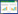中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 温继龙, 彭琦, 赵欣, 张杰, 宋福平. 2019
- Jilong Wen, Qi Peng, Xin Zhao, Jie Zhang, Fuping Song. 2019
- 苏云金芽胞杆菌BkdR和CcpA对bkd基因簇的转录调控
- bkd gene cluster is regulated by BkdR and CcpA in Bacillus thuringiensis
- 微生物学报, 59(11): 2229-2239
- Acta Microbiologica Sinica, 59(11): 2229-2239
-
文章历史
- 收稿日期:2019-01-23
- 修回日期:2019-03-11
- 网络出版日期:2019-03-18
2. 中国农业科学院植物保护研究所, 植物病虫害生物学国家重点实验室, 北京 100193
2. State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China
在低GC含量的革兰氏阳性细菌中,含有一种分解代谢物控制蛋白A (catabolite control protein A,CcpA),属于LacI-GalR家族的转录因子[1]。CcpA作为一个多效调控因子,通过与启动子上的保守cre (catabolite repression element)序列结合[2],在细菌中调控许多重要的生理功能,如在变异链球菌(Streptococcus mutans)中,Cid/Lrg操纵子系统与细胞自溶、生物膜形成、葡萄糖基转移酶表达和氧化应激反应相关,CcpA通过与cid和lrg启动子的结合从而调控该菌株的这些生理功能[3-4];枯草芽胞杆菌(Bacillus subtilis,Bs)中,GntR调控gntRKPZ操纵子,CcpA通过分别与gnt的启动子和gntR的启动子结合从而调控葡萄糖酸盐的代谢过程[5]。
Sigma因子是RNA聚合酶核心酶的亚基,在细菌的转录起始调控过程中具有重要的作用。Sigma因子按照功能可分为Sigma70和Sigma54两种,区别在于Sigma70识别启动子的–10和–35区,而Sigma54识别启动子的–12和–24区,并需要与特定的增强子结合蛋白(enhancer binding proteins,简称EBPs)共同作用,才能激活起始转录[6]。EBPs具有3个典型的结构域:N端的信号结构域(R domain),负责感应外界信号;中间的AAA+结构域(C domain),负责与Sigma54因子相互作用;C端的HTH结构域(D domain),通过与启动子上特定序列结合,从而起到调控作用[6]。许多细菌中含有多种EBPs,例如在大肠杆菌(Escherichia coli)中有13种EBPs,大多数与碳氮源代谢相关[6-7];恶臭假单胞菌(Pseudomonas putida)中有22种EBPs,参与的代谢途径很广泛,包括鞭毛合成、不同种碳氮源利用和藻朊酸盐生物合成等[8];枯草芽胞杆菌中有5种EBPs,调控精氨酸、支链氨基酸等碳氮源代谢[9-12]。这些转录因子在许多生理代谢途径中具有重要作用,如NtrC调控氮源代谢[13]、XylR调控芳香烃代谢[14]、PspF调控内膜压力[15]等。
苏云金芽胞杆菌(Bacillus thurigiensis,Bt)是革兰氏阳性细菌,其在芽胞形成的同时能够产生杀虫晶体蛋白(insecticidal crystal proteins,ICPs),对多种鳞翅目和鞘翅目等昆虫都有特异性的毒杀活性,是应用最为广泛的微生物杀虫剂[16-17]。本实验室前期研究发现,在Bt HD73菌株中有8种EBPs[18],调控多种代谢途径,包括γ-氨基丁酸代谢[19-20]、赖氨酸代谢[21]、肌氨酸代谢[22]等,其中BkdR调控bkd基因簇的转录,可能与支链氨基酸(亮氨酸、异亮氨酸和缬氨酸)的代谢相关[18]。
细菌中,支链氨基酸降解代谢形成的产物参与细胞重要的生理功能,例如支链氨基酸可降解成支链脂肪酸,在链霉菌(Streptomyces avermitilis)中[23],是聚酮化合物抗生素生产的一些前体的唯一来源;在Bs中,支链脂肪酸参与芽胞形成过程中细胞膜的形成[10, 24];在粪肠球菌(Enterococcus faecalis)中,参与ATP和能量代谢[25];在黄色粘球菌(Myxococcus xanthus)中参与细胞发育过程中的信号传导[26]。Bs中,从支链氨基酸降解成支链脂肪酸的代谢途径由bkd基因簇编码的酶系统催化,bkd基因簇的转录受Sigma54因子控制,并受转录因子BkdR和全局调控因子CodY的调控[10]。本实验室前期研究发现,Bt中bkd基因簇的基因组织和转录调控模式与Bs相似,bkd基因簇由8个基因组成,其中ptb-bkdB 7个基因形成一个转录单元,bkdR单独形成一个转录单元。bkd基因簇启动子Pptb的转录活性受Sigma54控制,并受BkdR正调控。Bt中bkdR基因的缺失对菌体生长、芽胞形成率和Cry1Ac蛋白产量无影响,但使菌体运动能力减弱[27]。本研究在此基础上,对bkd基因簇启动子Pptb的诱导转录活性的调控机制进行深入分析。
1 材料和方法 1.1 材料 1.1.1 菌株、质粒和细菌培养条件: 所用菌株和质粒见表 1。大肠杆菌(Escherichia coli,E. coli)的培养使用LB培养基,培养条件37 ℃、220 r/min;Bt的培养分别使用LB培养基、SSM培养基[28]和M9培养基(购自Sigma公司,货号:M6030),培养条件30 ℃、220 r/min。抗生素使用终浓度分别为:氨苄青霉素100 μg/mL,红霉素5 μg/mL,卡那霉素100 μg/mL。| Strains and plasmids | Characterization | Resource |
| Strains | ||
| HD73 | B. thuringiensis subsp. kurstaki carrying the cry1Ac gene | This lab |
| HD(ΔbkdR) | B. thuringiensis HD73 bkdR gene insertion mutant; KanR | [27] |
| HD(ΔccpA) | B. thuringiensis HD73 ccpA gene insertion mutant; KanR | This study |
| HD(Pptb) | HD73 strain containing plasmid pHTPptb | [27] |
| ΔbkdR(Pptb) | HD(ΔbkdR) strain containing plasmid pHTPptb | [27] |
| ΔccpA(Pptb) | HD(ΔccpA) strain containing plasmid pHTPptb | This study |
| E. coli TG1 | D(lac-proAB) supE thi hsd-5 (F' traD36 proA+ proB+ lacIq lacZDM15), general purpose cloning host | This lab |
| E. coli ET | F– dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1, for generation of unmethylated DNA | This lab |
| BL21(DE3) | E. coli B, F–, dcm, ompT hsdS(rB-mB-), gal, λ(DE3) | This lab |
| BL21(pETccpA) | BL21(DE3) strain containing plasmid pETccpA | [29] |
| BL21(pETbkdR) | BL21(DE3) strain containing plasmid pETbkdR | This study |
| BLpET | BL21 strain carrying pET21b | This lab |
| Plasmids | ||
| pMAD | AmpR, EmR shuttle vector, thermosensitive origin of replication | Institute Pasteur |
| pMADDbkdR | pMAD with bkdR insertion fragment | This study |
| pET21b | Expressional vector, Ampr, 5.4 kb | This lab |
| pETccpA | pET21b containing ccpA gene, Ampr | [29] |
| pETbkdR | pET21b containing bkdR gene, Ampr | This study |
| pHTPptb | pHT304-18Z carrying promoter upstream from ptb | [27] |
1.1.2 主要仪器和材料: 限制性内切酶、DNA聚合酶和DNA连接酶均购自宝生物工程(大连)有限公司和北京博迈德科技发展有限公司;质粒提取、DNA回收和PCR产物纯化试剂盒购自Axygen公司。镍亲和层析柱填料购自GE公司。poly(dI:dC)购自Sigma公司。Gel Shift Assay Systems购自Promega公司。 1.1.3 引物合成及序列测定: 根据Bt HD73基因组序列[30]设计引物,引物合成由生工生物工程(上海)股份有限公司北京合成部完成,序列测定由北京诺赛基因基因组研究中心有限公司完成,引物名称及序列见表 2。
| Primer name | Sequence (5 →3 ) |
| ccpA-A1 | CGGGATCCTCTGATGCAGCGCAACAAATG |
| ccpA-A2 | CTCAAATGGTTCGCTGTGAAACGTTCGCTTC |
| Kan-1 | GAAGCGAACGTTTCACAGCGAACCATTTGAG |
| Kan-2 | ACGGTGAGGTAAGATAAATTCCTCGTAGGC |
| ccpA-B1 | GCCTACGAGGAATTTATCTTACCTCACCGT |
| ccpA-B2 | CCGGAATTCGTACCATAATGCTACCTGCA |
| bkdR-F | CGGGATCCGATGAAACAAAAAGTATTAATTG |
| bkdR-R | ACGCGTCGACTTGCATGCTATTTTTTGCATG |
| Pptb-1 | GTGACAGAGTTTGAAGGG |
| Pptb-2 | ATTTTGTAATCAACCCTTTC |
1.2 ccpA突变菌株构建及筛选
为了研究CcpA对bkd基因簇的转录调控,利用同源重组的原理,构建了ccpA突变体,方法见参考文献[31]。简述如下:以Bt HD73基因组为模板,用引物ccpA-A1/ccpA-A2扩增ccpA基因上游片段(ccpA-A),大小为618 bp;用引物ccpA-B1/ccpA-B2扩增ccpA基因下游片段(ccpA-B),大小为646 bp。以△bkdR突变体[27]为模板,用引物Kan-1/Kan-2扩增卡那抗性基因(kan),大小为1503 bp。以ccpA-A、ccpA-B和kan为模板,用引物ccpA-A1/ccpA-B2通过重叠PCR扩增ccpA缺失突变盒(含有卡那霉素抗性基因)。PCR产物经限制性内切酶Bam HI和EcoR I双酶切后,连接到温敏穿梭载体pMAD,转化至大肠杆菌TG1菌株中,获得重组质粒命名为pMADΔccpA。将重组质粒转入ET去甲基化,再电击转入HD73菌株,转化方法见文献[32],获得具有红霉素抗性的HD (pMADΔccpA)菌株。该菌株进行38 ℃高温突变,筛选出有卡那霉素抗性并且没有红霉素抗性的菌株,进行PCR鉴定:以ccpA-A1/ccpA-B2为引物,以具有卡那霉素抗性并且无红霉素抗性的菌株为模板,获得的突变菌株命名为HD(ΔccpA)。
1.3 BkdR表达菌株构建根据GenBank中Bt HD73菌株(GenBank登录号:CP004069)的bkdR(HD73_4469)基因序列及pET21b质粒的酶切位点,设计扩增bkdR基因ORF的引物bkdR-F和bkdR-R (表 2),以HD73基因组为模板扩增bkdR基因(2040 bp),PCR产物纯化后经Bam HI和Sal I双酶切,连接含有His标签的pET21b质粒Bam HI和Sal I双酶切片段上,转化E. coli TG1菌株,获得重组质粒pETbkdR。重组质粒经PCR、酶切和测序鉴定,转化至E. coli BL21(DE3)菌株,获得表达菌株BL21(pETbkdR)。
1.4 蛋白表达纯化与凝胶迁移实验BkdR蛋白和CcpA蛋白的表达与纯化见文献[29]。以Bt HD73为模板,用带有FAM (羧基荧光素)标记的引物(表 2)扩增Pptb片段(345 bp)。凝胶阻滞实验(EMSA,Electrophoresis mobility shift assays)确定Pptb片段与BkdR蛋白和CcpA蛋白的结合,方法见文献[29]。
1.5 β-半乳糖苷酶活性测定Bt菌株过夜活化,1%转接至50 mL LB培养基,30 ℃、220 r/min振荡培养至对数生长期(OD600=2.0),取1 mL菌液加入到100 mL M9培养基(含有终浓度为17 mmol/L的葡萄糖),30 ℃、220 r/min振荡培养至OD600=0.1 (约3 h),分别加入终浓度为3 mmol/L的氨基酸(亮氨酸、异亮氨酸、缬氨酸),继续培养2 h后开始取样(记为A2),每2 h取样1次,共取9个点,每次取样10 mL,离心收集菌体,保留沉淀,β-半乳糖苷酶活性测定见参考文献[22],每组数据至少独立重复3次。
2 结果和分析 2.1 bkd基因簇的诱导转录活性分析为了研究bkd基因簇的诱导转录活性,构建了bkd基因簇的启动子Pptb融合lacZ基因的表达载体pHTPptb,分别电击转入Bt HD73菌株和bkdR突变体,获得菌株HD(Pptb)和ΔbkdR(Pptb)。β-半乳糖苷酶活性测定表明,在分别含有3 mmol/L亮氨酸、异亮氨酸和缬氨酸的M9培养基中,Pptb启动子在Bt HD73出发菌株中的转录活性明显高于不含氨基酸的M9培养基[图 1-A,HD(Pptb)],说明在基础培养基中,亮氨酸、异亮氨酸和缬氨酸可诱导Pptb的转录活性。而在含有3 mmol/L亮氨酸、异亮氨酸或缬氨酸的M9培养基中,Pptb启动子在bkdR突变体中的转录活性均显著下降[图 1-B,ΔbkdR(Pptb)],说明Pptb的诱导转录活性受BkdR的正调控。
|
| 图 1 Pptb的转录活性分析 Figure 1 Analysis of transcription of Pptb. A: Transcription of Pptb induced with different amino acid; B: comparison of transcription of HD(Pptb) and ΔbkdR(Pptb) induced with different amino acid at A4 point. The OD600 reached 1.0 (mid-exponential phase), that defined as A0, and An is n hours after A0. A4 is 4 hours after A0 |
2.2 BkdR对bkd基因簇的转录调控作用
为了进一步明确BkdR对bkd基因簇的转录调控作用,构建Bt HD73菌株的BkdR表达纯化载体,PCR扩增bkdR基因全长并与含有His标签的pET21b载体连接,转化E. coli TG1菌株,获得重组质粒pETbkdR,经PCR鉴定得到2040 bp的条带(图 2-A),经Bam HI和Sal I双酶切鉴定得到5.4 kb大小的载体条带和2040 bp左右大小的目的片段条带(图 2-A),经测序分析和序列比对表明重组质粒pETbkdR构建正确。重组质粒转化至E. coli BL21菌株,获得BL21(pETbkdR)表达菌株,经IPTG诱导表达,超声破碎细胞,离心收集可溶性组分,对BkdR-His融合蛋白进行Ni2+螯合琼脂糖亲和纯化,SDS-PAGE结果表明所得蛋白纯度较高,分子量大小约为77 kDa (图 2-B)。

|
| 图 2 BkdR表达载体构建及纯化 Figure 2 Construction and purification of BkdR. A: Identification of pETbkdR plasmid; M: DNA marker. B: purification of BkdR-His fusion protein; M: protein marker |
利用FAM标记的引物Pptb-1和Pptb-2扩增bkd基因簇的启动子片段(Pptb,345 bp),与纯化的BkdR-His蛋白进行体外结合实验,结果表明(图 3),凝胶底部的条带为20 ng带标记的自由DNA,上层为DNA与蛋白结合的条带,随着BkdR蛋白浓度的增加,底部未结合的DNA条带浓度越来越低,上层与蛋白结合的条带浓度逐渐升高,说明Pptb与BkdR蛋白有结合作用;而加入500倍浓度非标记的DNA可以与标记的DNA产生竞争作用(图 3,lane 7);阴性对照为20 ng cwlC基因[33] (cwlC基因编码母细胞水解酶,其转录受SigmaK因子控制)的启动子与0.37 μg BkdR蛋白无结合作用(图 3,条带8)。以上结果说明Pptb与BkdR蛋白具有特异性的结合作用,这些结果表明Pptb的转录受BkdR的直接调控。

|
| 图 3 BkdR与Pptb的结合 Figure 3 Binding of BkdR and Pptb. Lane 1: FAM-labeled DNA probe; lanes 2–6: incubation of the probe with increasing concentrations of purified BkdR indicated at the top of the figure; lane 7: incubation of labeled DNA and 500-fold unlabeled DNA with 0.37 μg BkdR; lane 8: negative control: incubation of 20 ng cwlC promoter with 0.37 μg BkdR |
2.3 CcpA对bkd基因簇的转录调控作用
前期研究表明,Pptb序列中包含一段序列(TTGAATGCGTTTTCA)与受CcpA调控的一致序列(WTGNAANCGNWNNCW)相似,为了明确CcpA是否对Pptb的转录有调控作用,构建了ccpA突变体,并将Pptb融合lacZ基因的表达载体pHTPptb电击转入ccpA突变体中,获得菌株ΔccpA(Pptb)。β-半乳糖苷酶活性测定表明,在SSM培养基中(图 4-A),HD(Pptb)菌株与ΔccpA(Pptb)菌株的转录活性无明显差异;而在含有葡萄糖(17 mmol/L)和3 mmol/L氨基酸的M9培养基中(图 4-B,C,D),ΔccpA(Pptb)菌株的转录活性显著高于HD(Pptb)菌株的活性,这些结果说明,在葡萄糖培养基中,Pptb的诱导转录活性受CcpA的负调控,而在普通培养基中,Pptb的基础转录活性不受CcpA的调控。
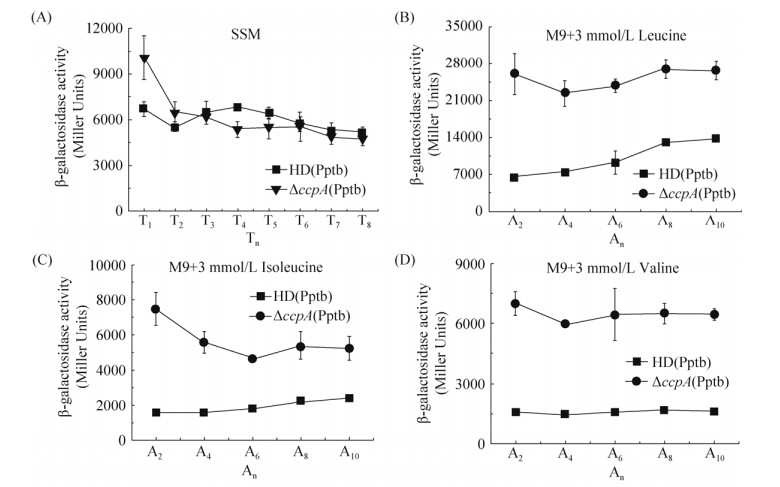
|
| 图 4 HD(Pptb)菌株与ΔccpA(Pptb)菌株的转录活性比较 Figure 4 Comparison of transcription of HD(Pptb) and ΔbkdR(Pptb). A: SMM; B: M9 with glucose and 3 mmol/L leucine; C: M9 with glucose and 3 mmol/L isoleucine; D: M9 with glucose and 3 mmol/L valcine. The OD600 reached 1.0 (mid-exponential phase), that defined as A0, and An is n hours after A0 |
为了进一步明确CcpA对Pptb是否存在直接调控作用,应用前期构建的CcpA表达菌株BL21(pETccpA)[29],经IPTG诱导表达和Ni2+螯合琼脂糖亲和纯化,获得CcpA-His融合蛋白。利用FAM标记的引物Pptb-1和Pptb-2扩增得到的bkd基因簇的启动子片段(Pptb,345 bp),与纯化的CcpA-His蛋白进行体外结合实验,结果表明(图 5),底部带标记的20 ng自由DNA条带浓度越来越低,上层DNA与蛋白结合的条带浓度逐渐升高,说明Pptb与CcpA蛋白有结合作用,而加入500倍浓度非标记的DNA可以与标记的DNA产生竞争结合作用(图 5,lane 7),说明Pptb与CcpA蛋白具有特异性的结合作用,这些结果表明Pptb的转录受CcpA的直接调控。

|
| 图 5 CcpA与Pptb的结合 Figure 5 Binding of CcpA and Pptb. Lane 1: FAM-labeled DNA probe; lanes 2–6: incubation of the probe with increasing concentrations of purified CcpA indicated at the top of the figure; lane 7: incubation of labeled DNA and 500-fold unlabeled DNA with 0.26 μg CcpA |
3 讨论
本实验室前期研究发现,Bt HD73菌株中bkd基因簇的启动子Pptb的转录受Sigma54因子的控制,并受BkdR的正调控[27],本研究进一步发现,Pptb的转录受亮氨酸、异亮氨酸和缬氨酸的诱导,该诱导活性受BkdR直接调控,这种调控模式与Bs中的bkd基因簇的转录相似[10]。本研究通过EMSA实验,进一步明确了纯化的BkdR蛋白和CcpA蛋白与Pptb启动子均存在结合作用,为bkd基因簇的转录调控机制提供了新的证据。BkdR蛋白是增强子结合蛋白,具有3个典型的结构域(图 6-A):N端的PAS信号结构域,可能与识别亮氨酸、异亮氨酸和缬氨酸信号有关;中间的AAA+结构域,具有保守氨基序列GAFTGA,可能与Sigma54因子进行相互作用;C端的HTH结构域,可能识别Pptb启动子上的回文序列(图 6-B)并与之相结合。Pptb启动子序列中存在一个典型的受Sigma54控制的保守–12/–24序列(图 6-B),Sigma54可能通过与该序列结合从而起始转录。Pptb启动子序列还存在一个CcpA识别的保守序列,CcpA可能通过与该序列结合从而起到调控功能。

|
| 图 6 BkdR氨基酸结构域分析(A)和Pptb启动子序列分析(B) Figure 6 Sequence analysis. A: Analysis of BkdR domain; B: Analysis of Pptb sequence |
本研究通过EMSA实验和β-半乳糖苷酶活性分析实验表明,在葡萄糖培养基中,bkd基因簇的诱导活性受CcpA的直接负调控,而在Bs中,没有相关报道显示bkd基因簇的转录与CcpA相关,通过在DBTBS数据库(http://dbtbs.hgc.jp/)中检索Bs 168菌株基因组中ptb基因上游启动子序列的转录调控因子结合位点,也没有发现与CcpA结合的cre位点。但是Bs中与支链氨基酸生物合成相关的ilv-leu操纵子受CcpA的调控。ilv-leu操纵子编码酶系统参与催化从丙酮酸到支链氨基酸的生物合成,将碳代谢与氨基酸合成偶联起来[34-35]。在Bt中,CcpA可能调控了支链氨基酸的合成及降解代谢过程。
| [1] | Lorca GL, Chung YJ, Barabote RD, Weyler W, Schilling CH, Saier MH, J r. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. Journal of Bacteriology, 2005, 187(22): 7826-7839. DOI:10.1128/JB.187.22.7826-7839.2005 |
| [2] | Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell, 2004, 118(6): 731-741. DOI:10.1016/j.cell.2004.08.027 |
| [3] | Kim HM, Waters A, Turner ME, Rice KC, Ahn SJ. Regulation of cid and lrg expression by CcpA in Streptococcus mutans. Microbiology, 2019, 165(1): 113-123. |
| [4] | Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology, 2010, 156(Pt 10): 3136-3147. |
| [5] | Majidian P, Kuse J, Tanaka K, Najafi H, Zeinalabedini M, Takenaka S, Yoshida KI. Bacillus subtilis GntR regulation modified to devise artificial transient induction systems. Journal of General and Applied Microbiology, 2017, 62(6): 277-285. |
| [6] | Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiology and Molecular Biology Reviews: MMBR, 2012, 76(3): 497-529. DOI:10.1128/MMBR.00006-12 |
| [7] | Studholme DJ, Dixon R. Domain architectures of sigma54-dependent transcriptional activators. Journal of Bacteriology, 2003, 185(6): 1757-1767. DOI:10.1128/JB.185.6.1757-1767.2003 |
| [8] | Cases I, Ussery DW, de Lorenzo V. The sigma54 regulon (sigmulon) of Pseudomonas putida. Environmental Microbiology, 2003, 5(12): 1281-1293. DOI:10.1111/j.1462-2920.2003.00528.x |
| [9] | Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Debarbouille M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. Journal of Bacteriology, 1994, 176(5): 1234-1241. DOI:10.1128/jb.176.5.1234-1241.1994 |
| [10] | Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. Journal of Bacteriology, 1999, 181(7): 2059-2066. |
| [11] | Debarbouille M, Martin-Verstraete I, Klier A, Rapoport G. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both sigma 54- and phosphotransferase system-dependent regulators. Proceedings of the National Academy of Sciences of the United States of America, 1991, 88(6): 2212-2216. DOI:10.1073/pnas.88.6.2212 |
| [12] | Ali NO, Bignon J, Rapoport G, Debarbouille M. Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. Journal of Bacteriology, 2001, 183(8): 2497-2504. DOI:10.1128/JB.183.8.2497-2504.2001 |
| [13] | Batchelor JD, Sterling HJ, Hong E, Williams ER, Wemmer DE. Receiver domains control the active-state stoichiometry of Aquifex aeolicus sigma54 activator NtrC4, as revealed by electrospray ionization mass spectrometry. Journal of Molecular Biology, 2009, 393(3): 634-643. DOI:10.1016/j.jmb.2009.08.033 |
| [14] | de Las Heras A, Martinez-Garcia E, Domingo-Sananes MR, Fraile S, de Lorenzo V. Rationally rewiring the connectivity of the XylR/Pu regulatory node of the m-xylene degradation pathway in Pseudomonas putida. Integrative Biology (Camb), 2016, 8(4): 571-576. DOI:10.1039/C5IB00310E |
| [15] | Osadnik H, Schopfel M, Heidrich E, Mehner D, Lilie H, Parthier C, Risselada HJ, Grubmuller H, Stubbs MT, Bruser T. PspF-binding domain PspA1-144 and the PspA.F complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins. Molecular Microbiology, 2015, 98(4): 743-759. DOI:10.1111/mmi.13154 |
| [16] | Jouzani GS, Valijanian E, Sharafi R. Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Applied Microbiology Biotechnology, 2017, 101(7): 2691-2711. DOI:10.1007/s00253-017-8175-y |
| [17] | Schnepf E, Crickmore N, van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiology and Molecular Biology Reviews: MMBR, 1998, 62(3): 775-806. |
| [18] | Peng Q, Wang G, Liu G, Zhang J, Song F. Identification of metabolism pathways directly regulated by sigma(54) factor in Bacillus thuringiensis. Frontiers in microbiology, 2015, 6: 407. |
| [19] | Zhu L, Peng Q, Song F, Jiang Y, Sun C, Zhang J, Huang D. Structure and regulation of the gab gene cluster, involved in the gamma-aminobutyric acid shunt, are controlled by a sigma54 factor in Bacillus thuringiensis. Journal of Bacteriology, 2010, 192(1): 346-355. DOI:10.1128/JB.01038-09 |
| [20] | Peng Q, Yang M, Wang W, Han L, Wang G, Wang P, Zhang J, Song F. Activation of gab cluster transcription in Bacillus thuringiensis by gamma-aminobutyric acid or succinic semialdehyde is mediated by the sigma 54-dependent transcriptional activator GabR. BMC Microbiology, 2014, 14: 306. DOI:10.1186/s12866-014-0306-3 |
| [21] | Zhang Z, Yang M, Peng Q, Wang G, Zheng Q, Zhang J, Song F. Transcription of the Lysine-2, 3-Aminomutase Gene in the kam locus of Bacillus thuringiensis subsp. kurstaki HD73 is controlled by both sigma54 and sigmaK factors. Journal of Bacteriology, 2014, 196(16): 2934-2943. DOI:10.1128/JB.01675-14 |
| [22] | Peng Q, Liu C, Wang B, Yang M, Wu J, Zhang J, Song F. Sox transcription in sarcosine utilization is controlled by sigma(54) and SoxR in Bacillus thuringiensis HD73. Scientific Reports, 2016, 6: 29141. DOI:10.1038/srep29141 |
| [23] | Denoya CD, Fedechko RW, Hafner EW, McArthur HA, Morgenstern MR, Skinner DD, Stutzman-Engwall K, Wax RG, Wernau WC. A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. Journal of Bacteriology, 1995, 177(12): 3504-3511. DOI:10.1128/jb.177.12.3504-3511.1995 |
| [24] | Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiological Reviews, 1991, 55(2): 288-302. |
| [25] | Ward DE, van der Weijden CC, van der Merwe MJ, Westerhoff HV, Claiborne A, Snoep JL. Branched-chain alpha-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. Journal of Bacteriology, 2000, 182(11): 3239-3246. DOI:10.1128/JB.182.11.3239-3246.2000 |
| [26] | Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Molecular Microbiology, 1995, 16(2): 171-175. DOI:10.1111/j.1365-2958.1995.tb02290.x |
| [27] |
Wang GN, Peng Q, Zheng QY, Li J, Zhang J, Song FP. Transcriptional regulation of bkd gene cluster in Bacillus thuringiensis. Acta Microbiologica Sinica, 2014, 54(10): 1129-1137.
(in Chinese) 王冠男, 彭琦, 郑庆云, 李杰, 张杰, 宋福平. 苏云金芽胞杆菌bkd基因簇的转录调控. 微生物学报, 2014, 54(10): 1129-1137. |
| [28] | Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proceedings of the National Academy of Sciences of the United States of America, 1965, 54(3): 704-711. DOI:10.1073/pnas.54.3.704 |
| [29] |
Cheng HJ, Peng Q, Zhang J, Song FP. Identification of genes regulated by Sigma54 and CcpA in Bacillus thuringiensis. Acta Microbiologica Sinica, 2018, 58(3): 380-390.
(in Chinese) 程海舰, 彭琦, 张杰, 宋福平. 苏云金芽胞杆菌Sigma54和CcpA共同调控的基因鉴定. 微生物学报, 2018, 58(3): 380-390. |
| [30] | Liu G, Song L, Shu C, Wang P, Deng C, Peng Q, Lereclus D, Wang X, Huang D, Zhang J, et al. Complete genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD73. Genome announcements, 2013, 1(2): e0008013. DOI:10.1128/genomeA.00080-13 |
| [31] | Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Applied and Environmental Microbiology, 2004, 70(11): 6887-6891. DOI:10.1128/AEM.70.11.6887-6891.2004 |
| [32] | Lereclus D, Arantes O, Chaufaux J, Lecadet M. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiology Letters, 1989, 51(1): 211-217. |
| [33] | Chen X, Gao T, Peng Q, Zhang J, Chai Y, Song F. Novel cell wall hydrolase CwlC from Bacillus thuringiensis is essential for mother cell lysis. Applied and Environmental Microbiology, 2018, 84(7). |
| [34] | Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Molecular Microbiology, 2005, 56(6): 1560-1573. DOI:10.1111/j.1365-2958.2005.04635.x |
| [35] | Ludwig H, Meinken C, Matin A, Stulke J. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. Journal of Bacteriology, 2002, 184(18): 5174-5178. DOI:10.1128/JB.184.18.5174-5178.2002 |
 2019, Vol. 59
2019, Vol. 59