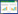中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 周卫国, 丁德文, 凌娟, 林显程, 杨清松, 张颖, Manzoor Ahmad, 张燕英, 董俊德. 2019
- Weiguo Zhou, Dewen Ding, Juan Ling, Xiancheng Lin, Qingsong Yang, Ying Zhang, Manzoor Ahmad, Yanying Zhang, Junde Dong. 2019
- 海草根际微生物与海草植株的互作效应
- Seagrass-microbial interactions in the rhizosphere
- 微生物学报, 59(11): 2117-2129
- Acta Microbiologica Sinica, 59(11): 2117-2129
-
文章历史
- 收稿日期:2018-11-27
- 修回日期:2019-01-22
- 网络出版日期:2019-03-13
2. 中国科学院大学, 北京 100049;
3. 中国科学院海南热带海洋生物实验站, 海南 三亚 572000
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Tropical Marine Biological Research Station in Hainan, Chinese Academy of Sciences, Sanya 572000, Hainan Province, China
海草是海洋中唯一开花的被子植物,生长于潮间带区域,由海草组成的海草床生态系统,同珊瑚礁、红树林生态系统一样,是海洋中生产力水平最高的生态系统之一[1]。海草床具有极其重要的生态服务价值,包括改变水向、营养物质循环以及食物链的结构;净化水质、稳固海床、减缓波浪和潮流;为很多海洋动物如海龟和经济鱼类提供主要的食物来源;固定沉积物并提供大量的有机碳[2-6]。微生物是海草床生态系统中重要的组成成分,它们附着于海草叶子上,或者生活于海草根际,参与生态系统中重要的生物地球化学循环过程,包括碳、氮、磷和硫等循环[7-9]。
植物根际是受植物根系分泌的化学物质如溶解有机碳直接影响的狭窄土壤区域(1-2 mm)[10],分布有数目庞大的微生物,并与植物间有密切的联系,其基因数量远超过植物本身的基因,因此,根际微生物组也被称为植物的第二套基因组。根际微生物在植物的生长发育过程中起着重要的作用,在一定程度上可以提高植物抗逆性和生产力、调控根际沉积物中的生源要素的生物地球化学循环,并可以通过调节根际微生物的生态环境来促进植物的生长[11-13]。
海草生长的沉积物处于一种极度厌氧、高度还原的环境,这主要是由沉积物中富集的植物性毒素H2S所引起的[14]。该硫化物是来自于硫还原细菌的活性产物,硫还原细菌利用硫酸盐作为电子受体进行有机物的再矿化作用[15]。大量的电子受体及海草的根部分泌物会促使海草根际发生大量的由微生物介导的氧化还原过程,从而塑造出各种复杂微生物群落的微环境,包含很多功能性群体如硫还原细菌、固氮细菌和产乙酸菌等[16-17]。
近些年来,分子生物学技术在海草根际微生物多样性研究中具有广泛的应用[18-20]。江玉凤等应用PCR-DGGE技术发现南海新村湾海草沉积物中变形菌门为主要的优势种群,其比例高达70.87%[21]。Cucio等通过16S rRNA基因扩增子测序的方式研究发现海草根际沉积物主要由参与硫循环的细菌所介导,并且其根际沉积物微生物结构组成在小尺度区域内由植物所塑造,而在大尺度区域内则由环境因子所主导[8]。Brodersen等应用16S rRNA基因高通量测序发现人工海草沉积物与自然海草沉积物中微生物群落组成是相似的,硫还原细菌和硫氧化细菌在海草根际沉积物中占据较高的丰度[20]。然而,关于海草室内培养培养过程中根际沉积物微生物的变化过程,以及海草对于高温处理后的沉积物的塑造,目前仍没有相关的研究。
因此,在本研究中,我们以泰来草为研究对象,采集原位海草和沉积物于实验室内进行模拟培养,定期收集泰来草根际沉积物,对其16S rRNA进行Illumina高通量测序和基因丰度研究,分析高温处理后的沉积物在海草定殖后其根际沉积物微生物结构组成的变化和响应过程,探索微生物群落变化的原因,为进一步研究根际微生物参与海草床中功能循环的作用,及保护海草床生态系统提供理论基础。
1 材料和方法 1.1 样品采集和分装泰来草及原位沉积物于2017年5月采集自三亚湾鹿回头珊瑚礁区域(18°12'19'' N,109°28'27'' E),该区域在夏季会受到冷水上升流的影响,引起营养盐浓度的上升[22]。在海水退潮后使用无菌铲采集泰来草和沉积物,将采集的一半海草沉积物于120 ℃下高温处理20 min,设置为实验处理组;将对照组和实验组沉积物分别分装于50 L培养缸中,分别设置两个生物学重复。随后移入采集的原位泰来草,保持每一培养缸中海草的数量和生长状况大致相同,于粗过滤后的原位海水(去除大型颗粒物)在中国科学院海南热带生物实验站培养。
于海草床处收集2 L海水经0.2 μm孔径滤膜(Millipore,USA)过滤,过滤后的海水根据国标法测定四项营养盐(硝酸盐、铵盐、亚硝酸盐、磷酸盐);原位海水处环境参数,包括pH、盐度、溶解氧和电导率,通过水质分析仪Orion Star A329 (Thermo Scientific,USA)进行测定。
1.2 室内海草培养体系构建泰来草培养体系于新环境适应2周后,移入实验室内,每一培养缸装配有水泵和12h:12h昼夜循环的LED灯,实验时,(1)定期使用水质分析仪监测培养缸内海水pH、盐度和溶解氧的变化;(2)培养初期(0)、7 d (7)和35 d (35) 3个时间段分别采集泰来草根际沉积物和收集海水,其中根际沉积物样品设置有5个生物学重复,海水样品设置2个生物学重复,共收集30个根际沉积物和12个海水营养盐样品,采集后的根际沉积物样品加入DNA/RNA保护剂后(TaKaRa,Japan)迅速于-80 ℃保存,0.2 μm过滤后的海水于-20 ℃储存用于后续营养盐测定;(3)泰来草室内培养35 d后,使用Diving-pam (Walz,Germany)测定海草叶的光合曲线。
1.3 根际沉积物DNA提取和16S rRNA荧光定量使用E.Z.N.A.®土壤DNA试剂盒(Omega,USA)提取1.0 g根际沉积物群落获得群落总DNA,通过NanoDrop 2000 (Thermo Scientific,USA)测定DNA浓度。使用带barcode的引物341F (5′-CCTAYGGGRBGCASCAG-3′)和806R (5′-GGA CTACNNGGGTATCTAAT-3′)[23]扩增16S rRNA基因的V3+V4区,PCR反应体系为25 μL:1 μL目的DNA,0.5 μL正向引物,0.5 μL反向引物,12.5 μL rTaq酶(TaKaRa,Japan),10 μL无菌水和0.5 μL 1% BSA (TaKaRa,Japan)。PCR反应条件为:96 ℃ 2 min;96 ℃ 30 s,53 ℃ 30 s,72 ℃ 2 min,35个循环;72 ℃ 7 min。
16S rRNA的基因拷贝数的定量是在荧光定量PCR仪Light Cycler 480 (Roche Diagnostics,Germany)上进行的。引物27F (5′-GAGTTTGATC MTGGCTCAG-3′)和518R (5′-ATTACCGCGGCTG CTGG-3′)用于16S rRNA基因的扩增,标准曲线用导入16S rRNA基因的质粒稀释浓度为103拷贝/μL到108拷贝/μL生成,生成的标准曲线扩增效率为1.97,每一样品设置3个重复,通过琼脂凝胶电泳(2%)来确认扩增产物的片段长度和扩增的特异性。样品的16S rRNA拷贝数通过荧光定量PCR仪自带的软件分析完成。
1.4 16S rRNA高通量测序和生物信息学分析基于Illumina Hiseq测序平台进行双末端测序获得原始序列后,去除序列两端的barcode和引物序列,使用FLASH软件[24]将双末端序列拼接,应用Qiime软件[25]和UCHIME算法[26]处理数据,从而获得有效序列(effective tags)。然后使用USEARCH进行聚类,将序列相似性达到97%的序列聚类成一个分类操作单元(OTU)。每一OTU的代表性序列通过Krona软件进行物种注释[27],并对比Silva数据库对OTU代表性序列进行系统学归类[28]。
1.5 数据分析基于OTU的分析结果,使用Qiime软件计算alpha多样性指标,包括Shannon、Simpson和Chao指数,用来反映微生物群落的多样性;应用软件PRIMER v.6 (PRIMER-E)进行beta多样性分析,其中,主坐标分析(PCoA)是为了揭示不同处理及不同时间作用下海草根际沉积物微生物群落的差异。主坐标典型分析图(CAP)是用来分析微生物群落组成和水体环境参数间的线性关系;采用SPSS v19.0 (IBM)进行方差分析,用于检测对照组和处理组不同时间段内海水营养盐间的差异以及根际沉积物16S rRNA基因丰度的显著性。
1.6 核苷酸序列登录号本研究中高通量测序得到的序列均提交到NCBI的SRA数据库中,登录号为PRJNA506337。
2 结果和分析 2.1 水体环境参数和泰来草生理指标我们测定了3个时间段内(0,7,35 d)海草培养体系中海水的营养盐参数,包括磷酸盐、硝酸盐、亚硝酸盐和铵盐。表 1中可以看出,高温处理组中海水的磷酸盐、亚硝酸盐和铵盐含量在培养过程中均显著高于对照组。海水中硝酸盐含量在高温处理组中,第一周发生明显下降,随后显著性地上升(P < 0.01)。同时,我们追踪了海草培养体系中海水pH和溶解氧的变化,从图 1中,我们看出海水中pH和溶解氧在培养过程中呈现动态的变化,高温处理组中海水pH明显高于对照组,两者中海水的溶解氧水平无明显的差异。
| Index | Control | Treatment | |||||
| Day 0 | Day 7 | Day 35 | Day 0 | Day 7 | Day 35 | ||
| Phosphate/(mg/L) | 0.006±0a | 0.025±0.006b | 0.026±0.002b | 0.006±0a | 0.034±0.004c | 0.034±0.008c | |
| Nitrate/(mg/L) | 0.050±0.001a | 0.044±0.001a | 0.047±0.006a | 0.050±0.001a | 0.006±0.003b | 0.725±0.022c | |
| Nitrite/(mg/L) | 0.001±0a | 0.010±0b | 0.005±0.002ab | 0.001±0a | 0.023±0.001c | 0.113±0.008d | |
| Ammonium/(mg/L) | 0.050±0.001a | 0.010±0.003b | 0.020±0.006ab | 0.005±0.001a | 0.030±0.011ab | 0.230±0.050c | |
| Different letters represent significant differences (P < 0.05). | |||||||
|
| 图 1 随着时间的变化泰来草培养体系中海水pH和RDO的变化 Figure 1 Variation of pH and RDO in seawater (n=6) in seagrass culturing system. RDO, dissolved oxygen. |
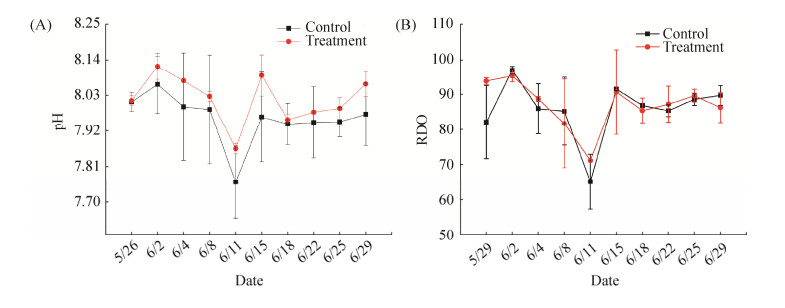
在培养泰来草35 d后,我们通过测定泰来草叶子的光合曲线来反映其光合作用能力,从图 2中可以看出,对照组泰来草叶的光合曲线斜率高于高温处理组泰来草,同时,对照组最大电子传递速率(ETRmax)最高,说明对照组泰来草对光的利用能力以及能吸收的最大光强要优于高温处理组泰来草。
|
| 图 2 泰来草叶光合有效辐射(PAR)VS电子传递速率(ETR)的光合曲线 Figure 2 Photosynthetically active radiation (PAR) vs. electron transport rate (ETR) plotted from light curves for Thalassia hemperichii (n=3) after culturing for 35 days (expressed as mean value and standard error, SE). |
2.2 泰来草根际沉积物16S rRNA基因丰度
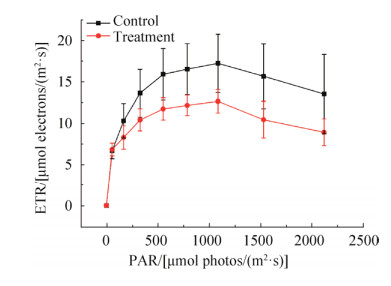
泰来草根际沉积物16S rRNA基因拷贝数分布为每克沉积物2.36×106到3.84×108拷贝数之间。从图 3可以看出,对照组泰来草根际沉积物16S rRNA基因拷贝数在培养一个月过程中没有发生显著变化,高温处理组泰来草根际沉积物16S rRNA基因拷贝数在培养第一周显著上升,随后又发生明显下降,培养35 d后的海草根际沉积物16S rRNA基因拷贝数要显著高于初始拷贝数。同时,高温处理组根际沉积物16S rRNA基因拷贝数均显著低于对照组。
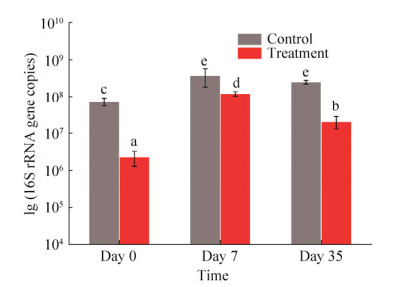
|
| 图 3 泰来草根际沉积物不同时间段内16S rRNA基因丰度 Figure 3 Abundance of 16S rRNA gene copy number from Thalassia hemperichii rhizosphere (n=6) in different time series. Different letters above the columns represent significant differences (P < 0.05). Day 0: at the beginning; Day 7: culturing for 7 days; Day 35: culturing for 35 days. |
2.3 泰来草根际沉积物微生物多样性和结构组成
V3+V4区测序的结果共获得2394088个有效数据。每一样品的OTU数目分布为3018到4106之间,其中初始时间高温处理沉积物组(T0)有着最少的OTU数。Shannon指数分布为6.76-9.07,Chao系数分布为2118.36-2648.28,Simpson指数分布在0.94-0.99之间。随着室内培养时间的增加,对照组的Shannon指数先上升随后降低,高温处理组的Shannon指数则不断增加。因此,对照组泰来草根际沉积物的微生物多样性要高于高温处理沉积物组,而且高温处理组沉积物在泰来草定殖后多样性在不断增加。
| Index | Control | Treatment | |||||
| C0 | C7 | C35 | T0 | T7 | T35 | ||
| OTU | 3952 | 4106 | 3623 | 3018 | 3441 | 3562 | |
| Shannon | 7.74 | 8.88 | 8.92 | 6.76 | 7.86 | 8.66 | |
| Chao | 2609.72 | 2490.03 | 2559.78 | 2118.36 | 2428.85 | 2463.41 | |
| Simpson | 0.97 | 0.99 | 0.99 | 0.94 | 0.97 | 0.99 | |
如图 4所示,从门水平上来看,初始阶段泰来草根际沉积物的优势种群为厚壁菌门(43.8%)和变形菌门(31.95%),随后变形菌门数量继续增加,厚壁菌门大幅度减少,由蓝细菌门和放线菌门取代占据主导地位;初始高温处理后的根际沉积物的微生物群体主要是厚壁菌门(32.4%)、梭杆菌门(27.21%)和变形菌门(22.92%),随着培养时间的增加,厚壁菌门和梭杆菌门数量在泰来草根际沉积物中逐渐缩小。
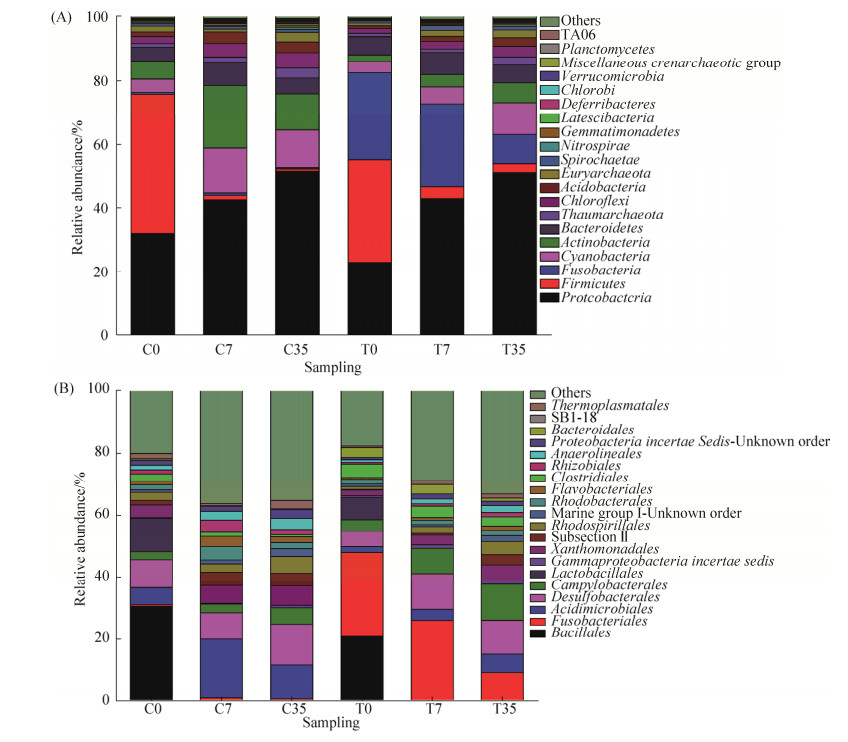
|
| 图 4 泰来草根际沉积物在门水平(A)和目水平下(B)微生物组成 Figure 4 Total 16S rRNA gene community composition at phylum level (A) and order level (B) in Thalassia hemperichii rhizosphere. C0: control at day 0; C7: control culturing for 7 days; C35: control culturing for 35 days; T0: treatment at day 0; T7: treatment culturing for 7 days; T35: treatment culturing for 35 days. |
从目水平上可以看出,不管是对照组还是高温处理组,一周后芽孢杆菌目丰度显著降低,随着培养时间的增加,对照组中酸微菌目、脱硫菌目和弯曲菌目丰度增加,乳杆菌目丰度降低,高温处理组有着高丰度的梭杆菌目,但培养过程中数目不断降低,由初始阶段的27.21%降低为35 d后的9.05%,酸微菌目、脱硫菌目、弯曲菌目、黄色单胞菌目和红螺菌目丰度增加,乳杆菌目丰度降低。
如图 5所示,在科水平上,链球菌科(Streptococcaceae)、内杆菌科(Carnobacteriaceae)、芽孢杆菌科(Bacillaceae)和动球菌科(Planococcaceae)仅存在于初始时间样品中,在高温处理组中,梭杆菌科(Fusobacteriaceae)随着培养的过程中不断减少,而螺杆菌科(Helicobacteraceae)和脱硫杆菌科(Desulfobacteraceae)丰度则不断增加(图 5)。
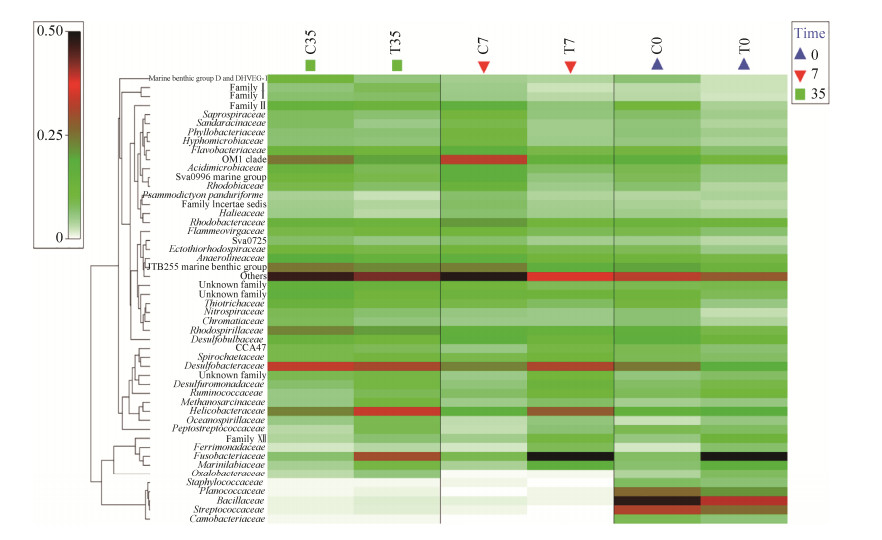
|
| 图 5 不同样品在科水平下的丰度热图 Figure 5 The heatmap plot depicts 16S rRNA gene abundance of each sample at the family level. The relative values for bacterial are depicted by color density with the legend indicated at the left of the figure. C0: control at day 0; C7: control culturing for 7 days; C35: control culturing for 35 days; T0: treatment at day 0; T7: treatment culturing for 7 days; T35: treatment culturing for 35 days. |
2.4 16S rRNA基因群落和环境因子间的关系
主坐标分析(PCoA)(图 6)表明泰来草根际沉积物16S rRNA基因群落根据不同的处理(原位和高温处理)和培养的时间(0、7、35 d)分成不同的簇。主坐标典型分析图(CAP)表明(图 7),35 d培养后的高温处理泰来草根际沉积物微生物群体与磷酸盐、硝酸盐、亚硝酸盐和铵盐呈正相关,pH同初始时期的高温处理组根际沉积物微生物群体呈正相关。
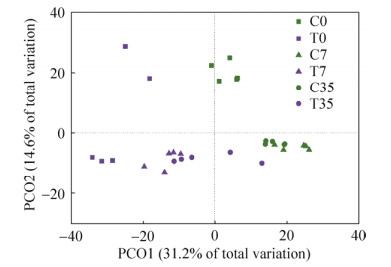
|
| 图 6 基于高通量测序数据的16S rRNA基因群落的主坐标分析(PCoA) Figure 6 Principal coordinates analysis (PCoA) of total 16S rRNA gene communities based on high-throughput sequencing data. The percentage of variation explained by each axis is shown. C0: control at day 0; C7: control culturing for 7 days; C35: control culturing for 35 days; T0: treatment at day 0; T7: treatment culturing for 7 days; T35: treatment culturing for 35 days. |
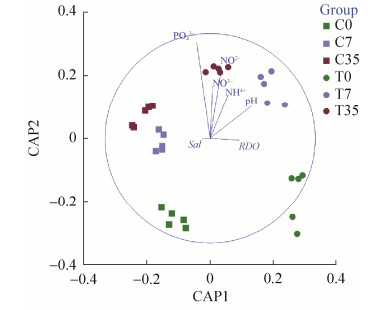
|
| 图 7 泰来草根际沉积物微生物群落组成和水体参数的主坐标典型分析图(CAP) Figure 7 CAP ordination biplot showing the relationships between environmental variables and total 16S rRNA gene communities in Thalassia hemperichii rhizosphere. RDO: dissolved oxygen. Sal: salinity. PO3-: phosphate. NO3-: nitrate. NO2-: nitrite. NH4+: ammonium. C0: control at day 0; C7: control culturing for 7 days; C35: control culturing for 35 days; T0: treatment at day 0; T7: treatment culturing for 7 days; T35: treatment culturing for 35 days. |
3 讨论
根际沉积物蕴含着丰富多样的微生物。本文研究目的在于海草定殖沉积物后其根际沉积物群落和丰度的变化。在本研究中,我们展示了高温作用下海草根际沉积物微生物多样性和结构组成,以及海草定殖后根际沉积物微生物群落的多样性、组成和结构的变化过程。本研究提高我们对海草床对其根际微生物的塑造的认识,对于海草生态系统的保护有着深刻的理解。
微生物群落能够通过改变其群落结构和组成来适应新的环境[29-31]。研究表明泰来草在室内暂养阶段厚壁菌门和变形菌门为根际沉积物中最丰富的微生物群体。高丰度的厚壁菌门主要是由沉积物中残留的动物排泄物所引起的,在本研究中主要是以链球菌科(Streptococcaceae)、内杆菌科(Carnobacteriaceae)、芽孢杆菌科(Bacillaceae)和动球菌科(Planococcaceae)的形式存在,是一种致病菌体。随着一段时间的培养,厚壁菌门大幅度减少,由变形菌门占据主导地位,由此可以看出海草的定殖能够显著地改变其根际沉积物的群落结构,并且能够显著降低病原菌的数量,有研究表明,由海草床所固定的沉积物对于疾病的改善起着至关重要的作用[32-33]。这与我们的研究结果是相一致的。
很多研究均有报道Delta变形菌纲和Gamma变形菌纲是很多海草和红树林根际沉积物中占主导的微生物群体[19, 34-35]。在本研究中,不管是对照组,还是高温处理组,泰来草根际沉积物微生物中具有优势地位的是Delta变形菌纲和Gamma变形菌纲,在室内培养35 d后,它们在对照组和高温处理组总微生物群落中分别占据32.31%和28.16%,说明它们在海草根际环境中起着非常重要的功能性作用。而且,本研究发现Delta变形菌纲中脱硫杆菌科(Desulfobacteraceae)随着培养时间的变化,其相对丰度在不断上升,脱硫杆菌科属于硫还原细菌,后者在硫酸盐还原、硫氧化和硝酸盐同化过程中起着尤为重要的作用[36-37]。隶属于Epison变形菌纲的螺杆菌科(Helicobacteraceae),在培养过程中其丰度在不断提高,螺杆菌科过去主要发现在较浅的冷泉沉积物中[38-39],属于硫氧化细菌[40],在海草生长过程中通过去除根际的H2S帮助海草免受毒素的损伤。我们的研究进一步加强了对硫还原细菌及硫氧化细菌在海草根际微生物组中占据的重要地位的认识。
与原位对照组相比较,高温处理组根际微生物中变形菌门、厚壁菌门和放线菌门丰度降低,放线菌门是陆地植物及海草根部常见的微生物类群,在抵抗病原体的过程中可能提供重要的保护机制,从而促进植物的免疫反应[41-42],高温处理后该类群丰度降低,意味着抵抗病原体的能力下降,随着海草定殖后,海草根际中放线菌门的丰度不断提高,从而能够提高海草的免疫防御能力;同时,高温处理组中具有独有的高丰度梭杆菌门,梭杆菌门是一种专性厌氧菌,在海湾口,特别是受过石油污染的沿岸沉积物中较为常见[43-44],厌氧的环境促使该类群丰度增加,而在海草定殖之后其数量又会显著下降。在以往的研究中,放线菌门、拟杆菌门在葡萄牙苏门答腊岛的大叶藻、海神草的根际沉积物中占有优势地位[8],蓝细菌门在南海新村湾海草沉积物微生物中属于第三大门类类群[21]。本研究中放线菌门、蓝细菌门和拟杆菌门在泰来草根际沉积物中也有着较高的分布。
qPCR的结果表明高温处理后的沉积物16S rRNA基因丰度显著性地低于对照组,在泰来草定殖后其微生物丰度呈现出先上升后降低的趋势。高温处理后的沉积物大幅度减少了沉积物中微生物的丰度,在海草定殖后,海草会塑造其根际微生物的群落结构[12, 45],使得原有的微生物数量短时间内大量增加,随后稳定维持在一定水平上。有研究表明,大叶藻生态系统沉积物中微生物基因拷贝数分布在(1.10±0.17)×109到(1.50±0.25)×109个拷贝数之间[45]。该结果要高于我们对照组,这可能是由于室内模拟原位环境培养过程中会降低沉积物中存在的微生物丰度。研究结果为模拟群落演替初期海草的定殖作用下根际沉积物微生物丰度变化的过程提供理论支持。
陆地植物的根际微生物结构受多重因素的影响,包括植物的品种、宿主的基因型、植物不同的发育期以及沉积物的类型等[46-48]。Cúcio研究表明在库拉特拉岛,沉积物的类型(粒径和有机质含量)、海草的品种并不是海草根际的决定性因子[8]。我们的研究结果发现,在泰来草室内培养35 d后,海水中营养盐指标在高温处理组中要显著高于对照组,这可能是由于高温处理后的沉积物被微生物降解后的结果,进而导致根际沉积物微生物群落发生变化。高温处理组中泰来草根际沉积物微生物群落同海水中硝酸盐、铵盐、亚硝酸盐和磷酸盐有着显著的正相关,其中,磷酸盐对根际沉积物微生物的群落组成具有最重要的效应,表明富营养化对微生物群落的变化有着显著性的影响。
致谢
感谢中国科学院海南热带生物实验站工作人员在海草的采集和室内的培养阶段提供的技术帮助。
| [1] | Costanza R, d'Arge R, de Groot R, Farber S, Grasso M, Hannon B, Limburg K, Naeem S, O'Neill RV, Paruelo J, Raskin RG, Sutton P, van den Belt M. The value of the world's ecosystem services and natural capital. Nature, 1997, 387(6630): 253-260. DOI:10.1038/387253a0 |
| [2] | Gacia E, Granata TC, Duarte CM. An approach to measurement of particle flux and sediment retention within seagrass (Posidonia oceanica) meadows. Aquatic Botany, 1999, 65(1/4): 255-268. |
| [3] | Cebrian J, Duarte CM. Detrital stocks and dynamics of the seagrass Posidonia oceanica (L.) delile in the Spanish mediterranean. Aquatic Botany, 2001, 70(4): 295-309. DOI:10.1016/S0304-3770(01)00154-1 |
| [4] | Orth RJ, Carruthers TJB, Dennison WC, Duarte CM, Fourqurean JW, Heck KL Jr, Hughes AR, Kendrick GA, Kenworthy WJ, Olyarnik S, Short FT, Waycott M, Williams SL. A global crisis for seagrass ecosystems. Bioscience, 2006, 56(12): 987-996. DOI:10.1641/0006-3568(2006)56[987:AGCFSE]2.0.CO;2 |
| [5] | Fourqurean JW, Duarte CM, Kennedy H, Marbà N, Holmer M, Mateo MA, Apostolaki ET, Kendrick GA, Krause-Jensen D, McGlathery KJ, Serrano O. Seagrass ecosystems as a globally significant carbon stock. Nature Geoscience, 2012, 5(7): 505-509. DOI:10.1038/ngeo1477 |
| [6] |
Liu SL, Jiang ZJ, Wu YC, Zhang JP, Huang XP. Nursery function of seagrass beds and its mechanisms. Acta Ecologica Sinica, 2015, 35(24): 7931-7940.
(in Chinese) 刘松林, 江志坚, 吴云超, 张景平, 黄小平. 海草床育幼功能及其机理. 生态学报, 2015, 35(24): 7931-7940. |
| [7] | Jensen SI, Kühl M, Priemé A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiology Ecology, 2007, 62(1): 108-117. DOI:10.1111/j.1574-6941.2007.00373.x |
| [8] | Cúcio C, Engelen AH, Costa R, Muyzer G. Rhizosphere microbiomes of european seagrasses are selected by the plant, but are not species specific. Frontiers in Microbiology, 2016, 7: 440. |
| [9] | Fahimipour AK, Kardish MR, Lang JM, Green JL, Eisen JA, Stachowicz JJ. Global-Scale structure of the eelgrass microbiome. Applied and Environmental Microbiology, 2017, 83(12): e03391-16. |
| [10] | Martens R. Contribution of rhizodeposits to the maintenance and growth of soil microbial biomass. Soil Biology and Biochemistry, 1990, 22(2): 141-147. DOI:10.1016/0038-0717(90)90078-E |
| [11] | Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends in Plant Science, 2012, 17(8): 478-486. DOI:10.1016/j.tplants.2012.04.001 |
| [12] | Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews, 2013, 37(5): 634-663. DOI:10.1111/1574-6976.12028 |
| [13] | Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, Hüttel B, McHardy AC, Vorholt JA, Schulze-Lefert P. Functional overlap of the Arabidopsis leaf and root microbiota. Nature, 2015, 528(7582): 364-369. DOI:10.1038/nature16192 |
| [14] | Borum J, Pedersen O, Greve TM, Frankovich TA, Zieman JC, Fourqurean JW, Madden CJ. The potential role of plant oxygen and sulphide dynamics in die-off events of the tropical seagrass, Thalassia testudinum. Journal of Ecology, 2005, 93(1): 148-158. DOI:10.1111/j.1365-2745.2004.00943.x |
| [15] | Nielsen LB, Finster K, Welsh DT, Donelly A, Herbert RA, de Wit R, Lomstein BA. Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environmental Microbiology, 2001, 3(1): 63-71. DOI:10.1046/j.1462-2920.2001.00160.x |
| [16] | Hansen JW, Udy JW, Perry CJ, Dennison WC, Lomstein BA. Effect of the seagrass Zostera capricorni on sediment microbial processes. Marine Ecology Progress Series, 2000, 199: 83-96. DOI:10.3354/meps199083 |
| [17] | Devereux R. Seagrass rhizosphere microbial communities//Kristensen E, Haese RR, Kostka JE. Interactions Between Macro- and Microorganisms in Marine Sediments. Washington, DC: American Geophysical Union, 2005, 60: 199-216. |
| [18] | James JB, Sherman TD, Devereux R. Analysis of bacterial communities in seagrass bed sediments by double-gradient denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA genes. Microbial Ecology, 2006, 52(4): 655-661. DOI:10.1007/s00248-006-9075-3 |
| [19] | Mejia AY, Rotini A, Lacasella F, Bookman R, Thaller MC, Shem-Tov R, Winters G, Migliore L. Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analyses. A study in Halophila stipulacea (Forsk.) Aschers meadows in the northern Red Sea. Ecological Indicators, 2016, 60: 1150-1163. DOI:10.1016/j.ecolind.2015.09.014 |
| [20] | Brodersen KE, Siboni N, Nielsen DA, Pernice M, Ralph PJ, Seymour J, Kühl M. Seagrass rhizosphere microenvironment alters plant-associated microbial community composition. Environmental Microbiology, 2018, 20(8): 2854-2864. DOI:10.1111/1462-2920.14245 |
| [21] |
Jiang YF, Ling J, Dong JD, Zhang YY. The bacterial communities constitution and distribution in sediment of seagrass in Xincun Bay, South China Sea. Journal of Biology, 2016, 33(6): 38-42.
(in Chinese) 江玉凤, 凌娟, 董俊德, 张燕英. 南海新村湾海草沉积物细菌群落组成及分布. 生物学杂志, 2016, 33(6): 38-42. DOI:10.3969/j.issn.2095-1736.2016.06.038 |
| [22] |
Li Y, Yu KF, Wang YH, Guo J, Huang XY, Pei JY, Luo YQ. Distribution characteristics of surface seawater nutrients in summer around Luhuitou Reef in Sanya. Tropical Geography, 2017, 37(5): 708-717.
(in Chinese) 李扬, 余克服, 王英辉, 郭靖, 黄学勇, 裴继影, 罗燕秋. 三亚鹿回头岸礁海域夏季表层海水营养盐年际变化特征. 热带地理, 2017, 37(5): 708-717. |
| [23] | Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnology and Bioengineering, 2005, 89(6): 670-679. DOI:10.1002/bit.20347 |
| [24] | Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 2011, 27(21): 2957-2963. DOI:10.1093/bioinformatics/btr507 |
| [25] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QⅡME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [26] | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [27] | Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics, 2011, 12: 385. DOI:10.1186/1471-2105-12-385 |
| [28] | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research, 2013, 41(D1): D590-D596. |
| [29] | Logue JB, Lindström ES. Species sorting affects bacterioplankton community composition as determined by 16S rDNA and 16S rRNA fingerprints. The ISME Journal, 2010, 4(6): 729-738. DOI:10.1038/ismej.2009.156 |
| [30] | Langenheder S, Székely AJ. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. The ISME Journal, 2011, 5(7): 1086-1094. DOI:10.1038/ismej.2010.207 |
| [31] | Liang YT, Jiang YJ, Wang F, Wen CQ, Deng Y, Xue K, Qin YJ, Yang YF, Wu LY, Zhou JZ, Sun B. Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. The ISME Journal, 2015, 9(12): 2561-2572. DOI:10.1038/ismej.2015.78 |
| [32] | Waycott M, Duarte CM, Carruthers TJB, Orth RJ, Dennison WC, Olyarnik S, Calladine A, Fourqurean J W, Heck KL Jr, Hughes AR, Kendrick GA, Kenworthy WJ, Short FT, Williams SL. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(30): 12377-12381. DOI:10.1073/pnas.0905620106 |
| [33] | Lamb JB, van de Water JAJM, Bourne DG, Altier C, Hein MY, Fiorenza EA, Abu N, Jompa J, Harvell CD. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science, 2017, 355(6326): 731-733. DOI:10.1126/science.aal1956 |
| [34] | Gomes NCM, Cleary DFR, Pires ACC, Almeida A, Cunha A, Mendonça-Hagler LCS, Smalla K. Assessing variation in bacterial composition between the rhizospheres of two mangrove tree species. Estuarine, Coastal and Shelf Science, 2014, 139: 40-45. DOI:10.1016/j.ecss.2013.12.022 |
| [35] | Alzubaidy H, Essack M, Malas TB, Bokhari A, Motwalli O, Kamanu FK, Jamhor SA, Mokhtar NA, Antunes A, Simões MF, Alam I, Bougouffa S, Lafi FF, Bajic VB, Archer JAC. Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene, 2016, 576(2): 626-636. |
| [36] | Asami H, Aida M, Watanabe K. Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture. Applied and Environmental Microbiology, 2005, 71(6): 2925-2933. DOI:10.1128/AEM.71.6.2925-2933.2005 |
| [37] | Castine SA, Bourne DG, Trott LA, McKinnon DA. Sediment microbial community analysis: establishing impacts of aquaculture on a tropical mangrove ecosystem. Aquaculture, 2009, 297(1/4): 91-98. |
| [38] | Mason OU, Case DH, Naehr TH, Lee RW, Thomas RB, Bailey JV, Orphan VJ. Comparison of Archaeal and Bacterial diversity in methane seep carbonate nodules and host Sediments, Eel River Basin and Hydrate Ridge, USA. Microbial Ecology, 2015, 70(3): 766-784. DOI:10.1007/s00248-015-0615-6 |
| [39] | Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, Takai K. Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing. Microbes and Environments, 2012, 27(4): 382-390. DOI:10.1264/jsme2.ME12032 |
| [40] | Inagaki F, Takai K, Nealson KH, Horikoshi K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa trough hydrothermal sediments. International Journal of Systematic and Evolutionary Microbiology, 2004, 54(5): 1477-1482. DOI:10.1099/ijs.0.03042-0 |
| [41] | Ting ASY, Tan SH, Wai MK. Isolation and characterization of Actinobacteria with antibacterial activity from soil and rhizosphere soil. Australian Journal of Basic and Applied Sciences, 2009, 3(4): 4053-4059. |
| [42] | Mendes R, Kruijt M, De Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science, 2011, 332(6033): 1097-1100. DOI:10.1126/science.1203980 |
| [43] | Orcutt BN, Joye SB, Kleindienst S, Knittel K, Ramette A, Reitz A, Samarkin V, Treude T, Boetius A. Impact of natural oil and higher hydrocarbons on microbial diversity, distribution, and activity in Gulf of Mexico cold-seep sediments. Deep Sea Research Part Ⅱ: Topical Studies in Oceanography, 2010, 57(21/23): 2008-2021. |
| [44] | Beazley MJ, Martinez RJ, Rajan S, Powell J, Piceno YM, Tom LM, Andersen GL, Hazen TC, Van Nostrand JD, Zhou JZ, Mortazavi B, Sobecky PA. Microbial community analysis of a coastal salt marsh affected by the Deepwater horizon oil spill. PLoS One, 2012, 7(7): e41305. DOI:10.1371/journal.pone.0041305 |
| [45] | Sun FF, Zhang XL, Zhang QQ, Liu FH, Zhang JP, Gong J. Seagrass (Zostera marina) Colonization promotes the accumulation of diazotrophic bacteria and alters the relative abundances of specific bacterial lineages involved in benthic carbon and sulfur cycling. Applied and Environmental Microbiology, 2015, 81(19): 6901-6914. DOI:10.1128/AEM.01382-15 |
| [46] | Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology, 2009, 68(1): 1-13. |
| [47] | Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. Defining the core Arabidopsis thaliana root microbiome. Nature, 2012, 488(7409): 86-90. DOI:10.1038/nature11237 |
| [48] | Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. The ISME Journal, 2014, 8(4): 790-803. DOI:10.1038/ismej.2013.196 |
 2019, Vol. 59
2019, Vol. 59