中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 许超, 熊亚茹, 黄桂媛, 王巧贞, 卢明倩, 张荣灿, 廖威, 张云开, 黄庶识. 2018
- Chao Xu, Yaru Xiong, Guiyuan Huang, Qiaozhen Wang, Mingqian Lu, Rongcan Zhang, Wei Liao, Yunkai Zhang, Shushi Huang. 2018
- 基于比较转录组分析海洋弧菌X511的褐藻胶代谢途径
- Analysis of the alginate metabolic pathways in the marine Vibrio X511 based on comparative transcriptome
- 微生物学报, 58(9): 1551-1562
- Acta Microbiologica Sinica, 58(9): 1551-1562
-
文章历史
- 收稿日期:2017-10-25
- 修回日期:2017-12-25
- 网络出版日期:2018-01-17
2. 广西大学生命科学与技术学院, 广西 南宁 530003;
3. 广西北部湾海洋研究中心, 广西 南宁 530007;
4. 广西职业技术学院, 广西 南宁 530226
2. College of Life Science and Technology, Guangxi University, Nanning 530003, Guangxi Zhuang Autonomous Region, China;
3. Guangxi Beibu Gulf Marine Research Center, Nanning 530007, Guangxi Zhuang Autonomous Region, China;
4. Guangxi Vocational and Technical College, Nanning 530226, Guangxi Zhuang Autonomous Region, China
Shushi Huang, Tel:+86-771-2503990, E-mail:hshushi@gxas.cn
褐藻胶是一种线性高分子多糖,经甘露醛糖酸(β-D-mannuronic acid,M)和古罗醛糖酸(α-L-guluronic acid,G)两种单糖以1, 4-糖苷键随机组合而成[1-2],由英国科学家Stanford于1881年率先自海带中提取获得[3]。褐藻胶在大型褐藻中的含量尤为丰富,约占总糖的30%–60% (W/W)[4],是最丰富的海洋生物多糖,也是世界上仅次于纤维素的生物高分子聚合物。以大型海洋藻类尤其是褐藻为原料的第三代生物乙醇技术的研发正在成为能源领域关注的热点,作为褐藻主要多糖成分的褐藻胶储量丰富,以其为原料转化为生物乙醇,是未来解决能源危机的潜在路径之一[5]。
在海洋和陆地,存在多种类能够降解和同化褐藻胶的降解菌,种类众多[6],这些微生物可以利用褐藻胶降解产物作为唯一碳源生长。但是,除了少数几个种类以外[5, 7],大部分褐藻胶降解菌并不能生产乙醇。由于褐藻胶及其降解产物不能被现有的工业产乙醇菌株利用,通过包括改造原始褐藻胶降解菌菌株的代谢途径,或者构建工程菌株生产乙醇,为大型褐藻生产燃料乙醇开辟了新的途径[8-10]。阐明藻胶降解菌代谢途径及其关键酶特性,是藻胶降解菌基因改造或者构建产乙醇工程菌的基础。
1962年,Preiss和Ashwell最早发现了假单胞菌褐藻胶裂解酶分解海藻酸的机制[11-12]。2010年,Takase等[13]对Sphingomonas A1菌株代谢褐藻胶的研究发现,该菌株通过细胞膜上的超级通道(superchannel)直接摄入褐藻胶到细胞体内,在Al-Ⅰ、Al-Ⅱ-Ⅲ三种裂解酶的作用下,褐藻胶降解成寡糖,进而被内切酶(Al-Ⅳ)降解为单糖,单糖经非酶促反应转变成4-脱氧-L-赤藓-糖醛酸(DEH),DEH被还原酶A1-R催化为2-酮-3-脱氧-D-葡萄糖酸(KDG),进入ED途径(Entner-Doudoroff pathway)。ED途径中,KDG在酶的作用下经两步催化反应被利用(图 1),第一步是在2-酮-3-脱氧葡糖酸激酶(Al-K)的催化下,KDG被磷酸化为2-酮-3-脱氧-6-磷酸-葡糖酸(KDPG);第二步是KDPG经醛缩酶(Al-A)催化发生分解反应,产生甘油醛-3-磷酸和丙酮酸,最后进入糖酵解和三羧酸循环。
我们早期筛选鉴定的海洋弧菌X511是一株新型的褐藻胶降解菌株,能够分泌高活性的海藻酸裂解酶。本研究在基于X511菌株全基因组测序基础上,分别对其在以葡萄糖和褐藻胶为唯一碳源培养下的转录组进行了比较研究,通过基因的转录差异快速定位目的基因及对应的代谢途径,为后续完全阐明X511菌株基本的遗传信息以及褐藻胶在该菌细胞内的代谢途径提供依据。
1 材料和方法 1.1 试剂和仪器海藻酸钠、硫酸镁、葡萄糖、氯化钠、七水硫酸镁、硫酸亚铁、硫酸铵均为市售分析纯。细菌总RNA提取试剂盒购自Transgen公司,反转录试剂盒PrimeScriptTM RT-PCR Kit及荧光定量试剂盒SYBR Premix EX Taq购自TaKaRa公司,超低温冰箱购自美国Themro公司,ABI 7500 Fast实时荧光定量PCR仪购自美国应用生物系统公司。
1.2 菌种与培养基供试X511菌株为本实验室筛选自广西北海涠洲岛海域。
种子培养基:海藻酸钠6 g,NaCl 15 g,蛋白胨5 g,磷酸氢二钾1 g,硫酸镁1 g,硫酸亚铁0.001 g,加去离子水定容到1 L,pH 7.5,121 ℃灭菌20 min。
海藻酸钠培养基:将种子培养基的蛋白胨换成等量的硫酸铵。
葡萄糖培养基:将海藻酸钠培养基中的海藻酸钠换成等量的葡萄糖。
1.3 样品处理将低温保藏的菌种接至种子培养基中摇床培养活化24 h,然后接种至海藻酸钠培养基培养至对数期(约8 h),按2%的接种量转接至葡萄糖培养基中培养至对数期,取样,编号X1;从葡萄糖培养基中取处于对数期的X511菌液,以2%的接种量接种至海藻酸钠培养基,待菌株X511生长至对数期,取样,编号X2;样品X1和X2均取3组平行,培养条件为30℃,200 r/min。
1.4 转录组测序和生物信息学分析采用天根公司细菌总RNA提取试剂盒提取菌株X511的总RNA,送北京诺禾致源生物信息科技有限公司基于Illumina HiSeq进行转录组测序。原始测序结果经质量评估、与相关物种参考序列进行比对分析、基因表达分析及基因差异表达分析后,采用GO、KEGG、COG、NR和Swiss-Prot等数据库进行生物信息学分析。
1.5 定量PCR验证转录组在相同条件下将X511分别培养至与样品X1和X2相同的状态,提取其总RNA,采用TaKaRa反转录试剂盒将总RNA反转录为cDNA。由于任何管家基因的稳定表达都有一定的适用范围[14-15],故挑选菌株X511中注释为recA、16S rRNA、fadL、rpoA、gyrB、GAPDH (3个)的共8个待选管家基因进行试验[16-19],以得到本供试菌株中表达最稳定的管家基因。为验证转录组结果的可靠性,拟挑选14个与褐藻胶及葡萄糖代谢相关的基因进行RT-PCR,利用Primer Premier 5软件设计产物长度为100–300 bp的引物(表 1),委托北京奥科公司合成。利用SYBR Premix EX Taq (Perfect Real time染料法实时荧光定量试剂盒)进行定量检测,采用2∆Ct的方法进行数据分析[20]。
| Gene ID | Gene name | R-primer | F-primer |
| GM004415 | recA | GTGATAACCGAGCAATG | CTTCTACGATACGACCC |
| GM001922 | 16S rRNA | AAAAGGGCAAGTGGGT | CGCAGGGTCAATAGCA |
| GM001598 | fadL | AGGTTCTTACCGTCTCA | TCCATTTCTGGGCTAT |
| GM003897 | rpoA | CTATGCCAGGTTGTGC | GCGTTGTCATCCGTTA |
| GM004299 | gyrB | GGCAAACAAGAGCAGT | GCGTTAGACGAGGAGT |
| GM001542 | GAPDH | CCGTATCGGTCGTTTCGTATT | CATTTAAGGTCCGCTGGGTT |
| GM002328 | GAPDH | TTTCTTCGTGTCGTGTAGATGTTG | GCTCGCAGTCGTAGTGGGTAA |
| GM004585 | GAPDH | GGCTGCGATGTTGTGATTGA | GGCTGTCACGATACGATGTTTT |
| GM001982 | algL | ACCTACATTCGCACTTGCTTCTG | TTGTCGTTACGGCTTTGTTGTG |
| GM001989 | algL | GTTGCGAATCAAGCAGGGAC | GGTTGCGTGGGTACGAGTGT |
| GM003561 | rpiB | GATGGAAAACAGCCAAGCAGC | AGTACCACAACCAGTTACCACGAA |
| GM003558 | kduD | CAAGGTGAAGGCGGCAAGA | GCCATGTAACCCGGAGCAA |
| GM003563 | kdgK | ATGCGGCGGCAAAATACT | TGTCGGTGACGCTGAGGAT |
| GM002020 | eda | TTTCGCAGCATCCCCTATC | TTCAACCGCCGCATCAT |
| GM003564 | eda | GGATGAAGCGATTGATGCG | TTGTTGACACCAGGGACGAT |
| GM000293 | PTS-Glc-EIIA | ATCAAGCCAGCAGGCAACA | CGATACCGAAGTGAACGAAAAG |
| GM000031 | pfk | TCATGGGTCGTCACTGTGGT | TGCGATGCCGTCTTGGA |
| GM003325 | fsaA | GTCAAAGTGCCAGCAACCG | CAATGCCGCAAGGAAACC |
| GM003942 | tpiA | ATGCGTCGTCCTGTAGTGATG | TGAGTGACCGATGATGATGTGA |
| GM000053 | PGAM | GCTATGGACCGTGACAACAACT | GGAATACCGCACGCTCAAA |
| GM001436 | pyk | GACTACCTTGCTATTTCGTTCCC | TTCTGCTCGTTCGACTTTGG |
| GM002414 | korA | AACGGCTCCGAATCTTGG | ACACGTCATCACGGGCATAG |
2 结果和分析 2.1 总RNA电泳结果
X511菌株的总RNA电泳结果如图 2,平行样品间的一致性较好,基本无蛋白质和DNA的污染,无明显的RNA降解现象,可作为转录组测序样品。

|
| 图 2 菌株X511的总RNA电泳结 Figure 2 Agarose gel electrophoresis of total RNAs from strain X511. M: Marker; 1–3: X1; 4–6: X2. |
2.2 RT-PCR验证 2.2.1 内参基因的选择: 选择菌株X511的8个管家基因分别进行RT-PCR,以得到在不同培养下转录最为稳定的内参基因(偏差C.V越小,说明表达越稳定),结果见表 2。分别在葡萄糖和褐藻胶培养下,菌株X511中转录最为稳定的基因为GM004585,其基因表达的产物为甘油醛-3-磷酸脱氢酶(GAPDH),故选择该基因为内参基因进行RT-PCR验证试验。
| Gene ID | Gene name | Product | C.V |
| GM004415 | recA | Recombination protein | 0.0132 |
| GM001922 | 16S rRNA | 16S rRNA processing protein | 0.0155 |
| GM001598 | fadL | Long-chain fatty acid transport protein | 0.0492 |
| GM003897 | rpoA | DNA-directed RNA polymerase subunit alpha | 0.0149 |
| GM004299 | gyrB | DNA gyrase subunit B | 0.0121 |
| GM001542 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | 0.0267 |
| GM002328 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | 0.0120 |
| GM004585 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | 0.0107 |
2.2.2 RT-PCR验证: 挑选14个与褐藻胶及葡萄糖代谢相关的基因进行RT-PCR,从表 3可以看出,基因RT-PCR的定量结果与转录组的差异转录倍数(FC)基本吻合,说明转录组测序结果的可信度较高,可作为生物信息学分析的参考。
| Gene ID | RT-PCR | FC | Gene ID | RT-PCR | FC |
| GM1989 | 5.270 | 44.067 | GM3942 | 0.160 | 0.74 |
| GM1982 | 1.488 | 153.190 | GM0031 | 0.223 | 0.50 |
| GM3561 | 3.620 | 1.970 | GM2414 | 0.350 | 0.35 |
| GM3558 | 1.100 | 2.330 | GM2020 | 0.450 | 0.80 |
| GM3564 | 1.960 | 4.120 | GM3325 | 0.510 | 0.59 |
| GM1436 | 1.870 | 4.080 | GM0053 | 0.250 | 0.56 |
| GM3563 | 0.720 | 3.870 | GM0293 | 0.990 | 0.71 |
| FC was the fold changes in the results of the transcriptome, RT-PCR was the fold changes in the quantitative results of the fluorescence quantitative PCR. | |||||
2.3 差异表达基因
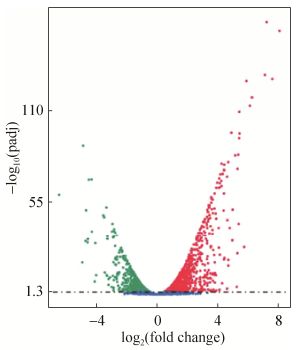
在不同培养下菌株X511的差异表达基因火山图(图 3),X2相对于X1(X2 vs X1,下同)有2024个基因的转录水平存在显著性差异,占该菌株总基因数目(5434个)的37.2%,其中1066个基因转录上调,958个基因下调,说明不同碳源的培养能显著影响该菌株的生长及代谢方式。
|
| 图 3 X2 vs X1差异表达基因的火山图 Figure 3 Speckle diagram of differentially expressed genes in X2 vs X1. |
2.4 差异基因的GO富集分析
样品X2相对X1的差异转录基因(P < 0.05) GO分析见图 4,根据功能将基因分为3类,分别为细胞组分(cellular component)、分子功能(molecular function)和生物学过程(biological process)。

|
| 图 4 X2 vs X1的差异基因GO富集分析 Figure 4 GO enrichment analysis of differentially expressed genes in X2 vs X1. |
细胞组分类别中存在显著差异的代谢途径总数最少,且发生上调和下调的代谢途径数量相当,与葡萄糖培养相比,以褐藻胶作为碳源的培养基对菌株X511的细胞组分产生较小的影响;上调基因明显较多的途径是“被膜组分(envelope)”和“表面封装结构组分(external encapsulating structure part)”,而“质子运输V型ATP酶复合体(proton-transporting V-type ATPase complex)”及“外膜组分(outer membrane)”则下调基因较多,可能是细菌因为应不同底物的吸收及代谢产物的分泌,导致这些细胞组分基因的上调和下调。
归类为分子功能的差异转录代谢途径数量其次,大部分的差异途径中上调基因总数多于下调基因,主要包括“磷酸转移传感器激酶活性(phosphorelay sensor kinase activity)”、“受体活性(receptor activity)”、“磷酸转移酶活性(phosphotransferase activity)”、“分子传导活性(molecular transducer activity)”以及“信号传导活性(signal transducer activity)”等代谢途径,磷酸转移酶、激酶、信号传递及分子运输相关的代谢途径均发生了不同程度的上调,上述上调的代谢途径与基因多与碳水化合物的利用有关[21-22],显示了菌株X511利用两种碳源的分子机制存在较大差异,褐藻胶作为高分子聚糖,其被利用的途径相较于葡萄糖更为复杂,褐藻胶分子在经一系列酶的作用后被细菌利用吸收的方式有待进一步研究。
在生物学过程类别中,除“细胞器组织(organelle organization)”和“单糖代谢过程(monosaccharide metabolic process)”上调基因数量略高于下调外,其余代谢途径如“ATP水解偶联质子转运(ATP hydrolysis coupled proton transport)”、“调节分解代谢过程(regulation of catabolic process)”、“自噬调节(regulation of autophagy)”、“代谢过程的积极调节(positive regulation of metabolic process)”、“共轭调节(regulation of conjugation)”及“GPI锚代谢过程(GPI anchor metabolic process)”等下调基因数量较多,说明在用褐藻胶培养该菌株时,其参与生物过程基因的活跃数量少于葡萄糖培养,推测其维持菌体自身稳态的能力可能发生了下降[23-24],对菌种保藏培养基的选择有一定的指导意义。
2.5 差异代谢途径KEGG富集分析利用KEGG数据库对各代谢通路的差异代谢途径(表 4)进行富集:相对于葡萄糖培养基而言,菌种在褐藻胶培养基中的基础代谢产能途径如柠檬酸循环(TCA cycle)和氧化磷酸化(oxidative phosphorylation)均发生了上调;其中TCA循环产生的一系列中间产物能作为其他诸多代谢途径的前体,如氨基酸和脂肪酸等大分子物质的合成及氧化磷酸化等,该代谢通路的极显著上调说明该菌株在褐藻胶培养下其体内的诸多代谢途径比葡萄糖培养时更为活跃;而氧化磷酸化则直接与电子传递链的产能方式相关,说明菌株在褐藻胶培养基中的生长繁殖更为活跃;此外,缬氨酸、亮氨酸和异亮氨酸降解(valine, leucine and isoleucine degradation)与丁酸代谢(butanoate metabolism)也发生了上调,推测该菌株的生长过程中可能有丁酸生成;值得一提的是,菌株中可能存在与光合作用(photosynthesis)相关的代谢途径且在褐藻胶培养下有一定的上调。代谢途径如泛醌等萜醌类生物合成(ubiquinone and other terpenoid-quinone biosynthesis)、核苷酸切除修复(nucleotide excision repair)及维生素B6代谢(vitamin B6 metabolism)的下调,以及核苷酸切除修复途径的下调,推测菌株X511在褐藻胶培养基中传代数过多易发生基因突变[25-26]。
| Pathway | Up (Down)-regulated gene numbers | Background gene numbers | P-value |
| TCA cycle(up) | 23 | 28 | 0.000791 |
| Valine, leucine and isoleucine degradation (up) | 19 | 31 | 0.020097 |
| Oxidative phosphorylation (up) | 23 | 41 | 0.022647 |
| Butanoate metabolism (up) | 20 | 35 | 0.028532 |
| Photosynthesis (up) | 7 | 8 | 0.048415 |
| Ubiquinone and other terpenoid-quinone biosynthesis (down) | 8 | 17 | 0.040378 |
| Nucleotide excision repair (down) | 5 | 8 | 0.047832 |
| Vitamin B6 metabolism (down) | 5 | 8 | 0.047832 |
2.6 菌株X511中与葡萄糖及褐藻胶代谢途径相关基因的转录组分析
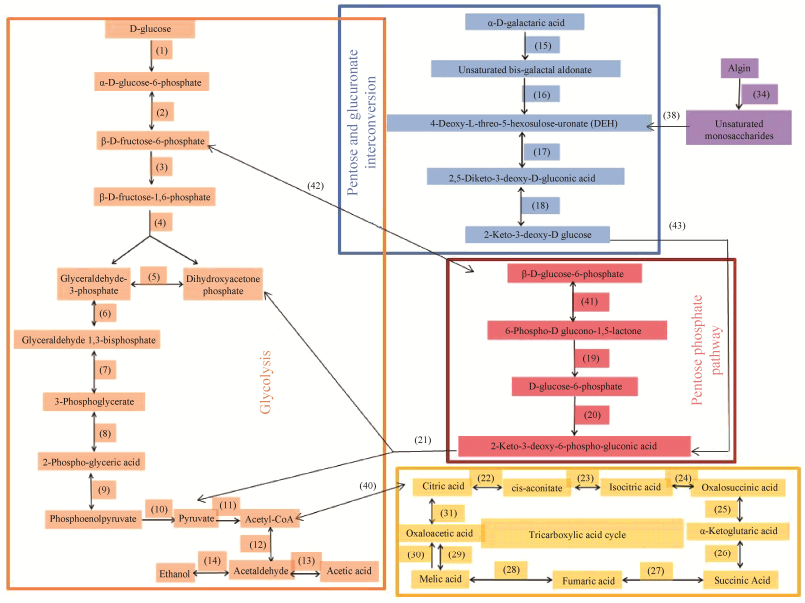
结合菌株X511的全基因组测序结果及相关文献对褐藻胶裂解酶产生菌代谢褐藻胶途径[27-28]的报道,推测并绘制该菌株代谢葡萄糖与褐藻胶的可能途径(图 5),葡萄糖经糖酵解、磷酸戊糖途径和三羧酸循环被菌株利用。褐藻胶在该菌株产生的褐藻胶裂解酶作用下被降解为褐藻胶寡糖,发生非酶促反应变成DEH,进入戊糖和葡萄糖醛酸互变途径,经过两步反应过程生成2-酮-3脱氧-D葡萄糖,被激酶(2-dehydro-3-deoxygluconokinase,KDGK)催化为2-酮-3脱氧-6磷酸-葡萄糖,在醛缩酶(2-dehydro- 3-deoxyphosphogluconate aldolase)的作用下发生分解反应,得到糖酵解途径的中间产物二羟丙酮磷酸和丙酮酸,经糖酵解途径和三羧酸循环合成该菌株生长和繁殖一系列的中间产物和能量。
|
| 图 5 菌株X511中葡萄糖及褐藻胶的代谢途径 Figure 5 Metabolic pathway of glucose and alginate in strain X511. |
各代谢途径中涉及的基因(对应图 5的序号)见表 5所示,相比于葡萄糖培养基,菌株X511在褐藻胶培养基的培养下,涉及催化褐藻胶代谢途径每一步的酶基因,包括褐藻胶裂解酶(序号34,Gene ID: GM001982,GM001986,GM001987,GM001989)、核糖5-磷酸异构酶(序号17,Gene ID: GM003561)、2-酮-D-葡萄糖酸3-脱氢酶(序号18,Gene ID: GM003558)、2-脱氢-3-脱氧葡萄糖激酶(序号43,Gene ID: GM001997,GM002016,GM003563)、2-脱氢-3-脱氧磷酸葡糖酸醛缩酶(序号21,Gene ID: GM003564),均发生了大幅度的上调,其中,编码褐藻胶裂解酶的4个基因(序号34)和2-脱氢-3-脱氧葡萄糖激酶的3个基因(序号43),当菌株X511从葡萄糖培养基转入褐藻胶培养基后,转录量上调明显,褐藻胶裂解酶4个基因差异转录倍数(FC)分别是153.19、61.36、26.50、44.06,2-脱氢-3-脱氧葡萄糖激酶3个基因差异转录倍数(FC)分别是26.32、1.77、3.87;同时,涉及三羧酸循的酶基因也呈现一定程度的上调(FC > 1),表明菌株在褐藻胶诱导下,菌株细胞为了获得生长所需的营养然后启动后续代谢途径不同关键酶基因的转录与表达,进而进入以褐藻胶为底物的生长模式。而葡萄糖代谢途径中所涉及酶基因(序号1–10对应的基因),在褐藻胶培养下的转录量都发生下调(FC < 1)。
| Number | Gene ID | Products | FC |
| 1 | GM000293 | Glucose-specific IIA component | 0.71 |
| 2 | GM004524 | Glucose-6-phosphate isomerase | 0.59 |
| 3 | GM000031 | 6-Phosphofructokinase | 0.50 |
| 4 | GM003325 | Fructose-6-phosphate aldolase | 0.59 |
| GM004461 | Fructose-bisphosphate aldolase | 0.89 | |
| 5 | GM003942 | Triosephosphate isomerase | 0.74 |
| 6 | GM004585 | Glyceraldehyde 3-phosphate dehydrogenase | 0.48 |
| 7 | GM004462 | Phosphoglycerate kinase | 0.61 |
| 8 | GM000053 | Phosphoglycerate mutase | 0.56 |
| 9 | GM004426 | Enolase | 0.77 |
| 10 | GM003820 | Pyruvate kinase | 0.72 |
| 11 | GM002414 | 2-Oxoglutarate ferredoxin oxidoreductase | 0.35 |
| 12 | GM000009 | Acetyl-CoA synthetase | 3.11 |
| 13 | GM001101 | Aldehyde dehydrogenase | 12.81 |
| GM002912 | Aldehyde dehydrogenase | 5.08 | |
| 14 | GM000869 | Alcohol dehydrogenase | 7.73 |
| GM004314 | Alcohol dehydrogenase | NC | |
| 15 | GM003567 | Pectate disaccharide-lyase | NC |
| 16 | GM003569 | Oligogalacturonate lyase | NC |
| 17 | GM003561 | Ribose 5-phosphate isomerase | 1.97 |
| 18 | GM003558 | 2-Deoxy-D-gluconate 3-dehydrogenase | 2.33 |
| 19 | GM001107 | 6-Phosphogluconolactonase | NC |
| 20 | GM002019 | Phosphogluconate dehydratase | NC |
| GM004187 | Phosphogluconate dehydratase | NC | |
| 21 | GM003564 | 2-Dehydro-3-deoxyphosphogluconate aldolase | 4.12 |
| 22, 23 | GM001041 | Aconitate hydratase | 5.46 |
| 24, 25 | GM000493 | Isocitrate dehydrogenase | 2.08 |
| 26 | GM000341 | 2-Oxoglutarate dehydrogenase | 4.40 |
| GM000342 | Dihydrolipoamide succinyltransferase | 3.874 | |
| GM000344 | Succinyl-CoA synthetase | 7.17 | |
| 27 | GM000043 | Fumarate reductase | 4.91 |
| 28 | GM001281 | Fumarate hydratase | 2.96 |
| 30 | GM002060 | Malate dehydrogenase | NC |
| 31 | GM000336 | Citrate synthase | 4.27 |
| 34 | GM001982 | Poly (beta-D-mannuronate) lyase | 153.19 |
| GM001986 | Alginate lyase | 61.36 | |
| GM001987 | Alginate lyase | 26.50 | |
| GM001989 | Alginate lyase | 44.06 | |
| 38 | Nonenzymatic reaction | ||
| 40 | GM000336 | Type Ⅱ citrate synthase | 4.40 |
| 41 | GM004524 | Glucose-6-phosphate isomerase | 0.59 |
| 42 | GM001543 | Glucose-6-phosphate 1-epimerase | 0.48 |
| GM004524 | Glucose-6-phosphate isomerase | 0.59 | |
| 43 | GM001997 | 2-Dehydro-3-deoxygluconokinase | 26.32 |
| GM002016 | 2-Dehydro-3-deoxygluconokinase | 1.77 | |
| GM003563 | 2-Dehydro-3-deoxygluconokinase | 3.87 | |
| “NC” represent no transcriptional differences. | |||
首先,值得关注的是该菌株的乙醇生成途径的几个关键酶基因乙酸辅酶A合成酶基因(序号12,Gene ID: GM000009)、乙醇脱氢酶基因(序号14,Gene ID: GM000869)以及乙醛脱氢酶(序号13,Gene ID: GM000869, GM001101,GM002912),在褐藻胶培养基条件下,其转录量及表达量都出现了一定程度的上调,我们后续研究中发现,该菌株发酵葡萄糖和褐藻胶的产物中存在少量乙醇,说明该菌株具备一定产乙醇的能力;同时,我们对菌株X511进行全基因组注释以及前期研究发现[27],该菌株除了可以直接利用褐藻胶和葡萄糖作为碳源产生乙醇外,还可以利用褐藻的其他组分包括甘露醇(占总糖5%–10%)、昆布多糖、岩藻糖;褐藻转化为液体燃料是大型海藻生物质能源发展的重要途径,但是褐藻多糖成分复杂,难以被工业微生物转化利用,野生菌中发现能同时发酵褐藻的主要成分的菌株不多[5],该菌株利用褐藻生物质为底物在常温下条件下发酵褐藻产乙醇,对于发展褐藻生物转化有潜在研究价值。第二,菌株X511代谢褐藻胶的途径与文献报道[13, 28]的路径基本一致,但是该菌株褐藻胶代谢途径中第17步的反应过程(图 5)尚未在弧菌中被报道,其具体的反应机理有待进一步研究。
3 结论本研究在菌株X511的全基因组测序结果基础上,通过比较菌株X511在葡萄糖和褐藻胶培养基培养下的基因转录差异,利用GO和KEGG数据库对差异转录基因进行了功能归类及代谢途径富集分析,快速定位出该菌株中可能与褐藻胶代谢相关的基因及代谢途径。结果显示,在两种培养条件下共存在2024个差异转录基因,其中有上调基因1066个,下调基因958个;KO功能注释中与“分子功能”相关的多个代谢途径如“磷酸转移酶”和“分子传导”等途径发生了上调,说明菌株可能以多种方式吸收和利用褐藻胶多糖的降解产物;KEGG富集时发现柠檬酸循环和氧化磷酸化途径发生了上调而核苷酸切除修复途径发生下调,说明褐藻胶培养基能使该菌株的生长代谢更为活跃但不适合菌种的长期保藏;依据全基因组测序以及转录组结果,推测绘制菌株X511代谢褐藻胶的途径,褐藻胶在菌株X511褐藻胶裂解酶作用下,产生不饱和的褐藻胶单糖,通过酶促反应变成DEH,经酮醇异构酶逆催化为2, 5-二酮-3-脱氧-D-葡萄糖酸(2, 5-Diketo-3-deoxy-D-gluconic acid),进而在2-脱氧-D-葡萄糖酸脱氢酶(kduD)的作用下以NADH为还原力,被还原成2-酮-3-脱氧-D葡萄糖酸(KDG),经KDG激酶催化,并消耗1 mol ATP,底物被磷酸化为2-酮-3-脱氧-磷酸葡糖酸(KDPG),被丙酮酸醛缩酶催化裂解为丙酮酸和甘油醛-3-磷酸,进入糖酵解途径从而被菌株利用;菌株X511代谢褐藻胶的途径关键酶基因的功能,将在后续的克隆研究中予以验证。
| [1] | Gacesa P. Enzymic degradation of alginates. International Journal of Biochemistry, 1992, 24(4): 545-2. DOI:10.1016/0020-711X(92)90325-U |
| [2] | Eftekhar F, Speert DP. Alginase treatment of mucoid Pseudomonas aeruginosa enhances phagocytosis by human monocyte-derived macrophages. Infection and Immunity, 1988, 56(11): 2788-2793. |
| [3] | Osawa T, Matsubara Y, Muramatsu T, Kimura M, Kakuta Y. Crystal structure of the alginate (poly α-L-guluronate) lyase from Corynebacterium sp. at 1.2 Ǻ resolution. Journal of Molecular Biology, 2005, 345(5): 1111-1118. DOI:10.1016/j.jmb.2004.10.081 |
| [4] | Chapman VJ. Seaweeds and Their Uses. 2nd ed. London: Methuen, 1970. |
| [5] | Ji SQ, Wang B, Lu M, Li FL. Direct bioconversion of brown algae into ethanol by thermophilic bacterium Defluviitalea phaphyphila. Biotechnology for Biofuels, 2016, 9: 81. DOI:10.1186/s13068-016-0494-1 |
| [6] |
Zhang X, Zhao L, Qian L, Huang SS, Irbsi C. Research progress of alginolytic bacteria. Chinese Bulletin of Life Sciences, 2012, 24(5): 475-482.
(in Chinese) 张绪, 赵琳, 钱龙, 黄庶识, 伊日布斯. 海藻酸分解菌研究进展. 生命科学, 2012, 24(5): 475-482. |
| [7] | Zhang W, Zhang J, Cui H. The isolation and performance studies of an alginate degrading and ethanol producing strain. Chemical and Biochemical Engineering Quarterly, 2014, 28(3): 391-398. DOI:10.15255/CABEQ |
| [8] | Takeda H, Yoneyama F, Kawai S, Hashimoto W, Murata K. Bioethanol production from marine biomass alginate by metabolically engineered bacteria. Energy & Environmental Science, 2011, 4(7): 2575-2581. |
| [9] | Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim P B, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. An engineered microbial platform for direct biofuel production from brown macroalgae. Science, 2012, 335(6066): 308-313. DOI:10.1126/science.1214547 |
| [10] | Enquist-Newman M, Faust AM, Bravo DD, Santos CN, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR, Peereboom L, Clark A, Martinez Y, Goldsmith J, Cho MY, Donohoue PD, Luo L, Lamberson B, Tamrakar P, Kim EJ, Villari JL, Gill A, Tripathi SA, Karamchedu P, Paredes CJ, Rajgarhia V, Kotlar HK, Bailey RB, Miller DJ, Ohler NL, Swimmer C, Yoshikuni Y. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature, 2014, 505(7482): 239-243. DOI:10.1038/nature12771 |
| [11] | Preiss J, Ashwell G. Alginic acid metabolism in bacteria. Ⅱ. The enzymatic reduction of 4-deoxy-L-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-D-gluconic acid. Journal of Biological Chemistry, 1962, 237: 317-321. |
| [12] | Preiss J, Ashwell G. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosac-charides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. The Journal of Biological Chemistry, 1962, 237: 309-316. |
| [13] | Takase R, Ochiai A, Mikami B, Hashimoto W, Murata K. Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2010, 1804(9): 1925-1936. DOI:10.1016/j.bbapap.2010.05.010 |
| [14] | Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta, 2008, 227(6): 1343-1349. DOI:10.1007/s00425-008-0706-4 |
| [15] |
Zhang JS, Li W, Que YX, Ruan MH, Zhang MQ, Chen RK. Cloning of Two House-keeping Genes from Erianthus arundinaceus and the Application in cDNA Microarray. Journal of Tropical and Subtropical Botany, 2007, 15(4): 277-283.
(in Chinese) 张积森, 李伟, 阙友雄, 阮妙鸿, 张木清, 陈如凯. 斑茅两个看家基因片段的克隆及其在基因芯片中的应用. 热带亚热带植物学报, 2007, 15(4): 277-283. DOI:10.3969/j.issn.1005-3395.2007.04.001 |
| [16] | Da Fonseca ÉL, Dos Santos Freitas F, Vicente ACP. Pc promoter from class 2 integrons and the cassette transcription pattern it evokes. Journal of Antimicrobial Chemotherapy, 2011, 66(4): 797-801. DOI:10.1093/jac/dkr011 |
| [17] | Bhowmick R, Ghosal A, Chatterjee NS. Effect of environmental factors on expression and activity of chitinase genes of vibrios with special reference to Vibrio cholerae. Journal of Applied Microbiology, 2007, 103(1): 97-108. DOI:10.1111/jam.2007.103.issue-1 |
| [18] | Guérout AM, Iqbal N, Mine N, Ducos-Galand M, van Melderen L, Mazel D. Characterization of the phd-doc and ccd Toxin-Antitoxin Cassettes from Vibrio Superintegrons. Journal of Bacteriology, 2013, 195(10): 2270-2283. DOI:10.1128/JB.01389-12 |
| [19] | Asakura H, Ishiwa A, Arakawa E, Makino SI, Okada Y, Yamamoto S, Igimi S. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environmental Microbiology, 2007, 9(4): 869-879. DOI:10.1111/emi.2007.9.issue-4 |
| [20] | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 2008, 3(6): 1101-1108. DOI:10.1038/nprot.2008.73 |
| [21] | Postma PW, Lengeler JW. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiological Reviews, 1985, 49(3): 232-269. |
| [22] | Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiology and Molecular Biology Reviews, 2006, 70(4): 939-1031. DOI:10.1128/MMBR.00024-06 |
| [23] | Turina P, Samoray D, Gr ber P. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1-liposomes. The EMBO Journal, 2003, 22(3): 418-426. DOI:10.1093/emboj/cdg073 |
| [24] | Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell, 2005, 120(2): 237-248. DOI:10.1016/j.cell.2004.11.046 |
| [25] | Kemp MG, Sancar A. DNA excision repair. Cell Cycle, 2012, 11(16): 2997-3002. DOI:10.4161/cc.21126 |
| [26] | De Laat WL, Jaspers NGJ, Hoeijmakers JHJ. Molecular mechanism of nucleotide excision repair. Genes & Development, 1999, 13(7): 768-785. |
| [27] |
Xu C, Xiong YR, Lu MQ, Liao W, Zhang YK, Huang SS. Screening and identification of a marine alginate-degrading bacterium and the utilization capacity of polysaccharide. Biotechnology Bulletin, 2017, 33(4): 198-204.
(in Chinese) 许超, 熊亚茹, 卢明倩, 廖威, 张云开, 黄庶识. 一株具有褐藻胶降解能力的海洋细菌的筛选鉴定及其多糖利用能力研究. 生物技术通报, 2017, 33(4): 198-204. |
| [28] | Takase R, Maruyama Y, Oiki S, Mikami B, Murata K, Hashimoto W. Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity. Proteins: Structure, Function, and Bioinformatics, 2016, 84(7): 934-947. DOI:10.1002/prot.25042 |
 2018, Vol. 58
2018, Vol. 58