性热泉中发现的Nanoarchaeota更为接近。
最新的一系列研究已经证实单细胞基因组与宏基因组相结合可以大大提高基因组拼接的完整性和连续性[75],在挖掘探索热泉生态系统中的微生物暗物质方面产生了实质性的进展(表 1)。Dodsworth等[7]通过单细胞基因组数据和宏基因组数据分别从热泉沉积物和纤维素降解富集培养物中获得了OP9的近完整的基因组,建议将其命名为“Atribacteria”,并预测了其代谢特征;Fadrosh等[30]在2016年从在全球收集的5.2 Tbp宏基因组数据中发现了一种专门生存于高温中性热泉的新型细菌门“Candidatus Kryptonia”,并通过宏基因组数据结合单细胞基因组数据组装得到其中4个属的基因组;Yu[76]等开发了一种改进后的单细胞-宏基因组技术,该技术首先将细胞分组,每组含有5–10个细胞,然后对每组细胞进行宏基因组测序分析,这样在保证单细胞的高分辨率的同时可以尽可能覆盖样品的多样性,通过这种技术他们从美国黄石国家公园的热泉样品中获得了29个新的基因组。
3.3 宏转录组和蛋白质组学证实潜在活性基因产物的存在基因的存在并不代表基因的表达,为了研究特定环境条件下的活性微生物,需要分析微生物群落的mRNA表达情况,即进行宏转录组学研究,比较不同环境条件下的差异表达基因和差异功能途径,从转录水平研究复杂微生物群落变化,揭示微生物对环境变化的响应机制,探索环境与微生物之间的互作机理(表 2)。通过转录组测序,学者们发现了热泉光合生物席在昼夜循环间的基因表达差异[21],并且识别了分别适应强光环境和弱光环境的专性基因[77]。前人研究表明通过热泉水、沉积物和气体的理化性质分析结合热动力学模型可以构建热泉中潜在的氧化还原过程[4],而宏转录组分析发现了参与甲烷、氮和硫的厌氧代谢的高表达基因[78],为其中部分过程提供了转录水平的证据。Agiarchaeota是近年来在基因组水平上解密的一类未培养微生物,宏转录组学结果证实其是一种具有自养潜力的好氧化能异养古菌,主要以氧气作为电子受体,并且具有显著的营养缺陷[29]。
| Research point | Combined method | Major discovery |
| Metatranscriptomics | ||
| Confirming transcription activity | Discovering high-expression genes involved in the anaerobic metabolism of methane, nitrogen and sulfur[78]. | |
| Proteomics | ||
| Confirming expression activity | Shotgun sequencing | 202 proteins encompassing 19 known functions from 12 known phyla were identified. Two key enzymes involving in the 3-hydroxypropionate CO2 fixation pathway were identified in uncultivated Roseiflexus spp., which are known photoheterotrophs[22]. |
不同于宏转录组学,蛋白质组学是直接研究在特定时空下的蛋白质种类和分布,这是研究微生物活性和功能更直接的手段,因为基因的表达只代表了转录活力,与蛋白质的存在并不是始终保持一致的[79]。Schaffert等[22]在美国黄石公园Octopus热泉生物席样品中发现了202种蛋白质,包括19类已知的功能,分属于12个微生物门。值得一提的是,他们识别出Roseiflexus spp.相关物种含有参与3-羟基丙酸循环的两种关键的酶,这为该类群能够通过固定CO2进行自养生长提供了更有力的证据(表 2),而此前的研究通过宏基因组测序仅仅对其自养代谢途径进行了预测[80],一直没有能够证明该物种进行碳固定的活性证据。目前蛋白质组的研究基本使用质谱分析,但其通量、灵敏度和数据解析率远远没有达到如DNA高通量测序那般的水平,严重限制了其应用的推广。
无论是通过宏基因组和单细胞基因组来预测代谢潜力,还是通过宏转录组和蛋白质组来识别相关基因产物的存在,这些手段的最大缺陷是无法直接衡量基因产物的功能,更不能直接量化物种的代谢活性。未来热泉地质微生物的研究工作将从群落多样性深入到物种-物种以及物种-环境的互作机理和关键功能群驱动元素循环的分子机制等方面,基于探针和同位素的技术将成为更主流、更有效的检测手段。
3.4 基于荧光原位杂交的技术识别功能微生物并验证其代谢活性荧光原位杂交技术的特色在于可以同时获得物种信息和其空间分布特征[81]。其原理是根据核酸碱基互补配对原则,用有荧光标记的特异性DNA或者RNA探针与细胞内经过变性的单链核酸序列互补配对,探测其中所有的同源核酸序列,其结果可直接在激光共聚焦显微镜或荧光显微镜下观察,无需单独分离DNA或RNA[82]。然而,FISH也存在着检测易受背景干扰、探针杂交效率低等问题[83]。催化报告沉积荧光原位杂交技术(Catalyzed reporter deposition-FISH,CARD-FISH)是FISH技术的升级,增加荧光信号强度的同时减少了背景干扰[84]。
这项技术在热泉地质微生物学研究中的应用较晚,但在检测特定微生物类群的丰度定量、空间分布特点和微生物间的相互关系方面贡献突出(表 3)。Ng等[9]通过FISH发现台北一热泉中古菌(探针ARC915)和硫酸盐还原菌(探针SRB385)细胞分别占微生物总细胞数(DAPI染色)的15.69%和7.16%。FISH在发现微生物相互作用关系(如联合代谢微生物群和共生关系)方面也有其独特优势。Kubo等[31]运用传统分子手段发现日本Nakabusa热泉微生物席中存在好氧硫氧化微生物、厌氧硫酸盐还原微生物和丝状不产氧光合微生物,并通过CARD-FISH技术展示了三种嗜热细菌群的垂直分布结构。这项研究说明具有不同功能属性的嗜热微生物通过在空间上有序排列共同完成硫代谢过程。据报道Nanoarchaeota是典型的共生微生物,Munson-McGee等[74]通过CARD-FISH在美国黄石公园高温酸性热泉中发现了Nanoarchaeota和Sulfolobales的共生关系。
| Research point | Combined method | Major discovery |
| Probing functional group | ||
| S-cycling | CARD-FISH | Revealing three thermophilic bacterial groups: aerobic chemolithotrophic sulfide-oxidizing species, anaerobic sulfate-reducing species, and filamentous anoxygenic photosynthetic species involving in mat sulfur cycling[31]. |
| Fe-cycling | qPCR 16S rRNA gene high-throughput sequencing | The isolation and characterization of the novel, Fe(II)- oxidizing, thermophilic, acidophilic organism Metallosphaera sp. strain MK1[87]. |
| Verifying microbial activity | ||
| N-cycling | Clone library RT-PCR MAR-FISH | The first thermophilic ammonia oxidizer was described, which belongs to the widely distributed group I.1b of the Crenarchaeota[86]. |
| Fe-cycling | μ-XAFS-FISH | Ferrihydrate was generated by Gallionella-related bacteria in the top surface of mat (10–15 μm)[85]. |
FISH技术具有良好的延伸性,与其他技术手段相结合可以在单细胞水平上研究特定物种对放射性标记物质的代谢能力,如显微放射自显影技术(Microautoradiography,MAR-FISH)、纳米级二次离子质谱技术(Nano-scale secondary ion mass spectrometry,FISH-Nano-SIMS)、拉曼光谱技术(Raman-FISH)和扩展X射线吸收精细结构分析(X-ray absorption fine structure,μ-XAFS-FISH)[85]。
目前仅MAR-FISH和μ-XAFS-FISH在热泉环境中有应用(表 3)。Nubel等[24]通过MAR-CARD- FISH获得了美国黄石公园光合生物席中的Chloroflexaceae相关微生物的垂向分布及异养代谢特征;Hatzenpichler等[86]利用该技术从热泉中发现了中度嗜热的氨氧化古菌,并证明了该古菌对氨的代谢活性。μ-XAFS-FISH是一项在高空间分辨率下同时监测微生物群落组成和元素分布特征的先进手段,可为微生物活动在生物矿化中的作用提供直接的证据。Mitsunobua等[85]通过FISH发现日本Sambe热泉生物席中表层(10–15μm)以Gallionella属相关细菌为主,推测其在铁氧化中发挥关键作用,而通过μ-XAFS得到的结果验证了他们的推测,在Gallionella富集的表层发现大量的无定形态水铁矿,并且发现了Fe-O6八面体共边的次生结构。这些新兴技术的发展和应用为深入研究热泉微生物在地球化学循环中的作用带来了新的契机。
3.5 基于同位素标记方法验证并量化功能类群的代谢活性稳定性同位素探针技术(Stable isotope probing,SIP)是最常用的用来验证功能类群或系统发育类群在特定生物地球化学过程中所发挥作用的同位素标记技术[8-9, 23, 31-33, 88]。其原理是添加稳定性同位素(如13C、15N和18O等)标记底物来培养环境样品,在细胞不断分裂、生长、繁殖的过程中合成含有稳定同位素的细胞物质。然后提取总标志物(如DNA、RNA和PLFA等),经过超高速密度梯度离心将“重”的(如被13C标记的)和“轻”的(如被12C标记的)细胞物质分离,进一步采用分子生物学技术对含有重的同位素标记的细胞物质进行分析[89]。
DNA-SIP技术近几年被用来分析热泉中参与代谢特定标记底物的活性微生物群落组成[8, 23, 32-33],即识别驱动热泉元素循环的关键微生物(表 4),这对探究地质历史时期的生命-环境演化过程具有重大意义。Sharp等[23]运用DNA-SIP与qPCR和16S rRNA基因扩增测序相结合的方法首次鉴定出具有活性的甲烷营养细菌在热泉中的存在;Fortney[8]等采用DNA-SIP、16S rRNA基因扩增测序以及宏基因组测序相结合的方法发现在美国Chocolate Pots的沉积物中存在大量异化铁还原菌,推测它们在铁循环中发挥着积极的作用。此外,同位素标记技术还能修正我们对微生物代谢方式的固有认识,Aquificales是普遍存在于高温碱性热泉中的微生物类群,最初学者们认为该类群主营自养生长,但最新通过同位素标记培养实验发现,该类群的代谢类型会随着碳源的改变而发生适应性变化,在添加外源有机碳源时可以通过异养或混合营养的方式获得能量[33]。这些研究结果为我们窥探热泉微生物在生物地球化学循环中的作用作出了一定贡献,同时也证实热泉微生物的特殊代谢能力,为挖掘其与环境的互作机制奠定了基础。
| Research point | Combined method | Major discovery |
| Confirming metabolic activity by DNA-SIP | ||
| Chemoautotrophy | Shotgun sequencing Stable 13C isotope | The extent and mechanisms of CO2 fixation were evaluated across a comprehensive set of microbial communities in Yellowstone National Park. The minimum fractions of autotrophic C in microbial biomass were > 50% in the majority of communities analyzed[32]. |
| Mixotrophy | HPLC-MS GC Stable 13C isotope | Streamer biofilm communities (SBC) can alternate their metabolism between autotrophy and heterotrophy depending on substrate availability[33]. |
| Dissimilatory iron reduction | 16S rRNA gene high-throughput sequencing Shotgun sequencing Stable 13C isotope | Fe(III)-reducing bacteria (FeRB) play an active role in Fe redox cycling within Fe-Si oxide-rich deposits located at the hot spring vent[88]. |
| Methane oxidation | 16S rRNA gene high-throughput sequencing qPCR Stable 13C isotope analyses | Methane oxidation was first measured at potential rates up to 141 μmol CH4 d-1g-1 sediment. Diverse methanotroph groups are adapted to warm environments[23]. |
| Determination of metabolic rate | ||
| Ammonia oxidation rate | qPCR FISH Clone library Stable 15N isotope | The ammonia oxidation rate of high temperature hot springs was obtained by means of stable isotope dilution method using 15N-NH4 and 15N-NO3[44]. |
| Carbon fixation rate | Radioactive 14C isotope RT-PCR Clone library DGGE | First determining the chemoautotrophic rate in high temperature acid hot spring, and the rate was comparable with the moderate temperature environment[91]. |
| 16S rRNA gene high-throughput sequencing Stable 13C isotope | Carbon uptake rates of photoautotrophic communities is greater than chemoautotrophic; filaments had the highest uptake rates whereas carbon fixation by stromatolites was significantly lower[92]. | |
| Sulfate-reduction rate | Clone library Radioactive 35S isotope | The significant sulfate respiration in the microbial mat was found in the low-sulfate thermal outflow[42]. |
除了能够识别驱动特定生物过程的关键微生物群,同位素标记还可以用于定量化研究特定生物地球化学过程的代谢速率(表 4)。早在2002年,Norris等[90]基于14C-NaHCO3标记的光合作用同化实验证明长期紫外照射的生物席的光合活性要高于无紫外照射的群落,但他们并没有定量化其速率。Boyd等[91]基于14C-NaHCO3首次测定了高温酸性热泉的化能自养速率(由细菌Hydrogenobaculum spp.执行),发现该速率甚至要高于此温度下碱性热泉中光合固碳(由细菌Synechococcus spp.执行)速率。近年来,出现了运用13C-NaHCO3稳定同位素标记来检测碳固定速率的研究并且得到较好的认可[32, 92],相比较放射性同位素,稳定同位素的成本较低而且操作简单、安全。本课题组和合作单位学者基于13C-NaHCO3对腾冲热泉的光合藻席的固碳速率进行了测定,发现其速率与温度有较强的相关性。除了碳循环,同位素标记技术也被用来检测氨氧化速率[44]和硫酸盐还原速率[41-42]。目前该技术在热泉中的应用还处于初步阶段,大部分针对特定生物过程的研究仅定性地描述相关功能基因的丰度,而想要定量化热泉微生物在元素循环中的贡献需要将基因的检测与代谢速率的测定相结合,所以同位素标记技术的应用将成为热泉微生物功能与活性验证的关键手段。
4 总结和展望得益于分子生物学检测技术的发展和革新,一方面热泉微生物多样性得到了极大的扩充,尤其是组学技术的应用使得热泉微生物“暗物质”不断被解密,例如Marsarchaeota的发现就是富集培养和多组学手段(宏基因组测序、单细胞测序和反转录实验)联合使用的产物[93];另一方面,随着基于探针和同位素的一系列新技术的应用,热泉微生物特殊的能量代谢方式不断被发现和验证,使得我们对其在元素循环和生物成矿等方面发挥的作用有了更深的认识。但通过对本课题组和其他学者们的研究进行梳理,我们发现仍存在以下待解决的问题。
(1) 在挖掘热泉微生物功能方面,虽然早有报道热泉中存在多样的潜在功能过程,并且在功能基因水平上得到了验证,但绝大多数都缺乏定量化的功能活性的证据;
(2) 目前的研究主要集中在原核微生物,热泉真核微生物和病毒多样性及代谢能力的研究明显滞后。但是真核微生物和病毒在调节能量流动和物质循环方面起着重要作用。
以同位素标记手段为主的分子生物学、地球化学、矿物学的多元化研究手段是针对热泉地质微生物研究存在问题的有效的解决方案,未来应加强以下几方面的研究。
(1) 热泉真菌、原生生物和病毒的丰度和多样性及其生存的极限边界条件。
(2) 热泉原核微生物、真菌、原生生物、病毒之间的相互作用关系及其在调节能量流动中的作用。据报道在海洋中,真核生物通过食用细菌、古生菌和病毒,甚至食用其他真核微生物来建立复杂的相互作用网络,这样的互作关系对微生物群落起着重要的自上而下的控制作用,同时将能量和物质向更高营养级转移;而由病毒介导的分解可释放100亿t碳,这对全球碳循环有直接影响[94]。然而在热泉中,我们对此知之甚少。
(3) 驱动特定元素循环过程的关键功能群的相互作用关系以及同一类微生物调控多元素循环过程的耦联机制。微生物在环境中并非独立存在,而元素循环之间通常也有耦联,如C、N和S循环、S、Fe和As循环等,常常是牵一发而动全身,但其中的内在机制尚不清楚。
| [1] | Gold T. The deep, hot biosphere. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(13): 6045-6049. DOI:10.1073/pnas.89.13.6045 |
| [2] | Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews, 2001, 25(2): 175-243. DOI:10.1111/j.1574-6976.2001.tb00576.x |
| [3] | Hedlund BP, Murugapiran SK, Alba TW, Levy A, Dodsworth JA, Goertz GB, Ivanova N, Woyke T. Uncultivated thermophiles:current status and spotlight on 'Aigarchaeota'. Current Opinion in Microbiology, 2015, 25: 136-145. DOI:10.1016/j.mib.2015.06.008 |
| [4] | Shock EL, Holland M, Meyer-Dombard D, Amend JP, Osburn GR, Fischer TP. Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochimica et Cosmochimica Acta, 2010, 74(14): 4005-4043. DOI:10.1016/j.gca.2009.08.036 |
| [5] | Bryant DA, Costas AM, Maresca JA, Chew AG, Klatt CG, Bateson MM, Tallon LJ, Hostetler J, Nelson WC, Heidelberg JF, Ward DM. Candidatus Chloracidobacterium thermophilum:an aerobic phototrophic Acidobacterium. Science, 2007, 317(5837): 523-526. DOI:10.1126/science.1143236 |
| [6] | Miller SR, Strong AL, Jones KL, Ungerer MC. Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Applied and Environmental Microbiology, 2009, 75(13): 4565-4572. DOI:10.1128/AEM.02792-08 |
| [7] | Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PS, Scholz MB, Lo CC, Raymond J, Quake SR, Hedlund BP. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nature Communication, 2013, 4: 1854. DOI:10.1038/ncomms2884 |
| [8] | Fortney NW, He SM, Kulkarni A, Friedrich MW, Holz C, Boyd ES, Roden EE. Stable isotope probing for microbial iron reduction in Chocolate Pots hot spring, Yellowstone National Park. Applied and Environmental Microbiology, 2018, 84(11): e02894-17. |
| [9] | Ng CC, Chang CC, Shyu YT. Archaeal community revealed by 16s rRNA and fluorescence in situ hybridization in a sulphuric hydrothermal hot spring, Northern Taiwan. World Journal of Microbiology and Biotechnology, 2005, 21(6/7): 933-939. |
| [10] | Carere CR, Hards K, Houghton KM, Power JF, McDonald B, Collet C, Gapes DJ, Sparling R, Boyd ES, Cook GM, Greening C, Stott MB. Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. The ISME Journal, 2017, 11(11): 2599-2610. DOI:10.1038/ismej.2017.112 |
| [11] | Mall A, Sobotta J, Huber C, Tschirner C, Kowarschik S, Bačnik K, Mergelsberg M, Boll M, Hügler M, Eisenreich W, Berg IA. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science, 2018, 359(6375): 563-567. DOI:10.1126/science.aao2410 |
| [12] | Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, Mori K, Kato Y, Masayuki M, Shimamura S, Yanagawa K, Shuto A, Ohkouchi N, Fujita N, Takaki Y, Atomi H, Takai K. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science, 2018, 359(6375): 559-563. DOI:10.1126/science.aao3407 |
| [13] | Stahl DA, Lane DJ, Olsen GJ, Pace NR. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Applied and Environmental Microbiology, 1985, 49(6): 1379-1384. |
| [14] | Woese CR. Bacterial evolution. Microbiological Reviews, 1987, 51(2): 221-271. |
| [15] | Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. Journal of Bacteriology, 1998, 180(2): 366-376. |
| [16] | Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiology Reviews, 1990, 75(2/3): 105-115. |
| [17] | Ferris MJ, Muyzer G, Ward DM. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Applied and Environmental Microbiology, 1996, 62(2): 340-346. |
| [18] | Kvist T, Ahring BK, Westermann P. Archaeal diversity in Icelandic hot springs. FEMS Microbiology Reviews, 2007, 59(1): 71-80. DOI:10.1111/fem.2007.59.issue-1 |
| [19] | Bohorquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, Kolter R, Junca H, Baena S, Zambrano MM. In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the colombian andes. Evironmental Microbiology, 2012, 63(1): 103-115. |
| [20] | Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, de M Jennings R, Fouke BW, Reysenbach AL, Roberto F, Young M, Schwartz A, Boyd ES, Badger JH, Mathur EJ, Ortmann AC, Bateson M, Geesey G, Frazier M. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One, 2010, 5(3): e9773. DOI:10.1371/journal.pone.0009773 |
| [21] | Liu ZF, Klatt CG, Wood JM, Rusch DB, Ludwig M, Wittekindt N, Tomsho LP, Schuster SC, Ward DM, Bryant DA. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. The ISME Journal, 2011, 5(8): 1279-1290. DOI:10.1038/ismej.2011.37 |
| [22] | Schaffert CS, Klatt CG, Ward DM, Pauley M, Steinke L. Identification and distribution of high-abundance proteins in the Octopus Spring microbial mat community. Applied and Environmental Microbiology, 2012, 78(23): 8481-8484. DOI:10.1128/AEM.01695-12 |
| [23] | Sharp CE, Martínez-Lorenzo A, Brady AL, Grasby SE, Dunfield PF. Methanotrophic bacteria in warm geothermal spring sediments identified using stable-isotope probing. FEMS Microbiology Ecology, 2014, 90(1): 92-102. DOI:10.1111/fem.2014.90.issue-1 |
| [24] | Nübel U, Bateson MM, Vandieken V, Wieland A, Kühl M, Ward DM. Microscopic examination of distribution and phenotypic properties of phylogenetically diverse chloroflexaceae-related bacteria in hot spring microbial mats. Applied and Environmental Microbiology, 2002, 68(9): 4593-4603. DOI:10.1128/AEM.68.9.4593-4603.2002 |
| [25] | Yim LC, Hongmei J, Aitchison JC, Pointing SB. Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiology Reviews, 2006, 57(1): 80-91. DOI:10.1111/fem.2006.57.issue-1 |
| [26] | Reysenbach AL, Voytek M, Mancinelli R. Thermophiles:biodiversity, ecology, and evolution. New York: Kluwer Academic Publishers, 2001. |
| [27] | Brock TD, Freeze H. Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. Journal of Bacterioogy, 1969, 989(1): 289-297. |
| [28] | Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature, 2013, 499(7459): 431-437. DOI:10.1038/nature12352 |
| [29] | Beam JP, Jay ZJ, Schmid MC, Rusch DB, Romine MF, de M Jennings R, Kozubal MA, Tringe SG, Wagner M, Inskeep WP. Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous 'streamer' community. The ISME Journal, 2016, 10(1): 210-224. DOI:10.1038/ismej.2015.83 |
| [30] | Eloe-Fadrosh EA, Paez-Espino D, Jarett J, Dunfield PF, Hedlund BP, Dekas AE, Grasby SE, Brady AL, Dong HL, Briggs BR, Li WJ, Goudeau D, Malmstrom R, Pati A, Pett-Ridge J, Rubin EM, Woyke T, Kyrpides NC, Ivanova NN. Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nature Communication, 2016, 7: 10476. DOI:10.1038/ncomms10476 |
| [31] | Kubo K, Knittel K, Amann R, Fukui M, Matsuura K. Sulfur-metabolizing bacterial populations in microbial mats of the Nakabusa hot spring, Japan. Systematic and Applied Microbiology, 2011, 34(4): 293-302. DOI:10.1016/j.syapm.2010.12.002 |
| [32] | Jennings RM, Moran JJ, Jay ZJ, Beam JP, Whitmore LM, Kozubal MA, Kreuzer HW, Inskeep WP. Integration of metagenomic and stable carbon isotope evidence reveals the extent and mechanisms of carbon dioxide fixation in high-temperature microbial communities. Frontiers in Microbiology, 2017, 8: 88. |
| [33] | Schubotz F, Hays LE, Meyer-Dombard DR, Gillespie A, Shock EL, Summons RE. Stable isotope labeling confirms mixotrophic nature of streamer biofilm communities at alkaline hot springs. Frontiers in Microbiology, 2015, 6: 42. |
| [34] | Meyer-Dombard DR, Shock EL, Amend JP. Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology, 2005, 3(3): 211-227. DOI:10.1111/gbi.2005.3.issue-3 |
| [35] | Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Applied and Environmental Microbiology, 2008, 74(15): 4910-4922. DOI:10.1128/AEM.00233-08 |
| [36] | Nakagawa T, Fukui M. Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. Journal of General and Applied Microbiology, 2002, 48(4): 211-222. DOI:10.2323/jgam.48.211 |
| [37] | Everroad RC, Otaki H, Matsuura K, Haruta S. Diversification of bacterial community composition along a temperature gradient at a thermal spring. Microbes and Environments, 2012, 27(4): 374-381. DOI:10.1264/jsme2.ME11350 |
| [38] | Lau MCY, Aitchison JC, Pointing SB. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles, 2009, 13(1): 139-149. DOI:10.1007/s00792-008-0205-3 |
| [39] | Song ZQ, Chen JQ, Jiang HC, Zhou EM, Tang SK, Zhi XY, Zhang LX, Zhang CLL, Li WJ. Diversity of Crenarchaeota in terrestrial hot springs in Tengchong, China. Extremophiles, 2010, 14(3): 287-296. DOI:10.1007/s00792-010-0307-6 |
| [40] | Cousins CR, Fogel M, Bowden R, Crawford I, Boyce A, Cockell C, Gunn M. Biogeochemical probing of microbial communities in a basalt-hosted hot spring at Kverkfjöll volcano, Iceland. Geobiology, 2018, 16(5): 507-521. DOI:10.1111/gbi.2018.16.issue-5 |
| [41] | Fishbain S, Dillon JG, Gough HL, Stahl DA. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Applied and Environmental Microbiology, 2003, 69(6): 3663-3667. DOI:10.1128/AEM.69.6.3663-3667.2003 |
| [42] | Dillon JG, Fishbain S, Miller SR, Bebout BM, Habicht KS, Webb SM, Stahl DA. High rates of sulfate reduction in a low-sulfate hot spring microbial mat are driven by a Low Level of diversity of sulfate-respiring microorganisms. Applied and Environmental Microbiology, 2007, 73(16): 5218-5226. DOI:10.1128/AEM.00357-07 |
| [43] | Zhang CL, Ye Q, Huang ZY, Li WJ, Chen JQ, Song ZQ, Zhao WD, Bagwell C, Inskeep WP, Ross C, Gao L, Wiegel J, Romanek CS, Shock EL, Hedlund BP. Global occurrence of archaeal amoA genes in terrestrial hot springs. Applied and Environmental Microbiology, 2008, 74(20): 6417-6426. DOI:10.1128/AEM.00843-08 |
| [44] | Chen S, Peng XT, Xu HC, Ta KW. Nitrification of archaeal ammonia oxidizers in a high-temperature hot spring. Biogeosciences, 2016, 13(7): 2051-2060. DOI:10.5194/bg-13-2051-2016 |
| [45] | Loiacono ST, Meyer-Dombard DR, Havig JR, Poret-Peterson AT, Hartnett HE, Shock EL. Evidence for high-temperature in situ nifH transcription in an alkaline hot spring of Lower Geyser Basin, Yellowstone National Park. Environmental Microbiology, 2012, 14(5): 1272-1283. DOI:10.1111/emi.2012.14.issue-5 |
| [46] | Kraková L, Šoltys K, Budiš J, Grivalský T, Ďuriš F, Pangallo D, Szemes T. Investigation of bacterial and archaeal communities:novel protocols using modern sequencing by Illumina MiSeq and traditional DGGE-cloning. Extremophiles, 2016, 20(5): 795-808. DOI:10.1007/s00792-016-0855-5 |
| [47] |
Deng Y, Feng K, Wei ZY, Liu WZ, Liang YT, Jin DC. Recent studies and applications of metagenomics in environmental engineering. Chinese Journal of Environmental Engineering, 2016, 10(7): 3373-3382.
(in Chinese) 邓晔, 冯凯, 魏子艳, 刘文宗, 梁玉婷, 金德才. 宏基因组学在环境工程领域的应用及研究进展. 环境工程学报, 2016, 10(7): 3373-3382. |
| [48] | Colman DR, Thomas R, Maas KR, Takacs Vesbach CD. Detection and analysis of elusive members of a novel and diverse archaeal community within a thermal spring streamer consortium. Extremophiles, 2015, 19(2): 307-313. DOI:10.1007/s00792-014-0715-0 |
| [49] | Wang S, Hou WG, Dong HL, Jiang HC, Huang LQ, Wu G, Zhang CL, Song ZQ, Zhang Y, Ren HL, Zhang J, Zhang L. Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS One, 2013, 8(5): e62901. DOI:10.1371/journal.pone.0062901 |
| [50] | Wang S, Dong HL, Hou WG, Jiang HC, Huang QY, Briggs BR, Huang LQ. Greater temporal changes of sediment microbial community than its waterborne counterpart in Tengchong hot springs, Yunnan Province, China. Scientific Reports, 2014, 4: 7479. |
| [51] | Song ZQ, Wang FP, Zhi XY, Chen JQ, Zhou EM, Liang F, Xiao X, Tang SK, Jiang HC, Zhang CL, Dong H, Li WJ. Bacterial and archaeal diversities in Yunnan and Tibetan hot springs, China. Environmental Microbiology, 2013, 15(4): 1160-1175. DOI:10.1111/emi.2013.15.issue-4 |
| [52] | Hou WG, Wang S, Dong HL, Jiang HC, Briggs BR, Peacock JP, Huang QY, Huang LQ, Wu G, Zhi XY, Li WJ, Dodsworth JA, Hedlund BP, Zhang CL, Hartnett HE, Dijkstra P, Hungate BA. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One, 2013, 8(1): e53350. DOI:10.1371/journal.pone.0053350 |
| [53] | de León KB, Gerlach R, Peyton BM, Fields MW. Archaeal and bacterial communities in three alkaline hot springs in Heart Lake Geyser Basin, Yellowstone National Park. Frontiers in Microbiology, 2013, 4: 330. |
| [54] | Tekere M, Lötter A, Olivier J, Jonker N, Venter S. Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo, South Africa. African Journal of Biotechnology, 2011, 10(78): 18005-18012. |
| [55] | Menzel P, Gudbergsdóttir SR, Rike AG, Lin LB, Zhang Q, Contursi P, Moracci M, Kristjansson JK, Bolduc B, Gavrilov S, Ravin N, Mardanov A, Bonch-Osmolovskaya E, Young M, Krogh A, Peng X. Comparative metagenomics of eight geographically remote terrestrial hot springs. Microbical Ecology, 2015, 70(2): 411-424. DOI:10.1007/s00248-015-0576-9 |
| [56] | Peacock JP, Cole JK, Murugapiran SK, Dodsworth JA, Fisher JC, Moser DP, Hedlund BP. Pyrosequencing reveals high-temperature cellulolytic microbial consortia in Great Boiling Spring after in situ lignocellulose enrichment. PLoS One, 2013, 8(3). |
| [57] | Sharp CE, Brady AL, Sharp GH, Grasby SE, Stott MB, Dunfield PF. Humboldt's spa:microbial diversity is controlled by temperature in geothermal environments. The ISME Journal, 2014, 8(6): 1166-1174. DOI:10.1038/ismej.2013.237 |
| [58] |
Liu KH, Ding XW, Zhang B, Tang XF, Xiao M, Xian WD, Li WJ. High-throughput sequencing to reveal fungal diversity in hot springs of Rehai at Tengchong in Yunnan. Acta Microbiologica Sinica, 2017, 57(9): 1314-1322.
(in Chinese) 刘开辉, 丁小维, 张波, 唐小飞, 肖敏, 鲜文东, 李文均. 高通量测序分析云南腾冲热海热泉真菌多样性. 微生物学报, 2017, 57(9): 1314-1322. |
| [59] | Liu KH, Ding XW, Salam N, Zhang B, Tang XF, Deng BW, Li WJ. Unexpected fungal communities in the Rehai thermal springs of Tengchong influenced by abiotic factors. Extremophiles, 2018, 22(3): 525-535. DOI:10.1007/s00792-018-1014-y |
| [60] | Oliverio AM, Power JF, Washburne A, Cary SC, Stott MB, Fierer N. The ecology and diversity of microbial eukaryotes in geothermal springs. The ISME Journal, 2018, 12(8): 1918-1928. DOI:10.1038/s41396-018-0104-2 |
| [61] | Power JF, Carere CR, Lee CK, Wakerley GLJ, Evans DW, Button M, White D, Climo MD, Hinze AM, Morgan XC, McDonald IR, Cary SC, Stott MB. Microbial biogeography of 925 geothermal springs in New Zealand. Nature Communication, 2018, 9: 2876. DOI:10.1038/s41467-018-05020-y |
| [62] | Colman DR, Feyhl-Buska J, Robinson KJ, Fecteau KM, Xu HF, Shock EL, Boyd ES. Ecological differentiation in planktonic and sediment-associated chemotrophic microbial populations in Yellowstone hot springs. FEMS Microbiology Ecology, 2016, 92(9): fiw168. |
| [63] | Cole JK, Peacock JP, Dodsworth JA, Williams AJ, Thompson DB, Dong HL, Wu G, Hedlund BP. Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. The ISME Journal, 2013, 7(4): 718-729. DOI:10.1038/ismej.2012.157 |
| [64] | Cheng TW, Wang PL, Song SR, Lin LH. Segregated planktonic and bottom-dwelling archaeal communities in high-temperature acidic/sulfuric ponds of the Tatun Volcano Group, Northern Taiwan. Terrestrial, Atmospheric and Oceanic Sciences, 2013, 24(3): 345-356. DOI:10.3319/TAO.2013.01.11.01(TT) |
| [65] | Briggs BR, Brodie EL, Tom LM, Dong HL, Jiang HC, Huang QY, Wang S, Hou WG, Wu G, Huang LQ, Hedlund BP, Zhang CL, Dijkstra P, Hungate BA. Seasonal patterns in microbial communities inhabiting the hot springs of Tengchong, Yunnan Province, China. Environmental Microbiology, 2014, 16(6): 1579-1591. DOI:10.1111/emi.2014.16.issue-6 |
| [66] | Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, Jennings RD, Fouke BW, Reysenbach AL, Roberto F, Young M, Schwartz A, Boyd ES, Badger JH, Mathur EJ, Ortmann AC, Bateson M, Geesey G, Frazier M. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One, 2010, 5(3): e9773. DOI:10.1371/journal.pone.0009773 |
| [67] | Hua ZS, Qu YN, Zhu QY, Zhou EM, Qi YL, Yin YR, Rao YZ, Tian Y, Li YX, Liu L, Castelle CJ, Hedlund BP, Shu WS, Knight R, Li WJ. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nature Communications, 2018, 9: 2832. DOI:10.1038/s41467-018-05284-4 |
| [68] | Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of viral metagenomes from yellowstone hot springs. Applied and Environmental Microbiology, 2008, 74(13): 4164-4174. DOI:10.1128/AEM.02598-07 |
| [69] | Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. Journal of Virology, 2012, 86(10): 5562-5573. DOI:10.1128/JVI.07196-11 |
| [70] | Zablocki O, van Zyl L, Trindade M. Biogeography and taxonomic overview of terrestrial hot spring thermophilic phages. Extremophiles, 2018, 22(6): 827-837. DOI:10.1007/s00792-018-1052-5 |
| [71] | Gudbergsdottir SR, Menzel P, Krogh A, Young M, Peng X. Novel viral genomes identified from six metagenomes reveal wide distribution of archaeal viruses and high viral diversity in terrestrial hot springs. Environmental Microbiology, 2016, 18(3): 863-874. DOI:10.1111/1462-2920.13079 |
| [72] | Munson-McGee JH, Peng SY, Dewerff S, Stepanauskas R, Whitaker RJ, Weitz JS, Young MJ. A virus or more in (nearly) every cell:ubiquitous networks of virus-host interactions in extreme environments. The ISME Journal, 2018, 12(7): 1706-1714. DOI:10.1038/s41396-018-0071-7 |
| [73] | BrehmStecher BF, Johnson EA. Single-cell microbiology:tools, technologies, and applications. Microbiology and Molecular Biology Reviews, 2004, 68(3): 538-559. |
| [74] | Munson-McGee JH, Field EK, Bateson M, Rooney C, Stepanauskas R, Young MJ. Nanoarchaeota, their sulfolobales host, and nanoarchaeota virus distribution across Yellowstone National Park hot springs. Applied and Environmental Microbiology, 2015, 81(22): 7860-7868. DOI:10.1128/AEM.01539-15 |
| [75] | Xu Y, Zhao FQ. Single-cell metagenomics:challenges and applications. Protein & Cell, 2018, 9(5): 501-510. |
| [76] | Yu FQ, Blainey PC, Schulz F, Woyke T, Horowitz MA, Quake SR. Microfluidic-based mini-metagenomics enables discovery of novel microbial lineages from complex environmental samples. eLife, 2017, 6: e26580. DOI:10.7554/eLife.26580 |
| [77] | Olsen MT, Nowack S, Wood JM, Becraft ED, LaButti K, Lipzen A, Martin J, Schackwitz WS, Rusch DB, Cohan FM, Bryant DA, Ward DM. The molecular dimension of microbial species: 3. Comparative genomics of Synechococcus strains with different light responses and in situ diel transcription patterns of associated putative ecotypes in the Mushroom Spring microbial mat. Frontiers in Microbiology, 2015, 6: 604. |
| [78] | Tripathy S, Padhi SK, Mohanty S, Samanta M, Maiti NK. Analysis of the metatranscriptome of microbial communities of an alkaline hot sulfur spring revealed different gene encoding pathway enzymes associated with energy metabolism. Extremophiles, 2016, 20(4): 525-536. DOI:10.1007/s00792-016-0846-6 |
| [79] | Burg D, Ng C, Ting L, Cavicchioli R. Proteomics of extremophiles. Environmental Microbiology, 2011, 13(8): 1934-1955. DOI:10.1111/j.1462-2920.2011.02484.x |
| [80] | Klatt CG, Bryant DA, Ward DM. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environmental microbiology, 2007, 9(8): 2067-2078. DOI:10.1111/emi.2007.9.issue-8 |
| [81] |
Song LL, Zeng GM, Chen YN, Jiang R, Huang GH. Fluorescence in situ hybridization technology and its application in environmental micribial ecology. Journal of Microbiology, 2007, 27(1): 40-44.
(in Chinese) 宋琳玲, 曾光明, 陈耀宁, 蒋茹, 黄国和. 荧光原位杂交技术及其在环境微生物生态学中的应用研究. 微生物学杂志, 2007, 27(1): 40-44. DOI:10.3969/j.issn.1005-7021.2007.01.010 |
| [82] | Speicher MR, Carter NP. The new cytogenetics:Blurring the boundaries with molecular biology. Nature Reviews Genetics, 2005, 6(10): 782-792. DOI:10.1038/nrg1692 |
| [83] | Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 1995, 59(1): 143-169. |
| [84] |
Lu L. Catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) and its application in microbial ecology study. Journal of Microbiology, 2017, 37(6): 87-97.
(in Chinese) 路璐. CARD-FISH技术及其在微生物生态学研究中的应用. 微生物学杂志, 2017, 37(6): 87-97. DOI:10.3969/j.issn.1005-7021.2017.06.014 |
| [85] | Mitsunobu S, Shiraishi F, Makita H, Orcutt BN, Kikuchi S, Jorgensen BB, Takahashi Y. Bacteriogenic Fe(Ⅲ) (Oxyhydr) oxides characterized by synchrotron microprobe coupled with spatially resolved phylogenetic analysis. Environmental Science & Technology, 2012, 46(6): 3304-3311. |
| [86] | Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(6): 2134-2139. DOI:10.1073/pnas.0708857105 |
| [87] | Kozubal M, Macur RE, Korf S, Taylor WP, Ackerman GG, Nagy A, Inskeep WP. Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone National Park. Applied and Environmental Microbiology, 2008, 74(4): 942-949. DOI:10.1128/AEM.01200-07 |
| [88] | Fortney NW, He S, Converse BJ, Beard BL, Johnson CM, Boyd ES, Roden EE. Microbial Fe(Ⅲ) oxide reduction potential in Chocolate Pots hot spring, Yellowstone National Park. Geobiology, 2016, 14(3): 255-275. DOI:10.1111/gbi.2016.14.issue-3 |
| [89] |
Jia ZJ. Principle and application of DNA-based stable isotope probing-A review. Acta Microbiologica Sinica, 2011, 51(12): 1585-1594.
(in Chinese) 贾仲君. 稳定性同位素核酸探针技术DNA-SIP原理与应用. 微生物学报, 2011, 51(12): 1585-1594. |
| [90] | Norris TB, McDermott TR, Castenholz RW. The long-term effects of UV exclusion on the microbial composition and photosynthetic competence of bacteria in hot-spring microbial mats. FEMS Microbiology Ecology, 2002, 39(3): 193-209. |
| [91] | Boyd ES, Leavitt WD, Geesey GG. CO2 uptake and fixation by a thermoacidophilic microbial community attached to precipitated sulfur in a geothermal spring. Applied and Environmental Microbiology, 2009, 75(13): 4289-4296. DOI:10.1128/AEM.02751-08 |
| [92] | Schuler CG, Havig JR, Hamilton TL. Hot spring microbial community composition, morphology, and carbon fixation:implications for interpreting the ancient rock record. Frontiers in Earth Science, 2017, 5: 1-17. |
| [93] | Jay ZJ, Beam JP, Dlakić M, Rusch DB, Kozubal MA, Inskeep WP. Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats. Nature Microbiology, 2018, 3(6): 732-740. DOI:10.1038/s41564-018-0163-1 |
| [94] | Coutinho FH, Gregoracci GB, Walter JM, Thompson CC, Thompson FL. Metagenomics sheds light on the ecology of marine microbes and their viruses. Trends in Microbiology, 2018, 26(11): 955-965. DOI:10.1016/j.tim.2018.05.015 |
 2019, Vol. 59
2019, Vol. 59